Genome Mining of Fungal Unique Trichodiene Synthase-like Sesquiterpene Synthases
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains and Media
2.2. Collection and Phylogenetic Analysis of Reported Fungal STSs
2.3. Extraction and SSN Analysis of TDTSs from Fungi Genome Database
2.4. Construction of Expression Plasmids
2.5. Preparation of Protoplasts and Transformation of A. oryzae
2.6. Fermentation and Analysis of the Metabolites from A. oryzae Transformants
3. Results
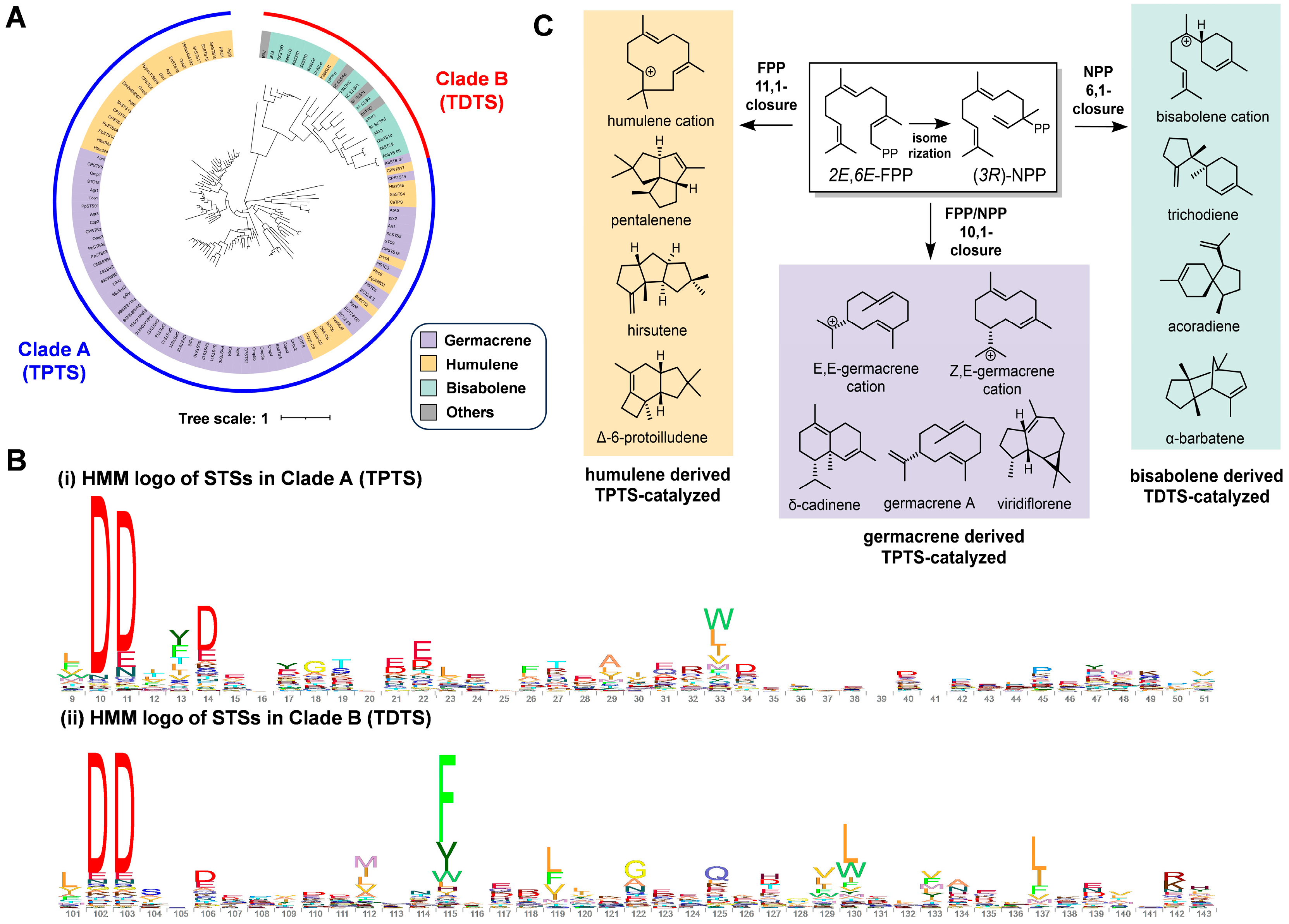
3.1. Identification of TDTSs through Phylogenetic Analysis of Fungal STSs
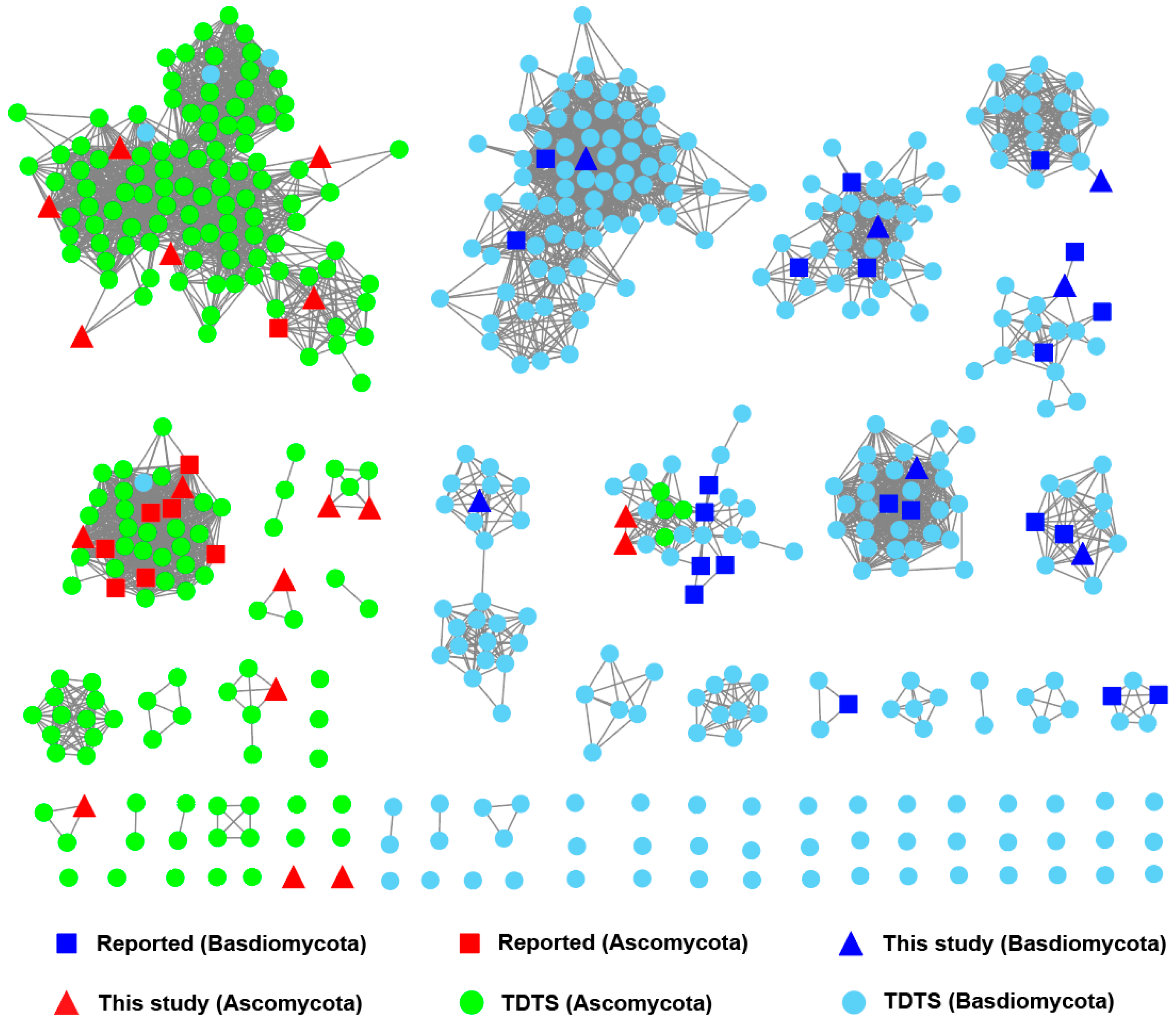
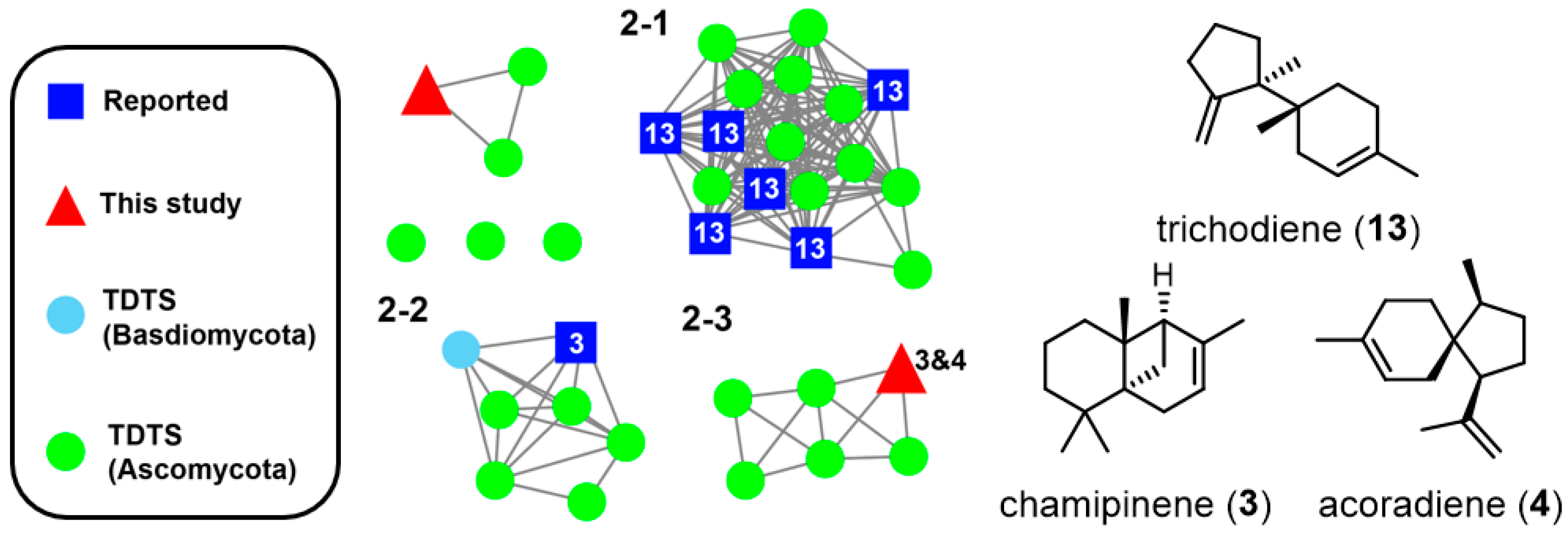
3.2. Extraction and SSN Analysis of TDTSs from a Fungal Genome Database
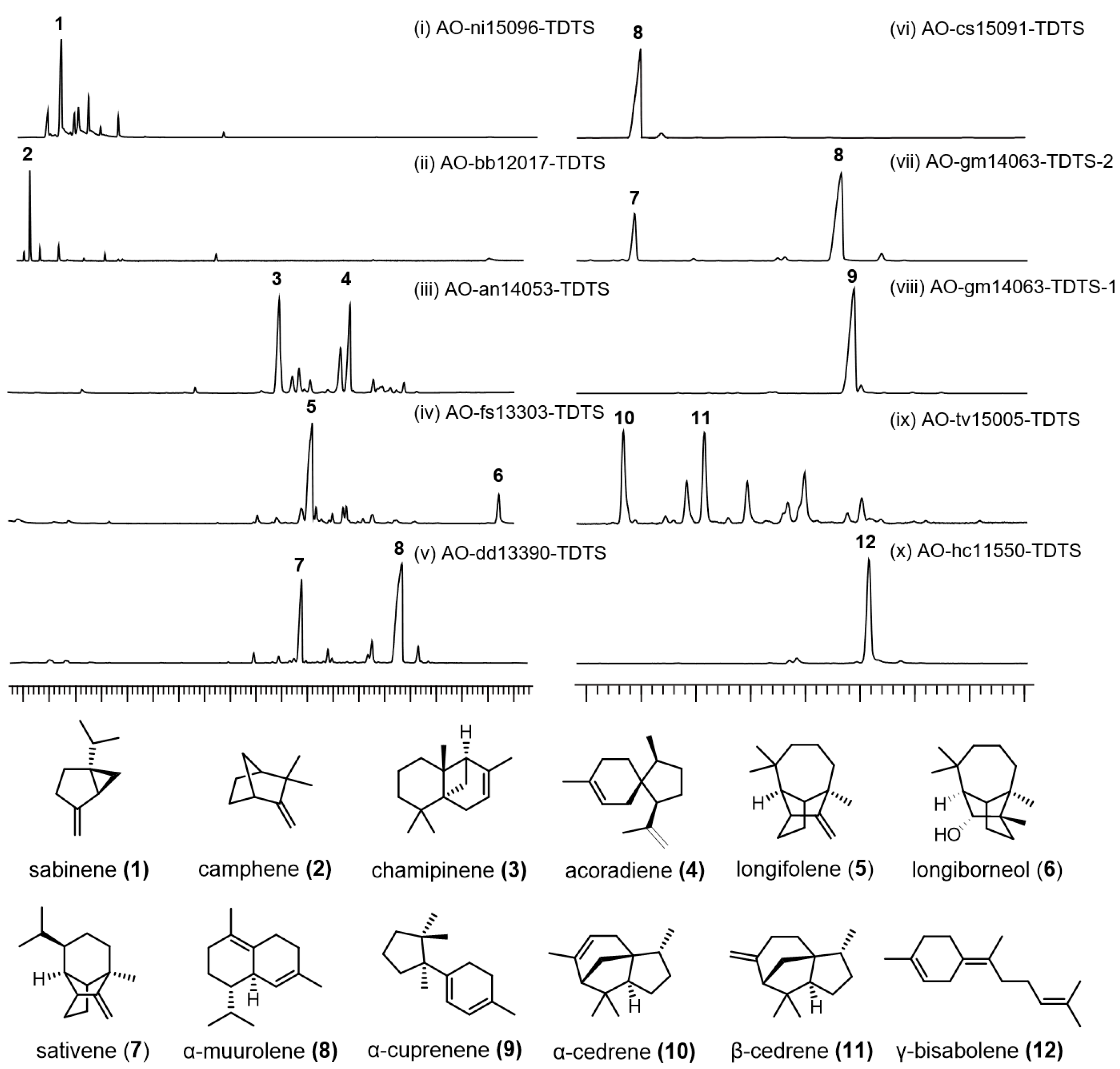
3.3. Heterologous Expression of TDTSs
3.4. Differences in Cyclization Paths of TDTSs in Families 1 and 2
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zeng, T.; Liu, Z.; Zhuang, J.; Jiang, Y.; He, W.; Diao, H.; Lv, N.; Jian, Y.; Liang, D.; Qiu, Y.; et al. TeroKit: A database-driven web server for terpenome research. J. Chem. Inf. Model. 2020, 60, 2082–2090. [Google Scholar] [CrossRef] [PubMed]
- Avalos, M.; Garbeva, P.; Vader, L.; van Wezel, G.P.; Dickschat, J.S.; Ulanova, D. Biosynthesis, evolution and ecology of microbial terpenoids. Nat. Prod. Rep. 2022, 39, 249–272. [Google Scholar] [CrossRef] [PubMed]
- Miller, D.J.; Allemann, R.K. Sesquiterpene synthases: Passive catalysts or active players? Nat. Prod. Rep. 2012, 29, 60–71. [Google Scholar] [CrossRef] [PubMed]
- Bergman, M.E.; Davis, B.; Phillips, M.A. Medically Useful Plant Terpenoids: Biosynthesis, Occurrence, and Mechanism of Action. Molecules 2019, 24, 3961. [Google Scholar] [CrossRef] [PubMed]
- Yin, Z.; Dickschat, J.S. Engineering fungal terpene biosynthesis. Nat. Prod. Rep. 2023, 40, 28–45. [Google Scholar] [CrossRef]
- Dai, Q.; Zhang, F.-L.; Feng, T. Sesquiterpenoids Specially Produced by Fungi: Structures, Biological Activities, Chemical and Biosynthesis (2015–2020). J. Fungi 2021, 7, 1026. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Chen, R.; Yuan, L.; Zhang, C.; Liang, D.; Qiao, J. Molecular and functional analyses of characterized sesquiterpene synthases in mushroom-forming fungi. J. Fungi 2023, 9, 1017. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Chen, X.; Orban, A.; Shukal, S.; Birk, F.; Too, H.P.; Ruhl, M. Agrocybe aegerita serves as a gateway for identifying sesquiterpene biosynthetic enzymes in higher fungi. ACS Chem. Biol. 2020, 15, 1268–1277. [Google Scholar] [CrossRef] [PubMed]
- Masunaga, N.; Kitaoka, T.; Ichinose, H. Biocatalyst collection and heterologous expression of sesquiterpene synthases from basidiomycetous fungi: Discovery of a novel sesquiterpene hydrocarbon. Microb. Biotechnol. 2022, 16, 632–644. [Google Scholar] [CrossRef]
- Nagamine, S.; Liu, C.; Nishishita, J.; Kozaki, T.; Sogahata, K.; Sato, Y.; Minami, A.; Ozaki, T.; Schmidt-Dannert, C.; Maruyama, J.; et al. Ascomycete aspergillus oryzae is an efficient expression host for production of basidiomycete terpenes by using genomic DNA sequences. Appl. Environ. Microbiol. 2019, 85, e00409–e00419. [Google Scholar]
- Zhang, T.; Feng, J.; He, W.; Rong, X.; Lv, H.; Li, J.; Li, X.; Wang, H.; Wang, L.; Zhang, L.; et al. Genomic and transcriptomic approaches povide a pedictive framework for sesquiterpenes bosynthesis in Desarmillaria tabescens CPCC 401429. J. Fungi 2023, 9, 481. [Google Scholar] [CrossRef]
- Proctor, R.H.; McCormick, S.P.; Gutierrez, S. Genetic bases for variation in structure and biological activity of trichothecene toxins produced by diverse fungi. Appl. Microbiol. Biotechnol. 2020, 104, 5185–5199. [Google Scholar] [CrossRef] [PubMed]
- Webb, E.C. Enzyme nomenclature 1992. In Recommendations of the Nomenclature Committee of the International Union of Biochemistry and Molecular Biology on the Nomenclature and Classification of Enzymes; Academic Press: New York, NY, USA, 1992; Volume 6. [Google Scholar]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Soding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef] [PubMed]
- Potter, S.C.; Luciani, A.; Eddy, S.R.; Park, Y.; Lopez, R.; Finn, R.D. HMMER web server: 2018 update. Nucleic Acids Res. 2018, 46, W200–W204. [Google Scholar] [CrossRef] [PubMed]
- Camacho, C.; Boratyn, G.M.; Joukov, V.; Vera Alvarez, R.; Madden, T.L. ElasticBLAST: Accelerating sequence search via cloud computing. BMC Bioinform. 2023, 24, 117. [Google Scholar] [CrossRef] [PubMed]
- Stanke, M.; Diekhans, M.; Baertsch, R.; Haussler, D. Using native and syntenically mapped cDNA alignments to improve de novo gene finding. Bioinformatics 2008, 24, 637–644. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Procter, J.B.; Carstairs, G.M.; Soares, B.; Mourao, K.; Ofoegbu, T.C.; Barton, D.; Lui, L.; Menard, A.; Sherstnev, N.; Roldan-Martinez, D.; et al. Alignment of Biological Sequences with Jalview. Methods Mol. Biol. 2021, 2231, 203–224. [Google Scholar] [CrossRef]
- Wheeler, T.J.; Clements, J.; Finn, R.D. Skylign: A tool for creating informative, interactive logos representing sequence alignments and profile hidden Markov models. BMC Bioinform. 2014, 15, 7. [Google Scholar] [CrossRef]
- Lu, H.N.; Ma, S.G.; Liu, Y.B.; Qu, J.; Li, Y.; Xu, S.; Zhu, H.; Yu, S.S. Sesquiterpenes from the roots of Illicium oligandrum. J. Asian Nat. Prod. Res. 2015, 17, 430–438. [Google Scholar] [CrossRef] [PubMed]
- Lou, T.; Li, A.; Xu, H.; Pan, J.; Xing, B.; Wu, R.; Dickschat, J.S.; Yang, D.; Ma, M. Structural insights into three sesquiterpene synthases for the biosynthesis of tricyclic sesquiterpenes and chemical space expansion by structure-based mutagenesis. J. Am. Chem. Soc. 2023, 145, 8474–8485. [Google Scholar] [CrossRef]
- Hong, Y.J.; Tantillo, D.J. Branching out from the bisabolyl cation. Unifying mechanistic pathways to barbatene, bazzanene, chamigrene, chamipinene, cumacrene, cuprenene, dunniene, isobazzanene, iso-gamma-bisabolene, isochamigrene, laurene, microbiotene, sesquithujene, sesquisabinene, thujopsene, trichodiene, and widdradiene sesquiterpenes. J. Am. Chem. Soc. 2014, 136, 2450–2463. [Google Scholar] [CrossRef]
- Cao, Y.; Zhang, H.; Liu, H.; Liu, W.; Zhang, R.; Xian, M.; Liu, H. Biosynthesis and production of sabinene: Current state and perspectives. Appl. Microbiol. Biotechnol. 2018, 102, 1535–1544. [Google Scholar] [CrossRef] [PubMed]
- Jorg, B.; Michael, P.; Vasanthi, R.; Sadanobu, K.; Croteau, R. cDNA cloning, characterization, and functional expression of four new monoterpene synthase members of the Tpsd gene family from grand fir (Abies grandis). Arch. Biochem. Biophys. 1999, 368, 232–243. [Google Scholar] [CrossRef]
- Druzhinina, I.S.; Seidl-Seiboth, V.; Herrera-Estrella, A.; Horwitz, B.A.; Kenerley, C.M.; Monte, E.; Mukherjee, P.K.; Zeilinger, S.; Grigoriev, I.V.; Kubicek, C.P. Trichoderma: The genomics of opportunistic success. Nat. Rev. Microbiol. 2011, 9, 749–759. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Shao, J.; Fu, Y.; Chen, Y.; Wang, H.; Xu, Z.; Feng, H.; Xun, W.; Liu, Y.; Zhang, N.; et al. The volatile cedrene from Trichoderma guizhouense modulates Arabidopsis root development through auxin transport and signalling. Plant Cell Environ. 2022, 45, 969–984. [Google Scholar] [CrossRef]
- Lopez-Gallego, F.; Agger, S.A.; Abate-Pella, D.; Distefano, M.D.; Schmidt-Dannert, C. Sesquiterpene synthases Cop4 and Cop6 from Coprinus cinereus: Catalytic promiscuity and cyclization of farnesyl pyrophosphate geometric isomers. Chembiochem 2010, 11, 1093–1106. [Google Scholar] [CrossRef]
- Wawrzyn, G.T.; Quin, M.B.; Choudhary, S.; Lopez-Gallego, F.; Schmidt-Dannert, C. Draft genome of Omphalotus olearius provides a predictive framework for sesquiterpenoid natural product biosynthesis in Basidiomycota. Chem. Biol. 2012, 19, 772–783. [Google Scholar] [CrossRef]
- Bahadoor, A.; Schneiderman, D.; Gemmill, L.; Bosnich, W.; Blackwell, B.; Melanson, J.E.; McRae, G.; Harris, L.J. Hydroxylation of longiborneol by a Clm2-encoded CYP450 monooxygenase to produce culmorin in Fusarium graminearum. J. Nat. Prod. 2016, 79, 81–88. [Google Scholar] [CrossRef]
- Yee, D.A.; Kakule, T.B.; Cheng, W.; Chen, M.; Chong, C.T.Y.; Hai, Y.; Hang, L.F.; Hung, Y.S.; Liu, N.; Ohashi, M.; et al. Genome mining of alkaloidal terpenoids from a hybrid terpene and nonribosomal peptide biosynthetic pathway. J. Am. Chem. Soc. 2020, 142, 710–714. [Google Scholar] [CrossRef] [PubMed]
- Nosenko, T.; Zimmer, I.; Ghirardo, A.; Kollner, T.G.; Weber, B.; Polle, A.; Rosenkranz, M.; Schnitzler, J.P. Predicting functions of putative fungal sesquiterpene synthase genes based on multiomics data analysis. Fungal Genet. Biol. 2023, 165, 103779. [Google Scholar] [CrossRef] [PubMed]
- Proctor, R.H.; McCormick, S.P.; Kim, H.S.; Cardoza, R.E.; Stanley, A.M.; Lindo, L.; Kelly, A.; Brown, D.W.; Lee, T.; Vaughan, M.M.; et al. Evolution of structural diversity of trichothecenes, a family of toxins produced by plant pathogenic and entomopathogenic fungi. PLoS Pathog. 2018, 14, e1006946. [Google Scholar] [CrossRef] [PubMed]
- Ichinose, H.; Kitaoka, T. Insight into metabolic diversity of the brown-rot basidiomycete Postia placenta responsible for sesquiterpene biosynthesis: Semi-comprehensive screening of cytochrome P450 monooxygenase involved in protoilludene metabolism. Microb. Biotechnol. 2018, 11, 952–965. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Yao, B.; Zeng, H. Identification and characterization of a nerol synthase in fungi. J. Agric. Food Chem. 2023, 72, 416–423. [Google Scholar] [CrossRef] [PubMed]
- Ichinose, H.; Ukeba, S.; Kitaoka, T. Latent potentials of the white-rot basidiomycete Phanerochaete chrysosporium responsible for sesquiterpene metabolism: CYP5158A1 and CYP5144C8 decorate (E)-alpha-bisabolene. Enzyme Microb. Technol. 2022, 158, 110037. [Google Scholar] [CrossRef] [PubMed]
- Al-Salihi, S.A.A.; Dao, T.T.; Williams, K.; Bailey, A.M.; Foster, G.D. The biogenetic origin of the biologically active naematolin of Hypholoma species involves an unusual sesquiterpene synthase. Mol. Biotechnol. 2019, 61, 754–762. [Google Scholar] [CrossRef] [PubMed]
- Burkhardt, I.; Kreuzenbeck, N.B.; Beemelmanns, C.; Dickschat, J.S. Mechanistic characterization of three sesquiterpene synthases from the termite-associated fungus Termitomyces. Org. Biomol. Chem. 2019, 17, 3348–3355. [Google Scholar] [CrossRef]
- Lackner, G.; Peters, E.E.; Helfrich, E.J.; Piel, J. Insights into the lifestyle of uncultured bacterial natural product factories associated with marine sponges. Proc. Natl. Acad. Sci. USA 2017, 114, E347–E356. [Google Scholar] [CrossRef]
- Mischko, W.; Hirte, M.; Fuchs, M.; Mehlmer, N.; Bruck, T.B. Identification of sesquiterpene synthases from the Basidiomycota Coniophora puteana for the efficient and highly selective beta-copaene and cubebol production in E. coli. Microb. Cell Fact. 2018, 17, 164. [Google Scholar] [CrossRef] [PubMed]
- Yap, H.Y.; Muria-Gonzalez, M.J.; Kong, B.H.; Stubbs, K.A.; Tan, C.S.; Ng, S.T.; Tan, N.H.; Solomon, P.S.; Fung, S.Y.; Chooi, Y.H. Heterologous expression of cytotoxic sesquiterpenoids from the medicinal mushroom Lignosus rhinocerotis in yeast. Microb. Cell Fact. 2017, 16, 103. [Google Scholar] [CrossRef] [PubMed]
- Agger, S.; Lopez-Gallego, F.; Schmidt-Dannert, C. Diversity of sesquiterpene synthases in the basidiomycete Coprinus cinereus. Mol. Biotechnol. 2009, 72, 1181–1195. [Google Scholar] [CrossRef] [PubMed]
- Ntana, F.; Bhat, W.W.; Johnson, S.R.; Jorgensen, H.J.L.; Collinge, D.B.; Jensen, B.; Hamberger, B. A sesquiterpene synthase from the endophytic fungus Serendipita indica catalyzes formation of viridiflorol. Biomolecules 2021, 11, 898. [Google Scholar] [CrossRef] [PubMed]
- Zeng, H.; Yin, G.; Wei, Q.; Li, D.; Wang, Y.; Hu, Y.; Hu, C.; Zou, Y. Unprecedented [5.5.5.6]dioxafenestrane ring construction in fungal insecticidal sesquiterpene biosynthesis. Angew. Chem. Int. Ed. Engl. 2019, 58, 6569–6573. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Wang, Y.; Jiang, B.; Li, W.; Zheng, L.; Yang, X.; Bao, Y.; Sun, L.; Huang, Y.; Li, Y. Structure, cytotoxic activity and mechanism of protoilludane sesquiterpene aryl esters from the mycelium of Armillaria mellea. J. Ethnopharmacol. 2016, 184, 119–127. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Litzenburger, M.; Cheng, S.; Bian, G.; Hu, B.; Yan, P.; Cai, Y.; Deng, Z.; Bernhardt, R.; Liu, T. Sesquiterpenoids produced by combining two sesquiterpene cyclases with promiscuous myxobacterial CYP260B1. Chembiochem 2019, 20, 677–682. [Google Scholar] [CrossRef] [PubMed]
- Buchvaldt Amby, D.; Manczak, T.; Petersen, M.A.; Sundelin, T.; Weitzel, C.; Grajewski, M.; Simonsen, H.T.; Jensen, B. Role of the Colletotrichum acutatum sesquiterpene synthase CaTPS in the biosynthesis of sesquiterpenoids. Microbiology 2016, 162, 1773–1783. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Tran, W.; Taatjes, C.A.; Alonso-Gutierrez, J.; Lee, T.S.; Gladden, J.M. Rapid discovery and functional characterization of terpene synthases from four endophytic Xylariaceae. PLoS ONE 2016, 11, e0146983. [Google Scholar] [CrossRef]
- Hidalgo, P.I.; Ullan, R.V.; Albillos, S.M.; Montero, O.; Fernandez-Bodega, M.A.; Garcia-Estrada, C.; Fernandez-Aguado, M.; Martin, J.F. Molecular characterization of the PR-toxin gene cluster in Penicillium roqueforti and Penicillium chrysogenum: Cross talk of secondary metabolite pathways. Fungal Genet. Biol. 2014, 62, 11–24. [Google Scholar] [CrossRef]
- Burkhardt, I.; Siemon, T.; Henrot, M.; Studt, L.; Rçsler, S.; Tudzynski, B.; Christmann, M.; Dickschat, J.S. Mechanistic characterisation of two sesquiterpene cyclases from the plant pathogenic fungus Fusarium fujikuroi. Angew. Chem. Int. Ed. 2016, 55, 8748–8751. [Google Scholar] [CrossRef] [PubMed]
- Shaw, J.J.; Berbasova, T.; Sasaki, T.; Jefferson-George, K.; Spakowicz, D.J.; Dunican, B.F.; Portero, C.E.; Narvaez-Trujillo, A.; Strobel, S.A. Identification of a fungal 1,8-cineole synthase from Hypoxylon sp. with specificity determinants in common with the plant synthases. J. Biol. Chem. 2015, 290, 8511–8526. [Google Scholar] [CrossRef] [PubMed]
- De Sena Filho, J.G.; Quin, M.B.; Spakowicz, D.J.; Shaw, J.J.; Kucera, K.; Dunican, B.; Strobel, S.A.; Schmidt-Dannert, C. Genome of Diaporthe sp. provides insights into the potential inter-phylum transfer of a fungal sesquiterpenoid biosynthetic pathway. Fungal Biol. 2016, 120, 1050–1063. [Google Scholar] [CrossRef]
- Sun, X.; Cai, Y.S.; Yuan, Y.; Bian, G.; Ye, Z.; Deng, Z.; Liu, T. Genome mining in Trichoderma viride J1-030: Discovery and identification of novel sesquiterpene synthase and its products. Beilstein J. Org. Chem. 2019, 15, 2052–2058. [Google Scholar] [CrossRef] [PubMed]
- Brock, N.L.; Huss, K.; Tudzynski, B.; Dickschat, J.S. Genetic dissection of sesquiterpene biosynthesis by Fusarium fujikuroi. Chembiochem 2013, 14, 311–315. [Google Scholar] [CrossRef]
- Moraga, J.; Dalmais, B.; Izquierdo-Bueno, I.; Aleu, J.; Hanson, J.R.; Hernandez-Galan, R.; Viaud, M.; Collado, I.G. Genetic and molecular basis of botrydial biosynthesis: Connecting cytochrome P450-encoding genes to biosynthetic intermediates. ACS Chem. Biol. 2016, 11, 2838–2846. [Google Scholar] [CrossRef] [PubMed]
- Proctor, R.H.; Hohn, T.M. Aristolochene synthase. Isolation, characterization, and bacterial expression of a sesquiterpenoid biosynthetic gene (Ari1) from Penicillium roqueforti. J. Biol. Chem. 1993, 268, 4543–4548. [Google Scholar] [CrossRef] [PubMed]
- Cane, D.E.; Kang, I. Aristolochene synthase: Purification, molecular cloning, high-level expression in Escherichia coli, and characterization of the Aspergillus terreus cyclase. Arch. Biochem. Biophys. 2000, 376, 354–364. [Google Scholar] [CrossRef]
- Murai, K.; Lauterbach, L.; Teramoto, K.; Quan, Z.; Barra, L.; Yamamoto, T.; Nonaka, K.; Shiomi, K.; Nishiyama, M.; Kuzuyama, T.; et al. An unusual skeletal rearrangement in the biosynthesis of the sesquiterpene trichobrasilenol from Trichoderma. Angew. Chem. Int. Ed. Engl. 2019, 58, 15046–15050. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cong, Z.; Yin, Q.; Tian, K.; Mukoma, N.J.; Ouyang, L.; Hsiang, T.; Zhang, L.; Jiang, L.; Liu, X. Genome Mining of Fungal Unique Trichodiene Synthase-like Sesquiterpene Synthases. J. Fungi 2024, 10, 350. https://doi.org/10.3390/jof10050350
Cong Z, Yin Q, Tian K, Mukoma NJ, Ouyang L, Hsiang T, Zhang L, Jiang L, Liu X. Genome Mining of Fungal Unique Trichodiene Synthase-like Sesquiterpene Synthases. Journal of Fungi. 2024; 10(5):350. https://doi.org/10.3390/jof10050350
Chicago/Turabian StyleCong, Zhanren, Qiang Yin, Kunhong Tian, Njeru Joe Mukoma, Liming Ouyang, Tom Hsiang, Lixin Zhang, Lan Jiang, and Xueting Liu. 2024. "Genome Mining of Fungal Unique Trichodiene Synthase-like Sesquiterpene Synthases" Journal of Fungi 10, no. 5: 350. https://doi.org/10.3390/jof10050350
APA StyleCong, Z., Yin, Q., Tian, K., Mukoma, N. J., Ouyang, L., Hsiang, T., Zhang, L., Jiang, L., & Liu, X. (2024). Genome Mining of Fungal Unique Trichodiene Synthase-like Sesquiterpene Synthases. Journal of Fungi, 10(5), 350. https://doi.org/10.3390/jof10050350






