Photoreactivation Activities of Rad5, Rad16A and Rad16B Help Beauveria bassiana to Recover from Solar Ultraviolet Damage
Abstract
:1. Introduction
2. Materials and Methods
2.1. Microbial Strains and Media
2.2. Bioinformatic Analysis of Rad5 and Rad16 Homologs in Ascomycetes
2.3. Subcellular Localization of Rad5, Rad16A and Rad16B in B. bassiana
2.4. Y2H Assays for Protein–Protein Interactions
2.5. Generation of rad5, rad16A and rad16B Mutants
2.6. Experiments for Lifecycle-Related Phenotypes
2.7. Assays for the Indices of NER and Photorepair Activities
2.8. Data Analysis
3. Results
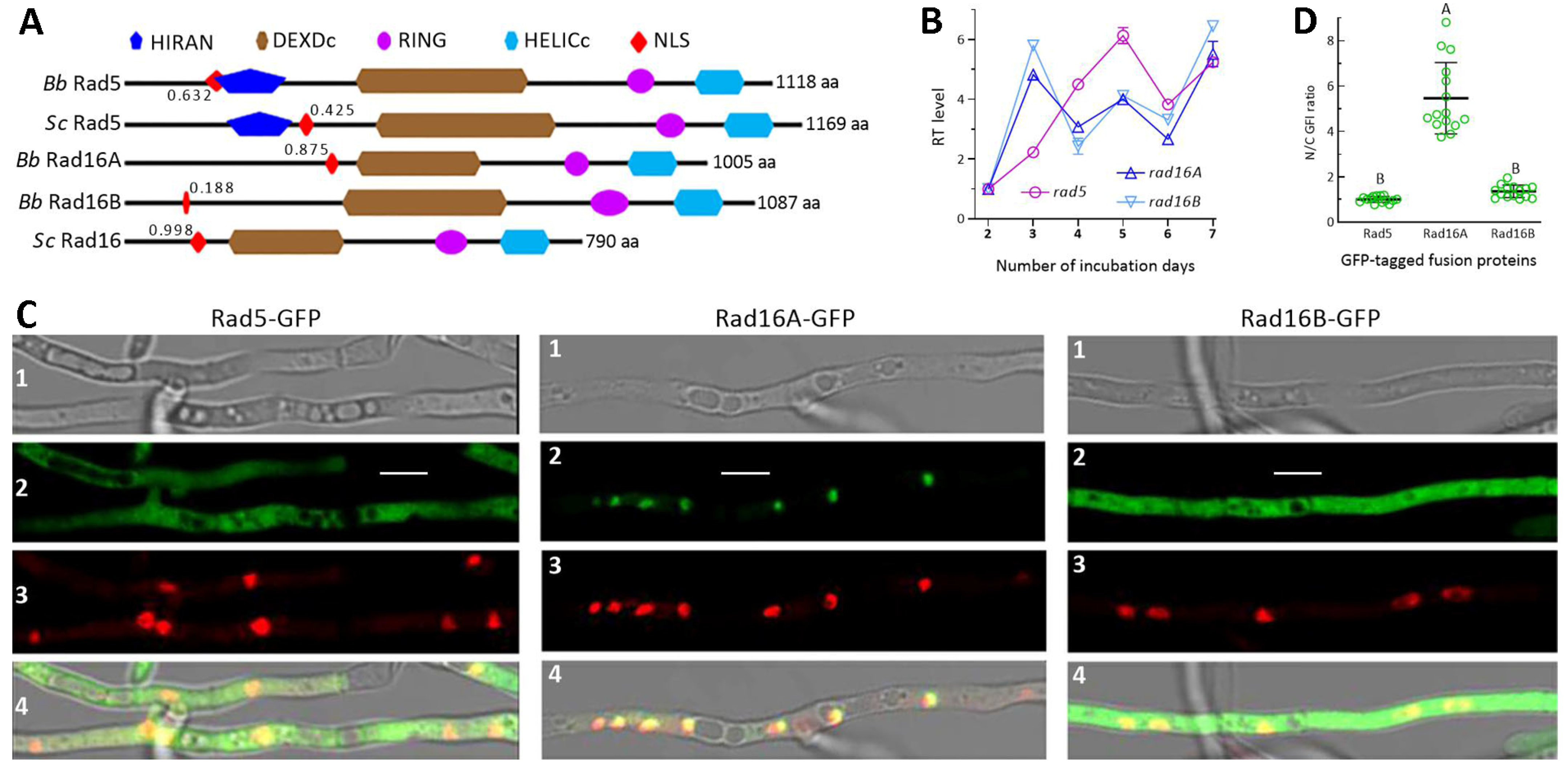
3.1. Recognition and Domain Architectures of Fungal Rad5 and Rad16 Homologs
3.2. Transcriptional and Posttranslational Features of Rad5, Rad16A and Rad16B
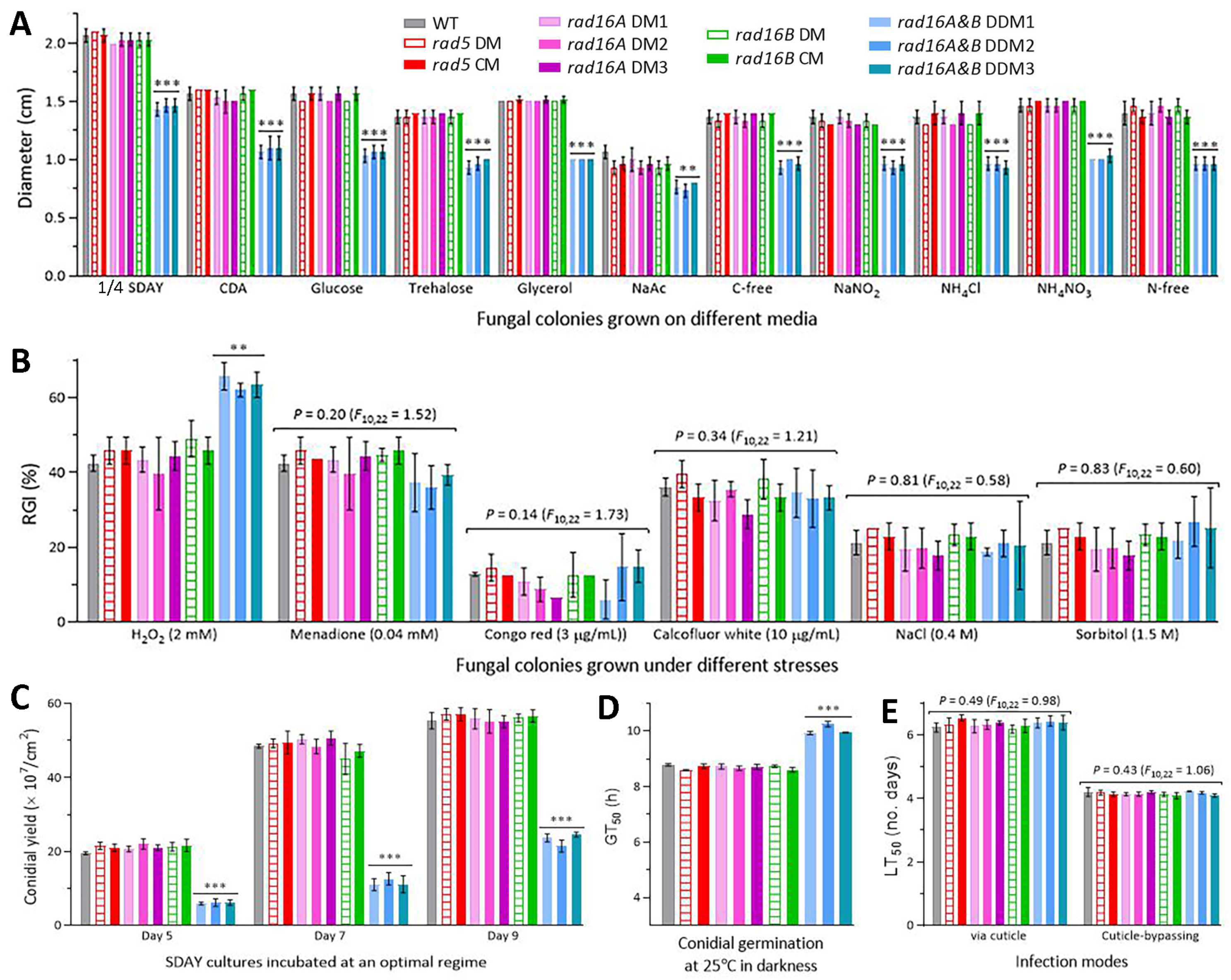
3.3. Negligible Roles of Rad5, Rad16A and Rad16B in Fungal Lifecycles
3.4. Low Activities of Rad5, Rad16A and Rad16B in Dark Reactivation
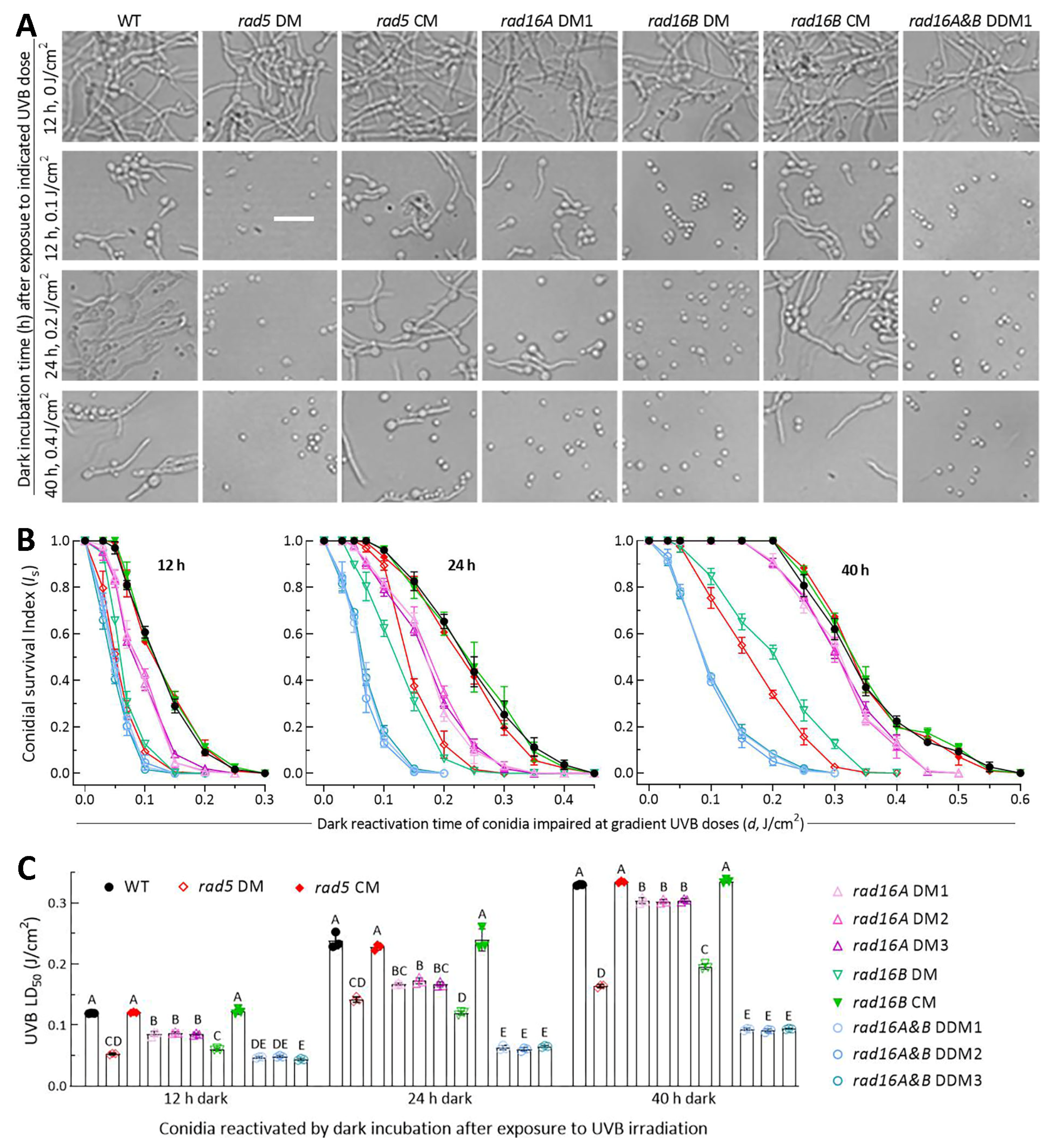
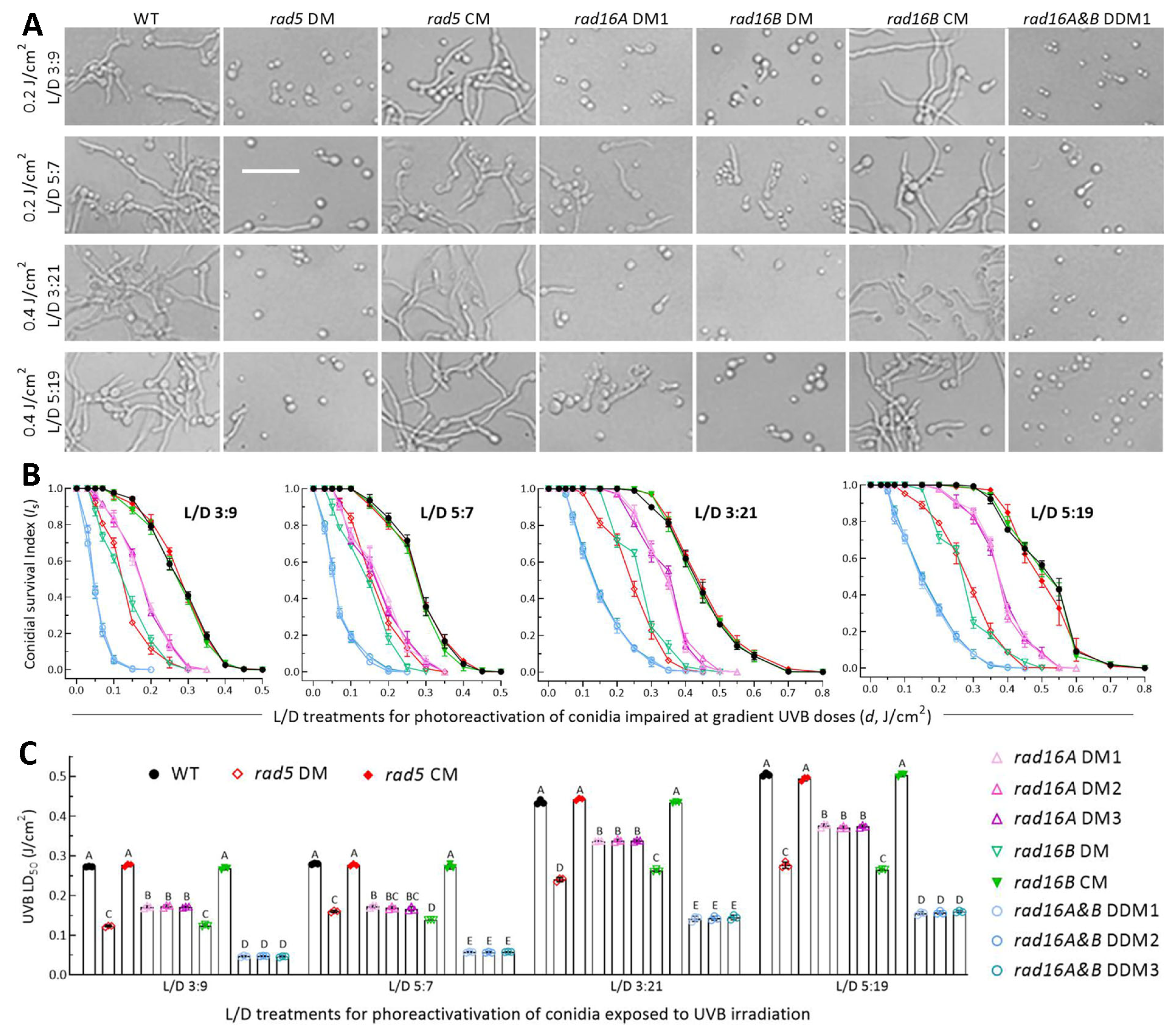
3.5. High Activities of Rad5, Rad16A and Rad16B in Photoreactivation
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Yu, L.; Xu, S.Y.; Tong, S.M.; Ying, S.H.; Feng, M.G. Optional strategies for low–risk and non–risk applications of fungal pesticides to avoid solar ultraviolet damage. Pest Manag. Sci. 2022, 78, 4660–4667. [Google Scholar] [CrossRef] [PubMed]
- Madronich, S. UV radiation in the natural and perturbed atmosphere. In UV–B Radiation and Ozone Depletion; Tevini, M., Ed.; Lewis: Boca Raton, FL, USA, 1993; pp. 17–69. [Google Scholar]
- Griffiths, H.R.; Mistry, P.; Herbert, K.E.; Lunec, J. Molecular and cellular effects of ultraviolet light–induced genotoxicity. Crit. Rev. Clin. Lab. Sci. 1998, 35, 189–237. [Google Scholar] [CrossRef] [PubMed]
- Braga, G.U.L.; Flint, S.D.; Miller, C.D.; Anderson, A.J.; Roberts, D.W. Both solar UVA and UVB radiation impair conidial culturability and delay germination in the entomopathogenic fungus Metarhizium anisopliae. Photochem. Photobiol. 2001, 74, 734–739. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, E.K.K.; Rangel, D.E.N.; Moraes, A.M.L.; Bittencourt, V.R.E.P.; Roberts, D.W. Variability in tolerance to UV–B radiation among Beauveria spp. isolates. J. Invertebr. Pathol. 2007, 96, 237–243. [Google Scholar] [CrossRef] [PubMed]
- Yao, S.L.; Ying, S.H.; Feng, M.G.; Hatting, J.L. In vitro and in vivo responses of fungal biocontrol agents to the gradient doses of UV–B and UV–A irradiation. BioControl 2010, 55, 413–422. [Google Scholar] [CrossRef]
- Yasui, A.; Eker, A.P.M.; Yasuhira, S.; Yajima, H.; Kobayashi, T.; Takao, M.; Oikawa, A. A new class of DNA photolyases present in various organisms including aplacental mammals. EMBO J. 1994, 13, 6143–6151. [Google Scholar] [CrossRef] [PubMed]
- Sancar, A. No “End of History” for Photolyases. Science 1996, 272, 48–49. [Google Scholar] [CrossRef]
- Sancar, A. Structure and function of DNA photolyase and cryptochrome blue–light photoreceptors. Chem. Rev. 2003, 103, 2203–2238. [Google Scholar] [CrossRef] [PubMed]
- de Laat, W.L.; Jaspers, N.G.J.; Hoeijmakers, J.H.J. Molecular mechanism of nucleotide excision repair. Genes Dev. 1999, 13, 768–785. [Google Scholar] [CrossRef] [PubMed]
- Brancini, G.T.P.; Hallsworth, J.E.; Corrochano, L.M.; Braga, G.U.L. Photobiology of the keystone genus Metarhizium. J. Photochem. Photobiol. B–Biol. 2022, 226, 112374. [Google Scholar] [CrossRef] [PubMed]
- Prakash, S.; Sung, P.; Prakash, L. DNA repair genes and proteins of Saccharomyces cerevisiae. Annu. Rev. Genet. 1993, 27, 33–70. [Google Scholar] [CrossRef]
- Smerdon, M.J.; Thoma, F. Site–specific DNA–repair at the nucleosome level in a yeast minichromosome. Cell 1990, 61, 675–684. [Google Scholar] [CrossRef] [PubMed]
- Suter, B.; Wellinger, R.E.; Thoma, F. DNA repair in a yeast origin of replication: Contributions of photolyase and nucleotide excision repair. Nucleic Acids Res. 2000, 28, 2060–2068. [Google Scholar] [CrossRef] [PubMed]
- Gong, F.; Fahy, D.; Smerdon, M.J. Rad4–Rad23 interaction with SWI/SNF links ATP–dependent chromatin remodeling with nucleotide excision repair. Nat. Struct. Mol. Biol. 2006, 13, 902–907. [Google Scholar] [CrossRef] [PubMed]
- Gödderz, D.; Giovannucci, T.; Laláková, J.; Benito, V.M.; Dantuma, N.P. The deubiquitylating enzyme Ubp12 regulates Rad23–dependent proteasomal degradation. J. Cell Sci. 2017, 130, 3336–3346. [Google Scholar] [CrossRef]
- Tsuchiya, H.; Ohtake, F.; Arai, N.; Kaiho, A.; Yasuda, S.; Tanaka, K.; Saeki, Y. In vivo ubiquitin linkage–type analysis reveals that the Cdc48–Rad23/Dsk2 axis contributes to K48–linked chain specificity of the proteasome. Mol. Cell 2017, 66, 488–502. [Google Scholar] [CrossRef] [PubMed]
- Boiteux, S.; Jinks–Robertson, S. DNA repair mechanisms and the bypass of DNA damage in Saccharomyces cerevisiae. Genetics 2013, 193, 1025–1064. [Google Scholar] [CrossRef] [PubMed]
- Den Dulk, B.; Sun, S.M.; De Ruijter, M.; Brandsma, J.A.; Brouwer, J. Rad33, a new factor involved in nucleotide excision repair in Saccharomyces cerevisiae. DNA Repair 2006, 5, 683–692. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, S.; Kiely, R.; McHugh, P.J. The Ino80 chromatin–remodeling complex restores chromatin structure during UV DNA damage repair. J. Cell Biol. 2010, 191, 1061–1068. [Google Scholar] [CrossRef] [PubMed]
- Bailly, V.; Sommers, C.H.; Sung, P.; Prakash, L.; Prakash, S. Specific complex formation between proteins encoded by the yeast DNA repair and recombination genes RAD1 and RAD10. Proc. Natl. Acad. Sci. USA 1992, 89, 8273–8277. [Google Scholar] [CrossRef] [PubMed]
- Tomkinson, A.E.; Bardwell, A.J.; Bardwell, L.; Tappe, N.J.; Friedberg, E.C. Yeast DNA repair and recombination proteins Rad1 and Rad10 constitute a single–stranded–DNA endonuclease. Nature 1993, 362, 860–862. [Google Scholar] [CrossRef] [PubMed]
- Bardwell, A.J.; Bardwell, L.; Tomkinson, A.E.; Friedberg, E.C. Specific cleavage of model recombination and repair intermediates by the yeast Rad1–Rad10 DNA endonuclease. Science 1994, 265, 2082–2085. [Google Scholar] [CrossRef] [PubMed]
- Davies, A.A.; Friedberg, E.C.; Tomkinson, A.E.; Wood, R.D.; West, S.C. Role of the Rad1 and Rad10 proteins in nucleotide excision repair and recombination. J. Biol. Chem. 1995, 270, 24638–24641. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, K.; Wang, Z.; Friedberg, E.C.; Tomkinson, A.E. Identification of functional domains within the RAD1•RAD10 repair and recombination endonuclease of Saccharomyces cerevisiae. J. Biol. Chem. 1996, 271, 20551–20558. [Google Scholar] [CrossRef] [PubMed]
- Evans, E.J.; Moggs, G.; Hwang, J.R.; Egly, J.M.; Wood, R.D. Mechanism of open complex and dual incision formation by human nucleotide excision repair factors. EMBO J. 1997, 16, 6559–6573. [Google Scholar] [CrossRef] [PubMed]
- Staresincic, L.; Fagbemi, A.F.; Enzlin, J.H.; Gourdin, A.M.; Wijgers, N.; Dunand–Sauthier, I.; Giglia–Mari, G.; Clarkson, S.G.; Vermeulen, W.; Schaerer, O.D. Coordination of dual incision and repair synthesis in human nucleotide excision repair. EMBO J. 2009, 28, 111–1120. [Google Scholar] [CrossRef]
- Lafrance–Vanasse, J.; Arseneault, G.; Cappadocia, L.; Chen, H.T.; Legault, P.; Omichinski, J.G. Structural and functional characterization of interactions involving the Tfb1 subunit of TFIIH and the NER factor Rad2. Nucleic Acids Res. 2012, 40, 5739–5750. [Google Scholar] [CrossRef] [PubMed]
- Verhage, R.; Zeeman, A.M.; De Groot, N.; Gleig, F.; Bang, D.D.; Vandeputte, P.; Brouwer, J. The RAD7 and RAD16 genes, which are essential for pyrimidine dimer removal from the silent mating type loci, are also required for repair of the nontranscribed strand of an active gene in Saccharomyces cerevisiae. Mol. Cell. Biol. 1994, 14, 6135–6142. [Google Scholar] [PubMed]
- Guzder, S.N.; Sung, P.; Prakash, L.; Prakash, S. Yeast Rad7–Rad16 complex, specific for the nucleotide excision repair of the nontranscribed DNA strand, is an ATP–dependent DNA damage sensor. J. Biol. Chem. 1997, 272, 21665–21668. [Google Scholar] [CrossRef] [PubMed]
- Guzder, S.N.; Sung, P.; Prakash, L.; Prakash, S. Synergistic interaction between yeast nucleotide excision repair factors NEF2 and NEF4 in the binding of ultraviolet–damaged DNA. J. Biol. Chem. 1999, 274, 24257–24262. [Google Scholar] [CrossRef] [PubMed]
- Reed, S.H.; You, Z.; Friedberg, E.C. The yeast RAD7 and RAD16 genes are required for postincision events during nucleotide excision repair. In vitro and in vivo studies with rad7 and rad16 mutants and purification of a Rad7/Rad16–containing protein complex. J. Biol. Chem. 1998, 273, 29481–29488. [Google Scholar] [CrossRef] [PubMed]
- Reed, S.H.; Akiyama, M.; Stillman, B.; Friedberg, E.C. Yeast autonomously replicating sequence binding factor is involved in nucleotide excision repair. Genes Dev. 1999, 13, 3052–3058. [Google Scholar] [CrossRef] [PubMed]
- Teng, Y.; Liu, H.; Gill, H.W.; Yu, Y.; Waters, R.; Reed, S.H. Saccharomyces cerevisiae Rad16 mediates ultraviolet–dependent histone H3 acetylation required for efficient global genome nucleotide–excision repair. EMBO Rep. 2008, 9, 97–102. [Google Scholar] [CrossRef] [PubMed]
- Chaudhuri, S.; Wyrick, J.J.; Smerdon, M.J. Histone H3 Lys79 methylation is required for efficient nucleotide excision repair in a silenced locus of Saccharomyces cerevisiae. Nucleic Acids Res. 2009, 37, 1690–1700. [Google Scholar] [CrossRef] [PubMed]
- Ramsey, K.L.; Smith, J.J.; Dasgupta, A.; Maqani, N.; Grant, P.; Auble, D.T. The NEF4 complex regulates Rad4 levels and utilizes Snf2/Swi2–related ATPase activity for nucleotide excision repair. Mol. Cell. Biol. 2004, 24, 6362–6378. [Google Scholar] [CrossRef] [PubMed]
- Gillette, T.G.; Yu, S.; Zhou, Z.; Waters, R.; Johnston, S.A.; Reed, S.H. Distinct functions of the ubiquitin–proteasome pathway influence nucleotide excision repair. EMBO J. 2006, 25, 2529–2538. [Google Scholar] [CrossRef] [PubMed]
- Guzder, S.N.; Habraken, Y.; Sung, P.; Prakash, L.; Prakash, S. RAD26, the yeast homolog of human Cockayne’s syndrome group B gene, encodes a DNA–dependent ATPase. J. Biol. Chem. 1996, 271, 18314–18317. [Google Scholar] [CrossRef] [PubMed]
- Malik, S.; Chaurasia, P.; Lahudkar, S.; Durairaj, G.; Shukla, A.; Bhaumik, S.R. Rad26p, a transcription–coupled repair factor, is recruited to the site of DNA lesion in an elongating RNA polymerase II–dependent manner in vivo. Nucleic Acids Res. 2010, 38, 1461–1477. [Google Scholar] [CrossRef] [PubMed]
- Taschner, M.; Harreman, M.; Teng, Y.; Gill, H.; Anindya, R.; Maslen, S.L.; Skehel, J.M.; Waters, R.; Svejstrup, J.Q. A role for checkpoint kinase–dependent Rad26 phosphorylation in transcription–coupled DNA repair in Saccharomyces cerevisiae. Mol. Cell. Biol. 2010, 30, 436–446. [Google Scholar] [CrossRef] [PubMed]
- Ulrich, H.D.; Jentsch, S. Two RING finger proteins mediate cooperation between ubiquitin–conjugating enzymes in DNA repair. EMBO J. 2000, 19, 3388–3397. [Google Scholar] [CrossRef] [PubMed]
- Gangavarapu, V.; Haracska, L.; Unk, I.; Johnson, R.E.; Prakash, S.; Prakash, L. Mms2–Ubc13–dependent and –independent roles of Rad5 ligase in postreplication repair and translesion DNA synthesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 2006, 26, 7783–7790. [Google Scholar] [CrossRef] [PubMed]
- Pagès, V.; Bresson, A.; Acharya, N.; Prakash, S.; Fuchs, R.P.; Prakash, L. Requirement of Rad5 for DNA polymerase ζ–dependent translesion synthesis in Saccharomyces cerevisiae. Genetics 2008, 180, 73–82. [Google Scholar] [CrossRef] [PubMed]
- Jans, J.; Schul, W.; Sert, Y.G.; Rijksen, Y.; Rebel, H.; Eker, A.P.M.; Nakajima, S.; van Steeg, H.; de Gruijl, F.R.; Yasui, A.; et al. Powerful skin cancer protection by a CPD–photolyase transgene. Curr. Biol. 2005, 15, 105–115. [Google Scholar] [CrossRef] [PubMed]
- Chaves, I.; Pokorny, R.; Byrdin, M.; Hoang, N.; Ritz, T.; Brettel, K.; Essen, L.-O.; van der Horst, G.T.J.; Batschauer, A.; Ahmad, M. The cryptochromes: Blue light photoreceptors in plants and animals. Annu. Rev. Plant Biol. 2011, 62, 335–364. [Google Scholar] [CrossRef] [PubMed]
- Berrocal–Tito, G.M.; Esquivel–Naranjo, E.U.; Horwitz, B.A.; Herrera–Estrella, A. Trichoderma atroviride PHR1, a fungal photolyase responsible for DNA repair, autoregulates its own photoinduction. Eukaryot. Cell 2007, 6, 1682–1692. [Google Scholar] [CrossRef] [PubMed]
- Brych, A.; Mascarenhas, J.; Jaeger, E.; Charkiewicz, E.; Pokorny, R.; Boelker, M.; Doehlemann, G.; Batschauer, A. White collar 1–induced photolyase expression contributes to UV–tolerance of Ustilago maydis. MicrobiologyOpen 2016, 5, 224–243. [Google Scholar] [CrossRef] [PubMed]
- Garcia–Esquivel, M.; Esquivel–Naranjo, E.U.; Hernandez–Onate, M.A.; Ibarra–Laclette, E.; Herrera–Estrella, A. The Trichoderma atroviride cryptochrome/photolyase genes regulate the expression of blr1–independent genes both in red and blue light. Fungal Biol. 2016, 120, 500–512. [Google Scholar] [CrossRef]
- Cohrs, K.C.; Schumacher, J. The two cryptochrome/photolyase family proteins fulfill distinct roles in DNA photorepair and regulation of conidiation in the gray mold fungus Botrytis cinerea. Appl. Environ. Microbiol. 2017, 83, e00812. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.Y.; Fu, B.; Tong, S.M.; Ying, S.H.; Feng, M.G. Two photolyases repair distinct DNA lesions and reactivate UVB–inactivated conidia of an insect mycopathogen under visible light. Appl. Environ. Microbiol. 2019, 85, e02459-18. [Google Scholar] [CrossRef] [PubMed]
- Tong, S.M.; Feng, M.G. Molecular basis and regulatory mechanisms underlying fungal insecticides’ resistance to solar ultraviolet irradiation. Pest Manag. Sci. 2022, 78, 30–42. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.Y.; Mou, Y.N.; Tong, S.M.; Ying, S.H.; Feng, M.G. Photoprotective role of photolyase–interacting RAD23 and its pleiotropic effect on the insect–pathogenic fungus Beauveria bassiana. Appl. Environ. Microbiol. 2020, 86, e00287-20. [Google Scholar] [CrossRef] [PubMed]
- Peng, H.; Guo, C.T.; Tong, S.M.; Ying, S.H.; Feng, M.G. Two white collar proteins protect fungal cells from solar UV damage by their interactions with two photolyases in Metarhizium robertsii. Environ. Microbiol. 2021, 23, 4925–4938. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Xu, S.Y.; Luo, X.C.; Ying, S.H.; Feng, M.G. Rad1 and Rad10 tied to photolyase regulators protect insecticidal fungal cells from solar UV damage by photoreactivation. J. Fungi 2022, 8, 1124. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.Y.; Yu, L.; Luo, X.C.; Ying, S.H.; Feng, M.G. Co–regulatory roles of WC1 and WC2 in asexual development and photoreactivation of Beauveria bassiana. J. Fungi 2023, 9, 290. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.L.; Peng, H.; Ying, S.H.; Feng, M.G. Efficient photoreactivation of solar UV–injured Metarhizium robertsii by Rad1 and Rad10 linked to DNA photorepair–required proteins. Photochem. Photobiol. 2023, 99, 1122–1130. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Xu, S.Y.; Luo, X.C.; Ying, S.H.; Feng, M.G. Comparative roles of Rad4A and Rad4B in photoprotection of Beauveria bassiana from solar ultraviolet damage. J. Fungi 2023, 9, 154. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.L.; Peng, H.; Zhang, K.; Ying, S.H.; Feng, M.G. Divergent roles of Rad4 and Rad23 homologs in Metarhizium robertsii’s resistance to solar ultraviolet damage. Appl. Environ. Microbiol. 2023, 89, e0099423. [Google Scholar] [CrossRef] [PubMed]
- Peng, H.; Zhang, Y.L.; Ying, S.H.; Feng, M.G. Rad2, Rad14 and Rad26 recover solar UV damage by their links to photoreactivation–depending factors in Metarhizium robertsii. Microbiol. Res. 2023, 280, 127589. [Google Scholar]
- Yu, L.; Xu, S.Y.; Luo, X.C.; Ying, S.H.; Feng, M.G. High photoreactivation activities of Rad2 and Rad14 in recovering insecticidal Beauveria bassiana from solar UV damage. J. Photochem. Photobiol. B–Biol. 2024, 251, 112849. [Google Scholar] [CrossRef]
- Fang, W.G.; Zhang, Y.J.; Yang, X.Y.; Zheng, X.L.; Duan, H.; Li, Y.; Pei, Y. Agrobacterium tumefaciens–mediated transformation of Beauveria bassiana using an herbicide resistance gene as a selection marker. J. Invertebr. Pathol. 2004, 85, 18–24. [Google Scholar] [CrossRef] [PubMed]
- Huang, B.F.; Feng, M.G. Comparative tolerances of various Beauveria bassiana isolates to UV–B irradiation with a description of a modeling method to assess lethal dose. Mycopathologia 2009, 168, 145–152. [Google Scholar] [CrossRef] [PubMed]
- Broomfield, S.; Chow, B.L.; Xiao, W. MMS2, encoding a ubiquitin–conjugating–enzyme–like protein, is a member of the yeast error–free postreplication repair pathway. Proc. Natl. Acad. Sci. USA 1998, 95, 5678–5683. [Google Scholar] [CrossRef] [PubMed]
- Lejeune, D.; Chen, X.; Ruggiero, C.; Berryhill, S.; Ding, B.; Li, S. Yeast Elc1 plays an important role in global genomic repair but not in transcription coupled repair. DNA Repair 2009, 8, 40–50. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.Y.; Wen, Z.X.; Li, X.J.; Hu, E.Z.; Qi, D.Y.; Feng, M.G.; Tong, S.M. Timing of fungal insecticide application to avoid solar ultraviolet irradiation enhances field control of rice planthoppers. Insects 2023, 14, 307. [Google Scholar] [CrossRef]
- Qi, D.Y.; Xu, W.Y.; Shao, Y.Z.; Feng, J.R.; Feng, M.G.; Tong, S.M. Mycoinsecticides applied in late afternoon are more efficacious against rice leaf–rolling insect pests than those in the morning. Biol. Control 2023, 186, 105352. [Google Scholar] [CrossRef]

| Strain (Abbreviated) Name | Medium (Abbreviated) Name | Purpose |
|---|---|---|
| B. bassiana ARSEF2860 wild-type strain (WT) | Sabouraud dextrose agar (w/v: 4% glucose, 1% peptone and 1.5% agar) plus 1% yeast extract (SDAY), 1/4 SDAY (amended with 1/4 nutrition strength of SDAY) | Gene manipulation, growth and conidiation |
| Targeted gene deletion mutants (DMs) | Czapek-Dox agar (CDA; w/v: 3% sucrose, 0.3% NaNO3, 0.1% K2HPO4, 0.05% KCl, 0.05% MgSO4 and 0.001% FeSO4 plus 1.5% agar), CDAs amended with different carbon or nitrogen sources | Radial growth, stress response |
| Targeted gene complementation mutants (CMs) | Germination medium (GM; w/v: 2% sucrose, 0.5% peptone and 1.5% agar) | Germination after UVB irradiation |
| S. cerevisiae Y187 and Y2HGold | YPD (w/v: 1% yeast extract, 2% peptone, 2% glucose plus 0.04% adenine hemisulfate salt) | Yeast two-hybrid (Y2H) assays |
| Escherichia coli DH5α | Luria-Bertani medium containing ampicillin (100 mg/mL) or kanamycin (50 mg/mL) | Vector propagation |
| Agrobacterium tumefaciens AGL-1 | YEB (w/v: 0.5% sucrose, 0.1% yeast extract, 1% peptone, 0.05% MgSO4, 1.5% agar) | Fungal transformation |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Luo, X.-C.; Yu, L.; Xu, S.-Y.; Ying, S.-H.; Feng, M.-G. Photoreactivation Activities of Rad5, Rad16A and Rad16B Help Beauveria bassiana to Recover from Solar Ultraviolet Damage. J. Fungi 2024, 10, 420. https://doi.org/10.3390/jof10060420
Luo X-C, Yu L, Xu S-Y, Ying S-H, Feng M-G. Photoreactivation Activities of Rad5, Rad16A and Rad16B Help Beauveria bassiana to Recover from Solar Ultraviolet Damage. Journal of Fungi. 2024; 10(6):420. https://doi.org/10.3390/jof10060420
Chicago/Turabian StyleLuo, Xin-Cheng, Lei Yu, Si-Yuan Xu, Sheng-Hua Ying, and Ming-Guang Feng. 2024. "Photoreactivation Activities of Rad5, Rad16A and Rad16B Help Beauveria bassiana to Recover from Solar Ultraviolet Damage" Journal of Fungi 10, no. 6: 420. https://doi.org/10.3390/jof10060420
APA StyleLuo, X.-C., Yu, L., Xu, S.-Y., Ying, S.-H., & Feng, M.-G. (2024). Photoreactivation Activities of Rad5, Rad16A and Rad16B Help Beauveria bassiana to Recover from Solar Ultraviolet Damage. Journal of Fungi, 10(6), 420. https://doi.org/10.3390/jof10060420









