The Cysteine Protease CfAtg4 Interacts with CfAtg8 to Govern the Growth, Autophagy and Pathogenicity of Colletotrichum fructicola
Abstract
1. Introduction
2. Results
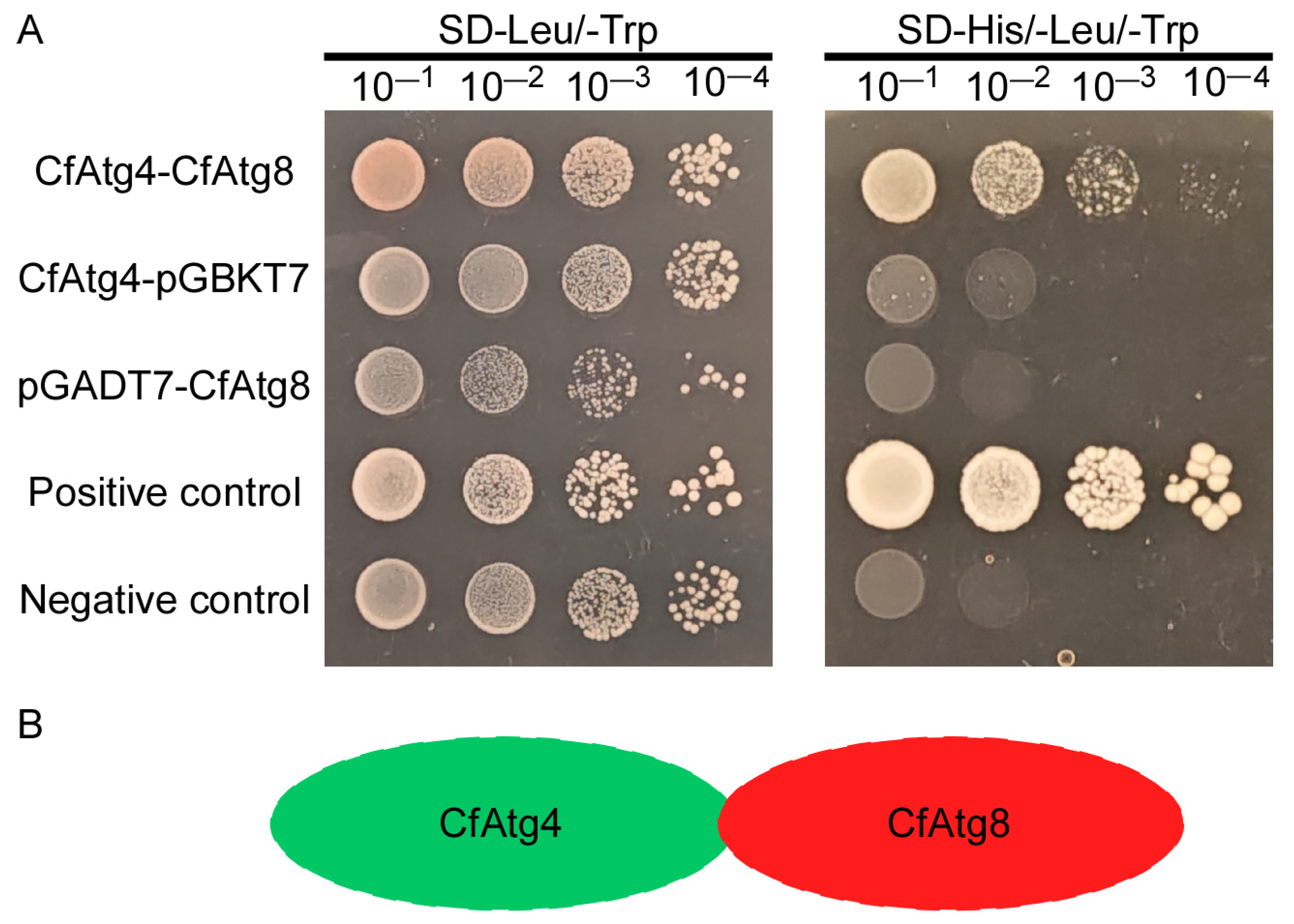
2.1. CfAtg4 Interacts with CfAtg8 in C. fructicola
2.2. The Acquisition of ΔCfatg4/CfAtg81–160 Strains
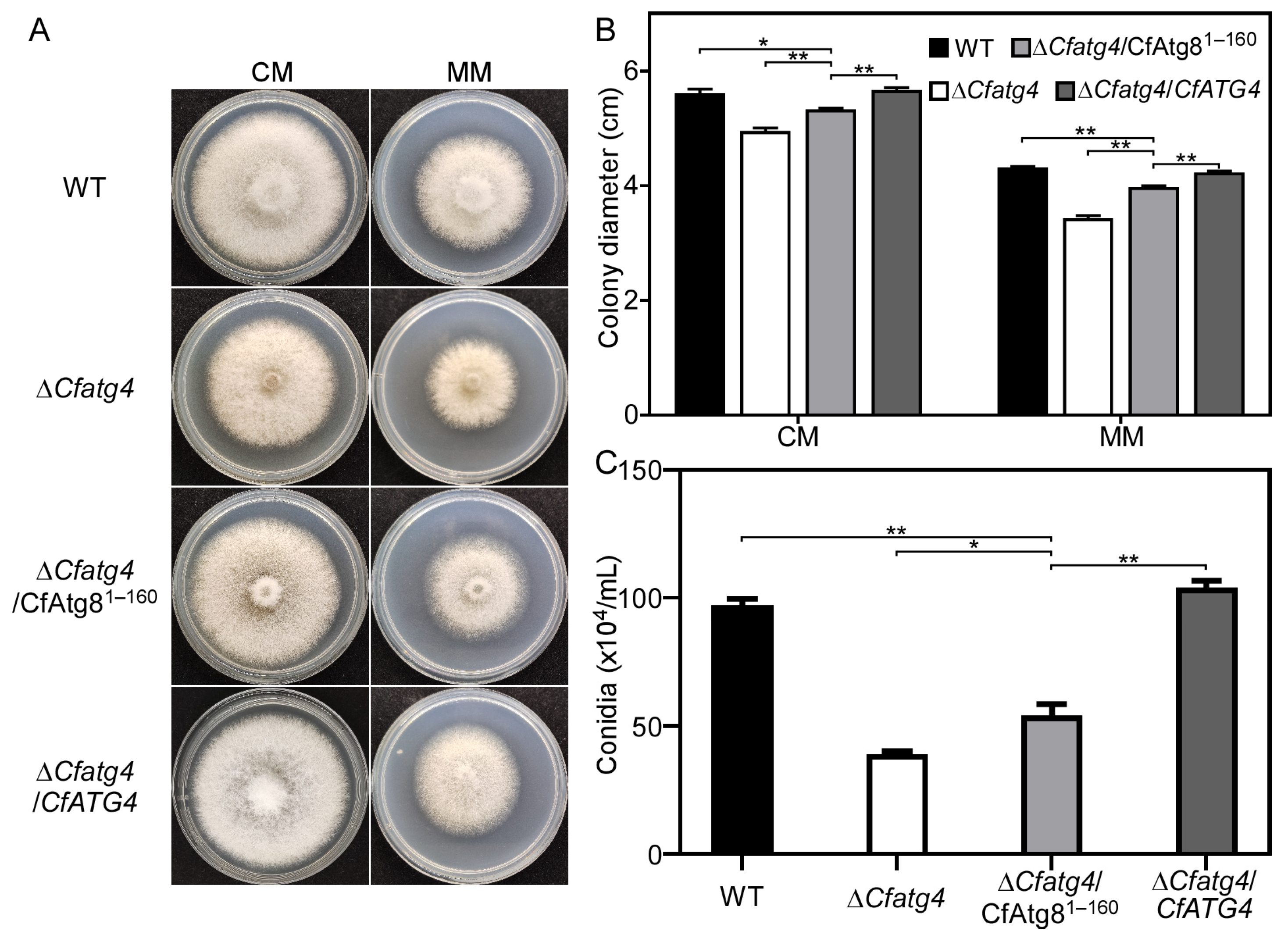
2.3. CfAtg81–160 Partially Restores Growth and Conidia Production of ΔCfatg4
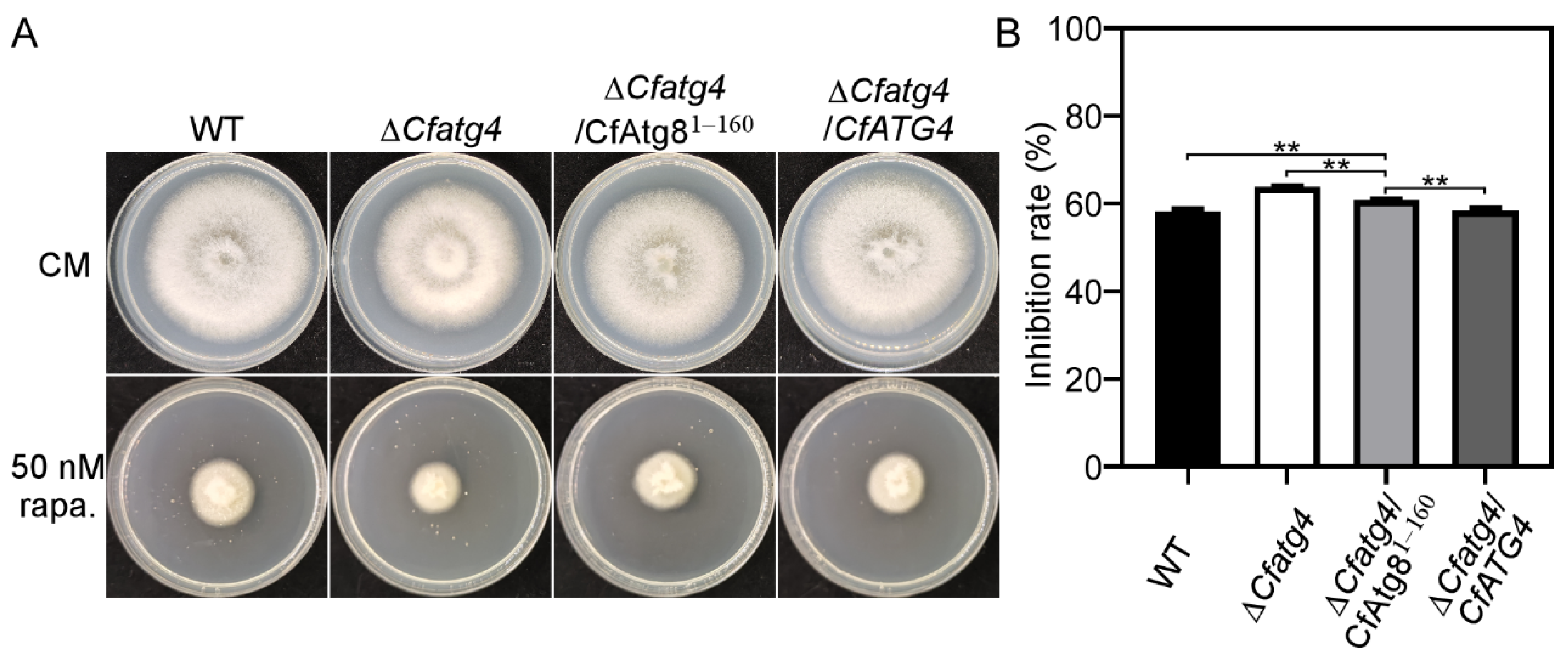
2.4. CfAtg81–160 Partially Restores the Resistance of ΔCfatg4 to Rapamycin
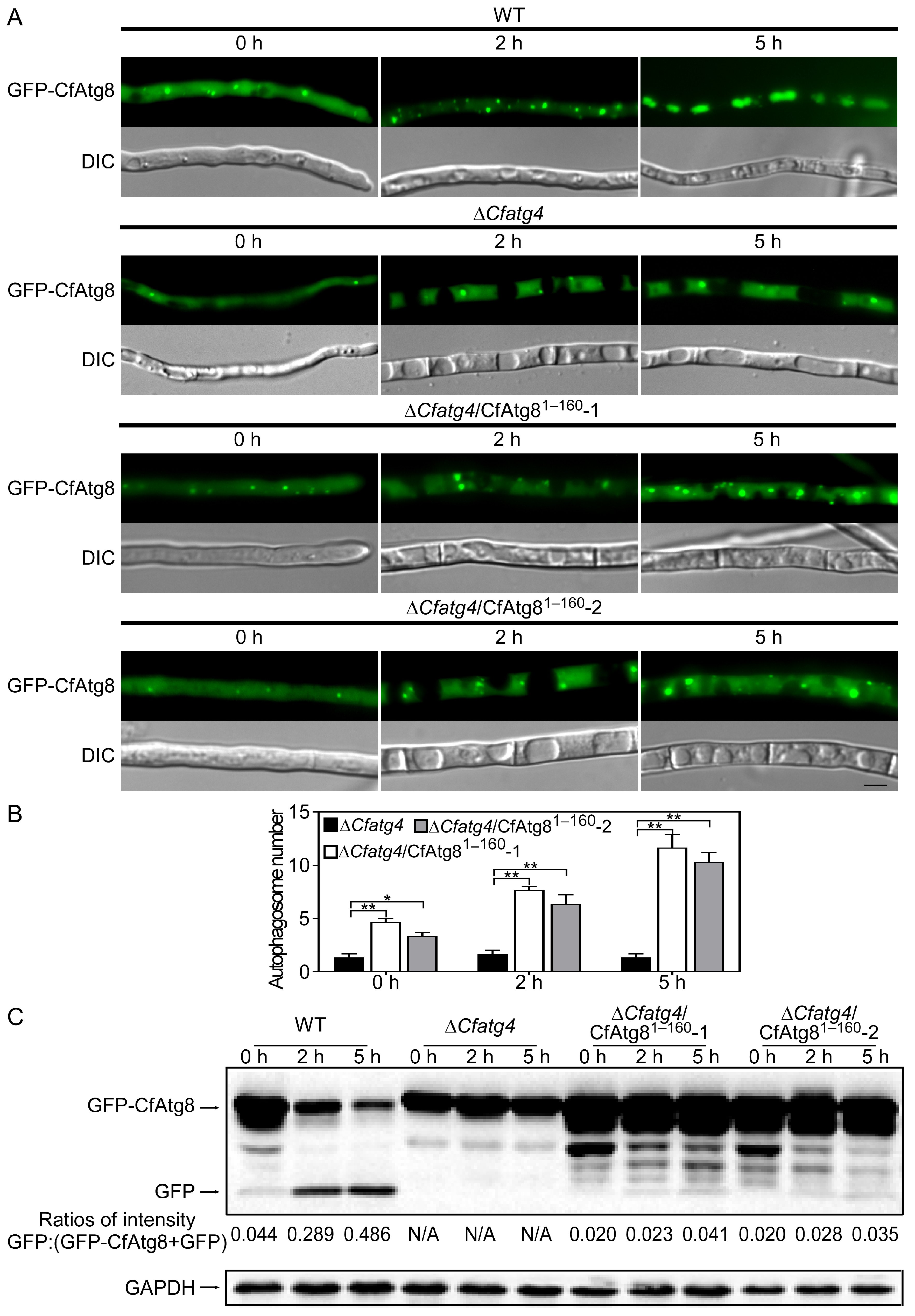
2.5. CfAtg81–160 Is Important for Autophagy
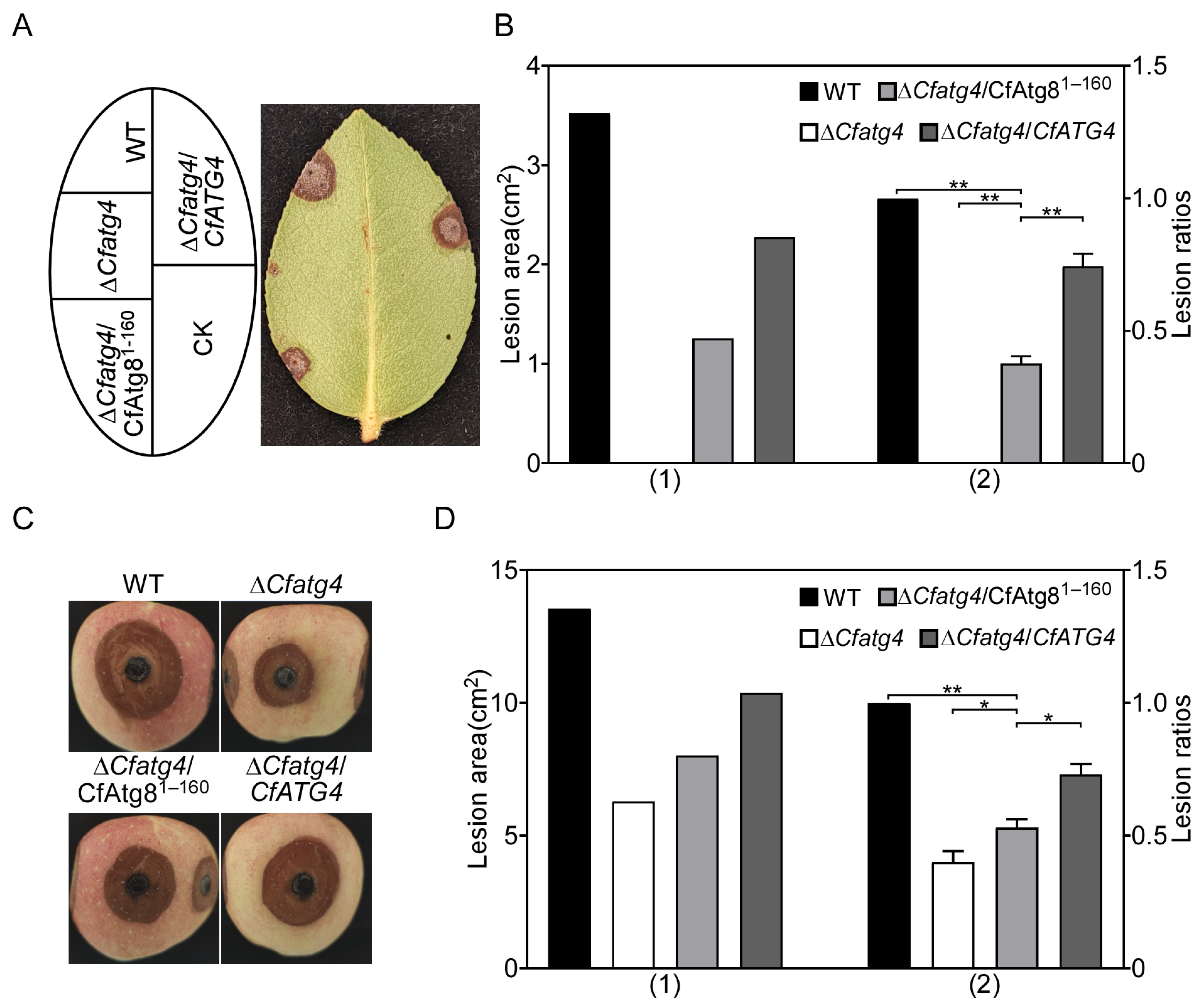
2.6. CfAtg81–160 Partially Restores the Pathogenicity Defects of ΔCfatg4
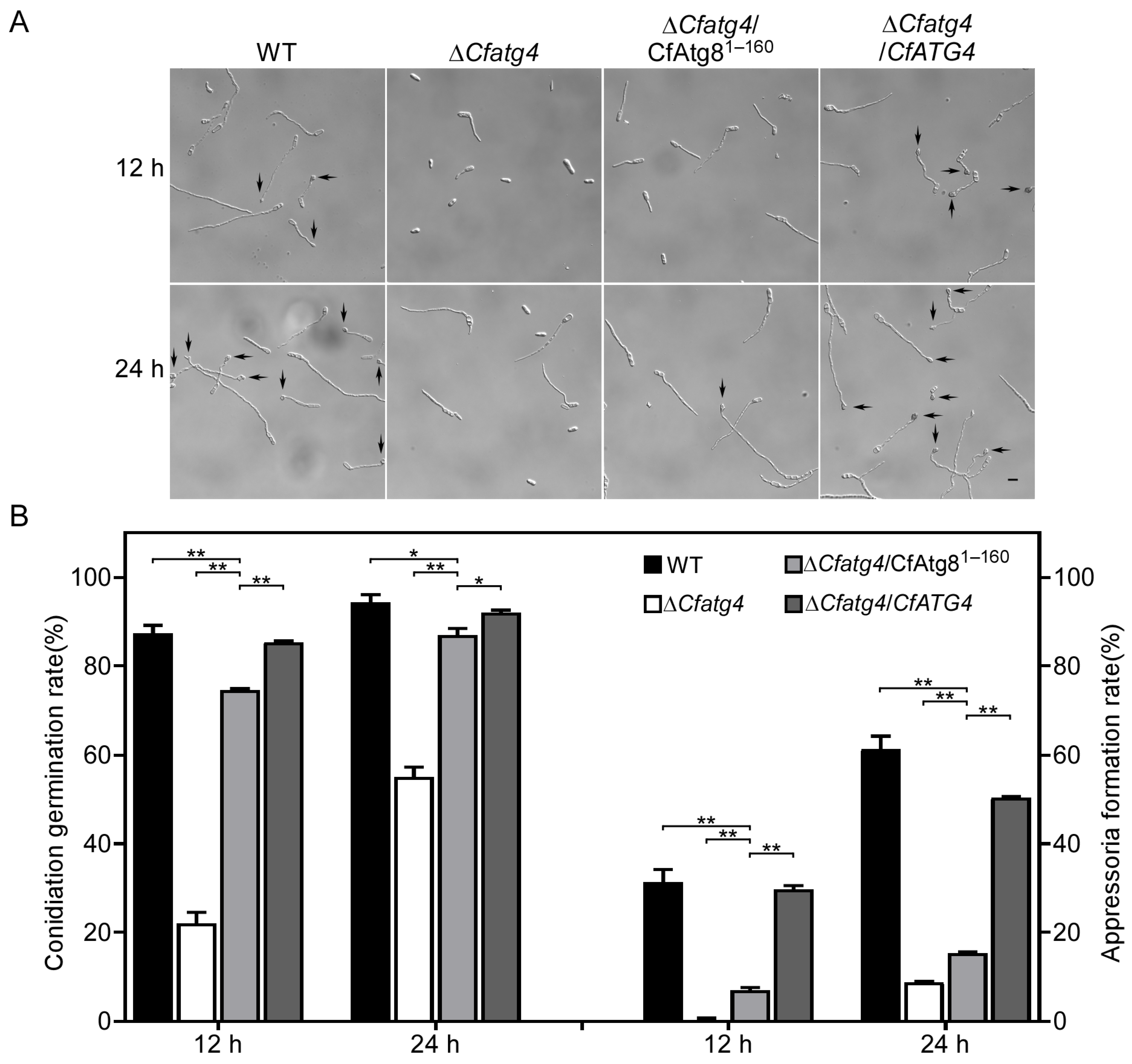
2.7. CfAtg81–160 Partially Restores Conidia Germination and Appressoria Formation in ΔCfatg4
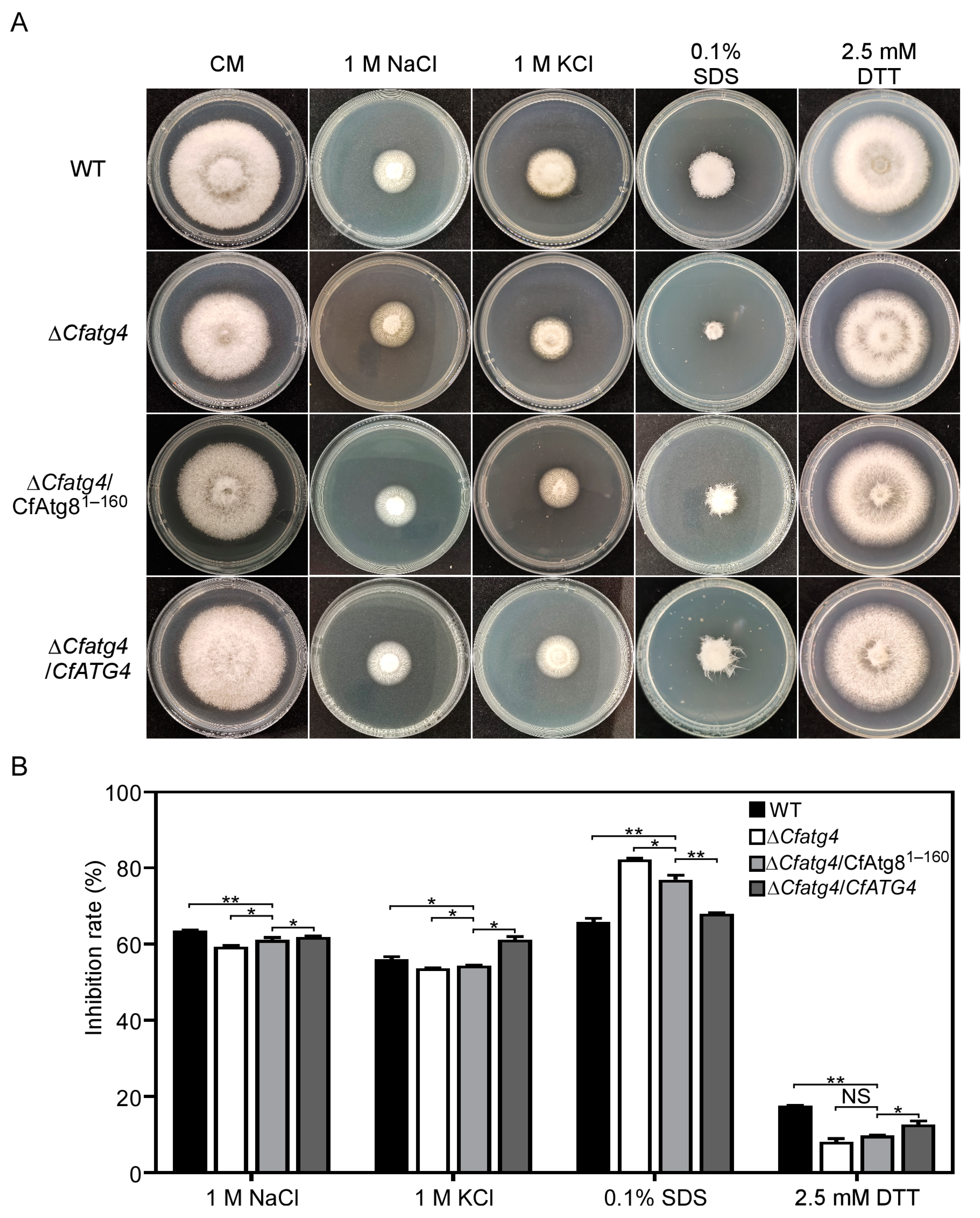
2.8. CfAtg81–160 Helps ΔCfatg4 Respond to External Environmental Stresses
3. Discussion
4. Materials and Methods
4.1. Strains and Culture Conditions
4.2. The Alignment of Atg8 Proteins
4.3. Yeast Two-Hybrid Assays
4.4. Growth, Conidiation and Appressoria Formation Assays
4.5. Stress Response Assays
4.6. Pathogenicity Assays
4.7. Autophagy Induction and Western Blotting Assays
4.8. Accession Number
4.9. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chen, Y.; Wang, B.; Chen, J.; Wang, X.; Wang, R.; Peng, S.; Chen, L.; Ma, L.; Luo, J. Identification of Rubisco rbcL and rbcS in Camellia oleifera and their potential as molecular markers for selection of high tea oil cultivars. Front. Plant Sci. 2015, 6, 131993. [Google Scholar] [CrossRef] [PubMed]
- Feás Sánchez, X.; Estevinho, L.M.; Salinero, C.; Vela, P.; Sainz Oses, M.J.; Vázquez Tato, M.d.P.; Seijas Vázquez, J.A. Triacylglyceride, antioxidant and antimicrobial features of virgin Camellia oleifera, C. reticulata and C. sasanqua oils. Molecules 2013, 18, 4573–4587. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zeng, Q.; del Mar Contreras, M.; Wang, L. Profiling and quantification of phenolic compounds in Camellia seed oils: Natural tea polyphenols in vegetable oil. Food Res. Int. 2017, 102, 184–194. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Zhou, G.-Y.; Liu, J.-A.; Xu, J. Population genetic analyses of the fungal pathogen Colletotrichum fructicola on tea-oil trees in China. PLoS ONE 2016, 11, e0156841. [Google Scholar] [CrossRef] [PubMed]
- Kershaw, M.J.; Talbot, N.J. Genome-wide functional analysis reveals that infection-associated fungal autophagy is necessary for rice blast disease. Proc. Natl. Acad. Sci. USA 2009, 106, 15967–15972. [Google Scholar] [CrossRef] [PubMed]
- Talbot, N.J.; Kershaw, M.J. The emerging role of autophagy in plant pathogen attack and host defence. Curr. Opin. Plant Biol. 2009, 12, 444–450. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.-M.; Li, L.; Wu, M.; Liang, S.; Shi, H.-B.; Liu, X.-H.; Lin, F.-C. Current opinions on autophagy in pathogenicity of fungi. Virulence 2019, 10, 481–489. [Google Scholar] [CrossRef] [PubMed]
- Veneault-Fourrey, C.; Barooah, M.; Egan, M.; Wakley, G.; Talbot, N.J. Autophagic fungal cell death is necessary for infection by the rice blast fungus. Science 2006, 312, 580–583. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.W.; Miao, P.F.; Lin, X.L.; Li, L.P.; Wu, C.X.; Chen, X.M.; Abubakar, Y.S.; Norvienyeku, J.; Li, G.P.; Zhou, J.; et al. Small GTPase Rab7-mediated FgAtg9 trafficking is essential for autophagy-dependent development and pathogenicity in Fusarium graminearum. PLoS Genet. 2018, 14, e1007546. [Google Scholar] [CrossRef]
- Chen, Y.; Jin, J.; Li, Y.; Jiao, H.; Luo, L.; Chen, Q.; Li, H.; Zhang, S. The CfAtg5 regulates the autophagy and pathogenicity of Colletotrichum fructicola on Camellia oleifera. Agronomy 2023, 13, 1237. [Google Scholar] [CrossRef]
- Zhang, S.; Guo, Y.; Li, S.; Li, H. Histone acetyltransferase CfGcn5-mediated autophagy governs the pathogenicity of Colletotrichum fructicola. mBio 2022, 13, e0195622. [Google Scholar] [CrossRef] [PubMed]
- Mizushima, N.; Komatsu, M. Autophagy: Renovation of cells and tissues. Cell 2011, 147, 728–741. [Google Scholar] [CrossRef] [PubMed]
- Ohsumi, Y. Historical landmarks of autophagy research. Cell Res. 2014, 24, 9–23. [Google Scholar] [CrossRef] [PubMed]
- Nakatogawa, H.; Ichimura, Y.; Ohsumi, Y. Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell 2007, 130, 165–178. [Google Scholar] [CrossRef]
- Ichimura, Y.; Kirisako, T.; Takao, T.; Satomi, Y.; Shimonishi, Y.; Ishihara, N.; Mizushima, N.; Tanida, I.; Kominami, E.; Ohsumi, M.; et al. A ubiquitin-like system mediates protein lipidation. Nature 2000, 408, 488–492. [Google Scholar] [CrossRef] [PubMed]
- Mizushima, N.; Yoshimori, T.; Ohsumi, Y. The role of Atg proteins in autophagosome formation. Annu. Rev. Cell Dev. Biol. 2011, 27, 107–132. [Google Scholar] [CrossRef] [PubMed]
- Ohsumi, Y. Molecular dissection of autophagy: Two ubiquitin-like systems. Nat. Rev. Mol. Cell Biol. 2001, 2, 211–216. [Google Scholar] [CrossRef]
- Mizushima, N.; Yoshimori, T.; Ohsumi, Y. Role of the Apg12 conjugation system in mammalian autophagy. Int. J. Biochem. Cell Biol. 2003, 35, 553–561. [Google Scholar] [CrossRef]
- Hirata, E.; Ohya, Y.; Suzuki, K. Atg4 plays an important role in efficient expansion of autophagic isolation membranes by cleaving lipidated Atg8 in Saccharomyces cerevisiae. PLoS ONE 2017, 12, e0181047. [Google Scholar] [CrossRef]
- Kirisako, T.; Ichimura, Y.; Okada, H.; Kabeya, Y.; Mizushima, N.; Yoshimori, T.; Ohsumi, M.; Takao, T.; Noda, T.; Ohsumi, Y. The reversible modification regulates the membrane-binding state of Apg8/Aut7 essential for autophagy and the cytoplasm to vacuole targeting pathway. J. Cell Biol. 2000, 151, 263–276. [Google Scholar] [CrossRef]
- Kim, J.; Huang, W.P.; Klionsky, D.J. Membrane recruitment of Aut7p in the autophagy and cytoplasm to vacuole targeting pathways requires Aut1p, Aut2p, and the autophagy conjugation complex. J. Cell Biol. 2001, 152, 51–64. [Google Scholar] [CrossRef] [PubMed]
- Maruyama, T.; Noda, N.N. Autophagy-regulating protease Atg4: Structure, function, regulation and inhibition. J. Antibiot. 2018, 71, 72–78. [Google Scholar] [CrossRef]
- Suzuki, K.; Kubota, Y.; Sekito, T.; Ohsumi, Y. Hierarchy of Atg proteins in pre-autophagosomal structure organization. Genes Cells 2007, 12, 209–218. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Han, L.; Huang, Y.; He, J.; He, F.; Dong, Y. Unveiling the significance of Target of Rapamycin (TOR) signalling in grafting. Veg. Res. 2024, 4, e004. [Google Scholar] [CrossRef]
- Dobrenel, T.; Caldana, C.; Hanson, J.; Robaglia, C.; Vincentz, M.; Veit, B.; Meyer, C. TOR signaling and nutrient sensing. Annu. Rev. Plant Biol. 2016, 67, 261–285. [Google Scholar] [CrossRef] [PubMed]
- Cheong, H.; Klionsky, D.J. Biochemical methods to monitor autophagy-related processes in yeast. Methods Enzymol. 2008, 451, 1–26. [Google Scholar]
- Fukada, F.; Kodama, S.; Nishiuchi, T.; Kajikawa, N.; Kubo, Y. Plant pathogenic fungi Colletotrichum and Magnaporthe share a common G1 phase monitoring strategy for proper appressorium development. New Phytol. 2019, 222, 1909–1923. [Google Scholar] [CrossRef]
- Malavazi, I.; Goldman, G.H.; Brown, N.A. The importance of connections between the cell wall integrity pathway and the unfolded protein response in filamentous fungi. Brief. Funct. Genom. 2014, 13, 456–470. [Google Scholar] [CrossRef] [PubMed]
- Tang, W.; Jiang, H.; Aron, O.; Wang, M.; Wang, X.; Chen, J.; Lin, B.; Chen, X.; Zheng, Q.; Gao, X.; et al. Endoplasmic reticulum-associated degradation mediated by MoHrd1 and MoDer1 is pivotal for appressorium development and pathogenicity of Magnaporthe oryzae. Environ. Microbiol. 2020, 22, 4953–4973. [Google Scholar] [CrossRef]
- Yin, Z.; Feng, W.; Chen, C.; Xu, J.; Li, Y.; Yang, L.; Wang, J.; Liu, X.; Wang, W.; Gao, C.; et al. Shedding light on autophagy coordinating with cell wall integrity signaling to govern pathogenicity of Magnaporthe oryzae. Autophagy 2020, 16, 900–916. [Google Scholar] [CrossRef]
- Ketelaar, T.; Voss, C.; Dimmock, S.A.; Thumm, M.; Hussey, P.J. Arabidopsis homologues of the autophagy protein Atg8 are a novel family of microtubule binding proteins. FEBS Lett. 2004, 567, 302–306. [Google Scholar] [CrossRef] [PubMed]
- Voigt, O.; Poggeler, S. Autophagy genes Smatg8 and Smatg4 are required for fruiting-body development, vegetative growth and ascospore germination in the filamentous ascomycete Sordaria macrospora. Autophagy 2013, 9, 33–49. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.B.; Liu, X.H.; Lu, J.P.; Zhang, L.; Min, H.; Lin, F.C. The cysteine protease MoAtg4 interacts with MoAtg8 and is required for differentiation and pathogenesis in Magnaporthe oryzae. Autophagy 2010, 6, 74–85. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Liu, X.; Xu, Y.; Gu, Y.; Zhang, X.; Zhang, M.; Wen, W.; Lee, Y.W.; Shi, J.; Mohamed, S.R.; et al. The autophagy-related proteins FvAtg4 and FvAtg8 are involved in virulence and fumonisin biosynthesis in Fusarium verticillioides. Virulence 2022, 13, 764–780. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.Q.; Ni, T.; Hong, B.; Wang, H.Y.; Jiang, F.J.; Zou, S.; Chen, Y.; Zheng, X.L.; Klionsky, D.J.; Liang, Y.; et al. Dual roles of Atg8-PE deconjugation by Atg4 in autophagy. Autophagy 2012, 8, 883–892. [Google Scholar] [CrossRef]
- Nair, U.; Yen, W.L.; Mari, M.; Cao, Y.; Xie, Z.; Baba, M.; Reggiori, F.; Klionsky, D.J. A role for Atg8-PE deconjugation in autophagosome biogenesis. Autophagy 2012, 8, 780–793. [Google Scholar] [CrossRef]
- Fukada, F.; Kubo, Y. Colletotrichum orbiculare regulates cell cycle G1/S progression via a two-component GAP and a GTPase to establish plant infection. Plant Cell 2015, 27, 2530–2544. [Google Scholar] [CrossRef]
- Ryder, L.S.; Cruz-Mireles, N.; Molinari, C.; Eisermann, I.; Eseola, A.B.; Talbot, N.J. The appressorium at a glance. J. Cell Sci. 2022, 135, jcs259857. [Google Scholar] [CrossRef]
- Levin, D.E. Regulation of cell wall biogenesis in Saccharomyces cerevisiae: The cell wall integrity signaling pathway. Genetics 2011, 189, 1145–1175. [Google Scholar] [CrossRef]
- Feng, W.Z.; Yin, Z.Y.; Wu, H.W.; Liu, P.; Liu, X.Y.; Liu, M.X.; Yu, R.; Gao, C.Y.; Zhang, H.F.; Zheng, X.B.; et al. Balancing of the mitotic exit network and cell wall integrity signaling governs the development and pathogenicity in Magnaporthe oryzae. PLoS Pathog 2021, 17, e1009080. [Google Scholar] [CrossRef]
- Bruno, K.S.; Tenjo, F.; Li, L.; Hamer, J.E.; Xu, J.R. Cellular localization and role of kinase activity of PMK1 in Magnaporthe grisea. Eukaryot. Cell 2004, 3, 1525–1532. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Li, H. The histone acetyltransferase CfGcn5 regulates growth, development, and pathogenicity in the anthracnose fungus Colletotrichum fructicola on the tea-oil tree. Front. Microbiol. 2021, 12, 680415. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, S.; Zhang, S. The Cysteine Protease CfAtg4 Interacts with CfAtg8 to Govern the Growth, Autophagy and Pathogenicity of Colletotrichum fructicola. J. Fungi 2024, 10, 431. https://doi.org/10.3390/jof10060431
Guo S, Zhang S. The Cysteine Protease CfAtg4 Interacts with CfAtg8 to Govern the Growth, Autophagy and Pathogenicity of Colletotrichum fructicola. Journal of Fungi. 2024; 10(6):431. https://doi.org/10.3390/jof10060431
Chicago/Turabian StyleGuo, Shufeng, and Shengpei Zhang. 2024. "The Cysteine Protease CfAtg4 Interacts with CfAtg8 to Govern the Growth, Autophagy and Pathogenicity of Colletotrichum fructicola" Journal of Fungi 10, no. 6: 431. https://doi.org/10.3390/jof10060431
APA StyleGuo, S., & Zhang, S. (2024). The Cysteine Protease CfAtg4 Interacts with CfAtg8 to Govern the Growth, Autophagy and Pathogenicity of Colletotrichum fructicola. Journal of Fungi, 10(6), 431. https://doi.org/10.3390/jof10060431




