Abstract
Candida albicans (Ca), a prominent opportunistic fungal pathogen in humans, has garnered considerable attention due to its infectious properties. Herein, we have identified and characterized CaCDAP1 (Ca orf19.1034), a homolog of ScDAP1 found in Saccharomyces cerevisiae. CaCDAP1 encodes a 183-amino acid protein with a conserved cytochrome b5-like heme-binding domain. The deletion of CaDAP1 renders Ca cells susceptible to caspofungin and terbinafine. CaDAP1 deletion confers resistance to Congo Red and Calcofluor White, and sensitivity to sodium dodecyl sulfate. The deletion of CaDAP1 results in a 50% reduction in chitin content within the cell wall, the downregulation of phosphorylation levels in CaMkc1, and the upregulation of phosphorylation levels in CaCek1. Notably, CaDAP1 deletion results in the abnormal hyphal development of Ca cells and diminishes virulence in a mouse systemic infection model. Thus, CaDAP1 emerges as a critical regulator governing cellular responses to antifungal drugs, the synthesis of cell wall chitin, and virulence in Ca.
1. Introduction
Candida albicans (Ca) stands out as the most prevalent opportunistic fungal pathogen in humans, existing as an innocuous commensal in approximately 70% of individuals [1,2]. However, in immunocompromised patients, it can transition to causing both superficial mucosal and potentially life-threatening systemic infections [3,4,5]. The pathogenicity of Ca is multifaceted, involving various factors and activities such as attachment to and penetration into host cells, the release of hydrolases, transition from yeast to hyphae, perception of contact, responsiveness to physical stimuli, alteration in phenotype, development of biofilms, adjustment to environmental acidity, flexibility in metabolism, efficient nutrient acquisition mechanisms, and reliable stress response systems [5,6].
The cell wall of Ca is composed of chitin, β-glucan, mannan, and cell wall proteins [7]. Chitin and β-glucan are located in the interior of the cell wall, and form the core skeleton structure. The outermost portion of the cell wall is mainly composed of mannan, which masks the internal β-glucan and reduces the recognition of Ca by the host immune system. The cell wall of Ca plays a key role in maintaining cell integrity, morphogenesis, response to changes in environmental conditions, interaction with the host, and pathogenesis [7,8]. Moreover, the cell wall of Ca is a prime target for antifungal drugs such as the echinocandins [9]. Despite its tough cell wall, Ca can flexibly change the relative levels of chitin, β-glucan, and mannan in response to environmental change [10,11]. Thus, this potential for cell wall remodeling is critical for maintaining Ca cell wall integrity (CWI) and is regulated by multiple signaling pathways, including the Mkc1, Hog1, and Cek1 mitogen-activated protein (MAP) kinase cascade [12]. Chitin is absent in humans and other vertebrates. Therefore, chitin synthesis is an excellent potential target for the development of antifungal drugs [13]. Ca regulates the expression of chitin synthase and the content of chitin in the cell wall, through HOG, PKC, and the Ca2+/calcineurin signaling pathways in response to environmental stress [13].
In the context of Saccharomyces cerevisiae, the heme-binding damage-resistance protein 1 (DAP1) is associated with cytochrome b5, and plays significant roles in iron metabolism and ergosterol biosynthesis [14,15,16,17,18]. In Schizosaccharomyces pombe, DAP1 positively regulates P450 enzymes and is essential for sterol biosynthesis [19]. In Aspergillus fumigatus, DapA influences the susceptibility to azoles by maintaining the stability of cytochrome P450 enzymes responsible for ergosterol synthesis [20]. Recognizing the significance of the heme-binding damage-resistance protein 1, we explored the Ca genome for genes homologous with S. cerevisiae ScDAP1. Our search revealed that the identity between Ca orf19.1034 (CaDAP1) and S. cerevisiae ScDAP1 (YPL170W) was 40%.
This study provides experimental evidence affirming the requirement of CaDAP1 (orf19.1034) for antifungal drug tolerance and chitin synthesis in the cell wall of Ca. Additionally, we demonstrate that the Cadap1/Cadap1 null mutant exhibits defective hyphal development and diminished virulence in a mouse systemic infection model. Our findings suggest that CaDap1 plays crucial roles in the morphogenesis and pathogenesis of Ca.
2. Materials and Methods
2.1. Strains, Media, Plasmids, and Primers
The Ca strains utilized in this investigation, including strains, plasmids and primers, are documented in Table 1 and Table 2. Strains were cultivated at 30 °C in either YPD medium or SD medium [21,22,23].

Table 1.
Strains and plasmids used in this study.

Table 2.
Primers used in this study.
2.2. Construction of the Cadap1/Cadap1 Mutant and the Re-Integrant Strain (RS)
For the deletion of the 1st CaDAP1 allele, a SAT1 flipper cassette harboring CaSAT1 was used for selection [25]. The SAT1 flipper cassette, derived from pSFS2 using the polymerase chain reaction (PCR) with primer pairs DAP1 − NAT − UP and DAP1 − NAT − DOWN, was introduced into the RM1000 strain. The resultant CaDAP1/Cadap1::SAT1-FLIP NatR transformant was chosen, and its genotypes were verified using PCR.
For the disruption of the second CaDAP1 allele, the HIS1 cassette was amplified from pGEM − HIS1 using primer pairs DAP1 − HIS − UP and DAP1 − HIS − DOWN, followed by transformation into the CaDAP1/Cadap1::SAT1 − FLIP strain. The resulting Cadap1::HIS1/Cadap1::SAT1 − FLIP NatR and HIS+ transformants were chosen, and their genotypes validated using PCR. The Cadap1::HIS1/Cadap1::SAT1 − FLIP NatR strain was grown on YPD medium supplemented with nourseothricin (25 μg/mL) to excise the NatR genetic biomarker, yielding the Cadap1::HIS1/Cadap1::FRT strain, whose genotype was verified by PCR using primers DAP1 − ORF − UP/DAP1 − ORF − DOWN and DAP1 − UP − CHECK/DAP1 − DOWN − CHECK (Figure 1).
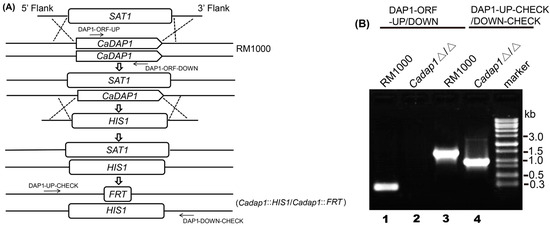
Figure 1.
Deletion of two CaDAP1 alleles. (A) Sequentially targeted disruption of two CaDAP1 alleles in WT RM1000 to generate Cadap1Δ/Δ (Cadap1::HIS1/Cadap1::FRT). Strain designations are depicted on the right, while primer sites are highlighted with arrows. (B) PCR verification of the homozygous mutant Cadap1Δ/Δ genotypes. Lane 1: A 0.25 kb fragment from RM1000 was amplified using the primers DAP1 − ORF − UP/DOWN. Lane 2: No fragment was amplified from Cadap1Δ/Δ (Cadap1::HIS1/Cadap1::FRT) using the primers DAP1 − ORF − UP/DOWN. Lane 3: A 1.4 kb fragment was amplified from RM1000 using primers DAP1 − UP − CHECK/DOWN-CHECK. Lane 4: The 0.98 kb and 2.6 kb fragments containing FRT and HIS1, respectively, were amplified from Cadap1Δ/Δ using the primers DAP1 − UP − CHECK/DOWN − CHECK.
To obtain a control, in this case a re-integrant strain (RS) for control purposes, the CaDAP1 open reading frame, along with 800 and 281 bp of its promoter and terminator, respectively, were amplified by PCR using SC5314 genomic DNA as a template, with primers DAP1−UP−CLO and DAP1−DOWN−CLO. The 1.36 kb products obtained were cloned into the SacI and NotI sites of CIp10. The resultant plasmids were digested with StuI and transformed into the Cadap1/Cadap1 mutant strain. Strain RM1000, transformed with StuI−digested CIp10, served as a positive control, with expression of URA3 from the RPS1 locus (Table 1).
2.3. Growth Phenotypic Analysis and Hyphal Formation Examination
Plate assays were conducted following established protocols [23]. In brief, cells were cultured in liquid medium at 30 °C overnight, serially diluted to 5 × 107, 5 × 106, 5 × 105, 5 × 104, and 5 × 103 cells/mL, and 2.5 µL samples grown on plates supplemented with terbinafine, caspofungin, fluconazole, SDS, CR, or CFW. Phenotypic observations were recorded after incubation at 30 °C for 48–72 h. Hyphal formation analysis was performed by culturing appropriately diluted cells in liquid YPD medium supplemented with 10% FBS, with shaking at 37 °C. Colony morphologies were assessed on YPD plates supplemented with 10% FBS. Approximately 20 cells of each strain were incubated on individual plates at 37 °C for 5–7 days before the photographic documentation of colonies.
2.4. Measurement of Cell Wall Composition
- (1)
- Chitin quantification [28]
Cells collected in the exponential growth phase were treated with calcifluor white (CFW) (50 μg/mL) for 10 min. After 3 washes with phosphate-buffered saline, their fluorescence was evaluated using a Varioskan Flash spectral-scanning multimode reader (Thermo Fisher Scientific Inc., Waltham, MA, USA) at excitation and emission wavelengths of 325 and 435 nm, respectively. The presented data were standardized to cell concentrations assessed by optical density measurements.
- (2)
- Determination of β-1,3-glucan concentrations [28,29]
Exponential phase cells were washed twice and resuspended in TE buffer to a final OD600 of 0.2–0.5. Subsequently, NaOH was added to a final concentration of 1.0 M, and the mixture was incubated at 80 °C for 30 min. Following the addition of 2.1 mL aniline blue solution (0.03% aniline blue, 0.18 M HCl and 0.49 M glycine/NaOH, pH9.5), the samples were incubated at 50 °C for 30 min, and then 30 min at room temperature for fluorochrome reaction and decolorization. Fluorescence was measured using the Varioskan Flash spectral-scanning multimode reader (Thermo Fisher Scientific Inc., Waltham, MA, USA) at the excitation and emission wavelengths of 400 and 460 nm, respectively.
- (3)
- Alcian blue staining and determination of phosphomannan content [28,30]
Exponential phase cells were harvested via centrifugation and washed with 1 mL HCl (0.02 M). The cells were resuspended in 1 mL Alcian blue (50 μg/mL in 0.02 M HCl), incubated at room temperature for 10 min, and centrifuged. The OD600 of the supernatant was measured using a spectrophotometer (Unico Instrument Co., Ltd., Shanghai, China).
2.5. CFW Staining of Chitin and Fluorescence Microscopy
The staining of the cell wall chitin with CFW was conducted following established procedures [31]. Exponential phase cells were fixed with neutral-buffered formalin (10% (v/v), Sigma-Aldrich) and stained with CFW (25 μg/mL).
2.6. Quantitative Reverse Transcription PCR Analysis of CaCHS1, CaCHS2, CaCHS3, and CaCHS8
Exponential phase cells were collected by centrifugation, washed twice with sterile water, and rapidly frozen in liquid nitrogen. Total RNA extraction was carried out using the MiniBEST Universal RNA extraction kit (TaKaRa, Dalian, China), following the manufacturer’s protocols, with DNaseI treatment to eliminate DNA contamination. The total RNA was utilized for cDNA synthesis using the PrimeScript RT master mix (TaKaRa). Quantitative PCR assays used TB-Green-Premix-Ex-Taq-II (TaKaRa). The expression levels of CaCHS1, CaCHS2, CaCHS3, and CaCHS8 were assessed using specific primer pairs (Table 2), while the CaACT1 transcript served as an internal control [18]. The change in gene expression was evaluated using the 2−△△CT approach [23].
2.7. Protein Extraction and Western Blotting
Ca protein extracts were prepared using a previously described protocol [32,33]. Lysates from OD600 = 1 of cells were subjected to SDS-PAGE and immunoblotting with the anti-phospho-p44/42MAPK (Thr202/Tyr204) antibody (Cell Signaling Technology (CST), Danvers, MA, USA) to concomitantly assess the phosphorylation levels of CaCek1 and CaMkc1. The probing of immunoblots using the anti-α-tubulin antibody (Novus Biologicals, Centennial, CO, USA) provided a loading control.
2.8. Virulence Assay
Virulence assays were conducted following established procedures [22,23]. Seven-week-old male BALB/c mice (weight = 20–22 g) were maintained in individually ventilated cages. The mice (n = 12/strain) were intravenously inoculated with 1 × 106 cells/mL in 0.9% (w/v) NaCl solution/mouse via the lateral tail vein. Animals (n = 2/strain) were randomly euthanized following 48 h of injection, and the CFUs in their kidneys were quantified on SD-URA plates. Mouse survival rates were determined over 30 days, and fungal infiltration in the mouse kidneys was assessed through histopathological evaluation, as described previously [22,23]. The histological analysis of the collected mouse kidneys was performed, and stained sections were analyzed using a Axio Imager 2 (ZEISS Microscopy, Jena, Germany). The mouse experiments were conducted following the protocols approved by the Ethics Committee of Huaibei Normal University, China (permission ECHN191016).
2.9. Statistical Analysis
Statistical tests were conducted using SPSS v19.0. p < 0.05 was deemed significant.
3. Results
3.1. Genomic Data Analysis
The sequences of Ca orf19.1034 came from the Ca genomic database (http://www.candidagenome.org (accessed on 21 March 2018)). orf19.1034 spans 552 base pairs and codes for a protein consisting of 183 amino acids. According to SMART analysis (http://smart.embl-heidelberg.de (accessed on 21 March 2018)), the protein encoded by orf19.1034 harbors one transmembrane region, commencing at position 13 and terminating at position 32 of the primary sequence, along with one cytochrome b5-like heme-binding domain, spanning from position 62 to 162 (Figure S1, Supplementary Materials). The protein encoded by orf19.1034 exhibited 27.6% and 31.6% sequence similarity to S. cerevisiae ScDap1 and Candida glabrata CgDap1, respectively. ScDap1 and CgDap1 also feature cytochrome b5-like heme-binding domains (Figure S1, Supplementary Materials). Consequently, orf19.1034 of Ca was designated as CaDAP1.
3.2. The Deletion of CaDAP1 Renders Ca Cells Susceptible to Caspofungin and Terbinafine
To assess the role of CaDAP1 in Ca, we obtained homozygous deletion mutants of CaDAP1 by sequentially replacing its two alleles with the SAT1-flipper and HIS1 cassette (Figure 1). We tested the sensitivity of the Cadap1/Cadap1 mutant to fluconazole, caspofungin, and terbinafine. Unlike the wild-type (WT) RM1000, the Cadap1/Cadap1 mutant was sensitive to 0.25 μg/mL caspofungin and 5 μg/mL terbinafine (Figure 2), but remained insensitive to 5 μg/mL fluconazole (Figure S2, Supplementary Materials). The reintroduction of CaDAP1 into the homozygous mutant cells reversed these sensitive phenotypes. Thus, it is suggested that CaDap1 plays critical roles in conferring resistance to terbinafine and caspofungin in Ca.
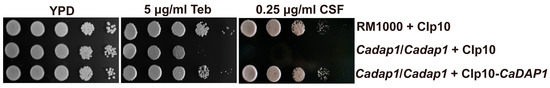
Figure 2.
CaDAP1 is involved in conferring resistance to terbinafine and caspofungin. Growth phenotypes of the WT, Cadap1/Cadap1 mutant, and CaDAP1 RS were evaluated following exposure to caspofungin (0.25 μg/mL) and terbinafine (5 μg/mL). Overnight cell cultures were then grown in YPD to the exponential phase at 30 °C. Tenfold serial dilutions were spotted onto YPD plates supplemented with caspofungin or terbinafine, and incubated at 30 °C for 48–72 h.
3.3. The Deletion of CaDAP1 Reduces the Chitin Content in Cell Walls, and Downregulates the Phosphorylation Levels of CaMkc1
The echinocandin drug inhibits β-1,3-glucan synthase, a pivotal enzyme tasked with synthesizing β-1,3-D-glucan, a primary component of cell walls [34,35]. The sensitivity of the Cadap1/Cadap1 mutant to caspofungin suggested the involvement of CaDAP1 in the cell wall stress response of Ca. We examined the phenotype of Ca cells lacking CaDAP1 following exposure to CR and CFW, as well as to SDS. Unlike the WT and re-integrant strain (RS), Ca cells lacking CaDAP1 displayed significant resistance to CR and CFW but sensitivity to SDS (Figure 3A). To elucidate the impact on the cell wall, we assessed the levels of cell-wall components, such as phosphomannan, β-1,3-glucan, and chitin. Compared with the WT, the chitin content was reduced by 50% (Figure 3B), while the phosphomannan content increased by 9% in the Cadap1/Cadap1 mutant (Figure 3C); the β-1,3-glucan content was unchanged (Figure S3, Supplementary Materials). These findings suggest that the loss of CaDAP1 alters the cell wall composition.
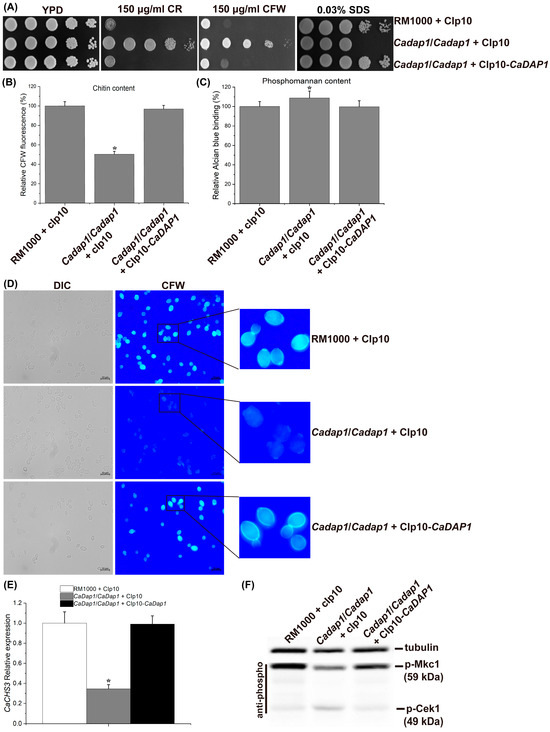
Figure 3.
The deletion of CaDAP1 leads to reduced chitin content in cell walls, accompanied by decreased phosphorylation levels of CaMkc1 and slightly increased phosphorylation levels of CaCek1. (A) The deletion of CaDAP1 renders Ca cells resistant to CR and CFW, but sensitive to SDS. Growth phenotypes of WT, Cadap1/Cadap1 mutant, and CaDAP1 RS following exposure to 150 μg/mL CR and 150 μg/mL CFW, along with 0.03% SDS. Knockout of CaDAP1 results in a 50% decrease in chitin content (B) and a 9% increase in phosphomannan content (C) of the cell wall. (D) The intensity of CFW fluorescence of WT, Cadap1/Cadap1 mutant, and CaDAP1 RS stained with 25 μg/mL CFW. (E) Loss of CaDAP1 downregulates the expression level of CaCHS3. The WT strain (white bar), Cadap1/Cadap1 mutant (gray bar), and RS (black bar) were cultured in YPD medium to the exponential phase at 30 °C. CaCHS3 expression is depicted as a fold increase compared to the WT strain. (F) The deletion of CaDAP1 downregulates the phosphorylation levels of CaMkc1 and slightly upregulates the phosphorylation levels of CaCek1. Exponential phase cultures were harvested, and protein extracts separated using SDS-PAGE, followed by immunoblotting with antibodies against phosphorylated CaMkc1 or CaCek1. Immunoblotting with antibodies against the α-tubulin served as a loading control. An asterisk (*) denotes a significant difference (p < 0.05) between the Cadap1/Cadap1 mutant and WT or RS.
To further explore the impact of the CaDAP1 knockout on chitin content in cell walls, Ca cells were stained with CFW to visualize chitin. The intensity of CFW fluorescence accurately reflects the relative chitin content [31,36]. Compared with the WT and RS, the intensity of CFW fluorescence was significantly decreased in the Cadap1/Cadap1 mutant (Figure 3D). This provides additional evidence that the CaDAP1 knockout reduces the chitin content in Ca cell walls.
To investigate whether the reduction in chitin content in the Cadap1/Cadap1 mutant corresponds with a decrease in the expression of chitin synthase genes, quantitative RT-qPCR was used to detect the expression of CaCHS1, CaCHS2, CaCHS3, and CaCHS8, responsible for synthesizing the chitin content of Ca cell walls [37]. Interestingly, CaDAP1 deletion decreased CaCHS3 expression by 65% compared to the WT and re-integrant strains (Figure 3E), while the expression of CaCHS1, CaCHS2, and CaCHS8 remained unchanged. These findings collectively suggest that CaDAP1 plays an essential role in maintaining chitin content within the cell wall of Ca.
Previous research has indicated the role of the PKC-CaMkc1 axis in regulating chitin biosynthesis [36]. Therefore, we investigated the impact of CaDAP1 deletion on CaMkc1-mediated CWI signaling. Compared to the WT and RS, the phosphorylation level of CaMkc1 was decreased in the Cadap1/Cadap1 mutants (Figure 3F). Additionally, the phosphorylation level of CaCek1 increased slightly in the Cadap1/Cadap1 mutants compared with the WT and RS (Figure 3F).
3.4. Impact of CaDAP1 Deletion on Hyphal Development and Colony Morphology
To explore the role of CaDAP1 in hyphal formation, filamentous growth was assayed in YPD medium supplemented with 10% FBS. Following induction with 10% FBS, the mean filament length of WT RM1000 was 41.9 μm (3 h induction; n = 174) (Figure 4A,B). The dap1/dap1 mutant exhibited an average filament length of only 28.3 μm (3 h induction; n = 163), whereas the revertant strain reached 41.7 μm (3 h induction; n = 159) (Figure 4A,B). These results suggest that CaDAP1 deletion impairs FBS-induced filamentation in Ca cells, a defect that can be reversed by reintroducing the CaDAP1 gene (Figure 4A,B). Moreover, on the solid YPD medium, the Cadap1/Cadap1 mutant formed colonies with a smoother surface compared to the wrinkled-surface colonies of the WT RM1000. The reintroduction of CaDAP1 into the homozygous mutant restored colony morphologies similar to the WT strain (Figure 4C). Thus, the deletion of CaDAP1 significantly affected filamentation and colony morphology in Ca.
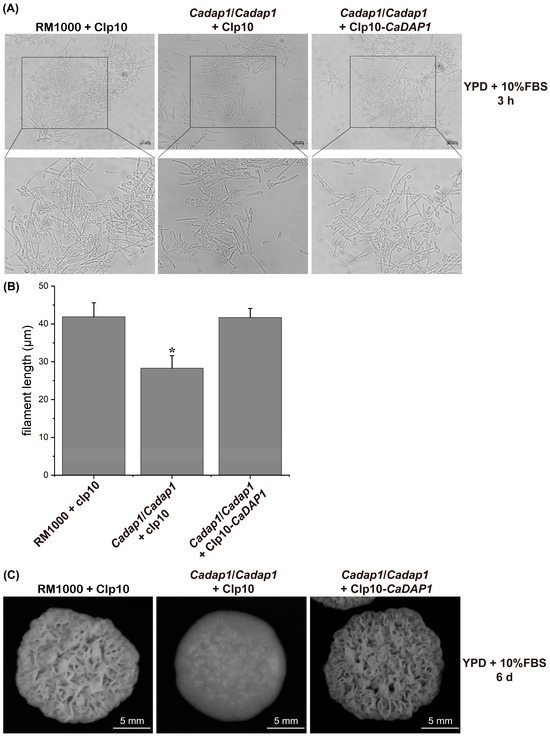
Figure 4.
The deletion of CaDAP1 affects hyphal development and colony morphology. Induction of filamentation was visualized in YPD medium supplemented with 10% FBS for both WT RM1000 and the Cadap1/Cadap1 mutant, which contained either the integrated CIp10 vector or CIp10-CaDAP1 plasmid (A). Filament length was quantified for each strain (B), and their colony-formation ability assessed (C). The difference in filament length between WT RM1000 and Cadap1/Cadap1 mutant (B) was significant. An asterisk (*) denotes a significant difference (p < 0.05) between the Cadap1/Cadap1 mutant and WT or RS.
3.5. The Deletion of CaDAP1 Affects the Virulence of Ca
To assess the involvement of CaDAP1 in the virulence of Ca cells, mice were intravenously injected with WT RM1000, Cadap1/Cadap1 mutant, or RS cells. At day 9, no survivors were observed among the groups of 10 mice injected with the WT strain, whereas for the Cadap1/Cadap1 mutant, no survivors were observed by day 14, and for the RS by day 10 (Figure 5A). The CFUs/g in wet kidney tissues of mice correlated with the virulent effects of Ca after 48 h of infection: 4.35 × 106 for the WT RM1000 (n = 2), 1.21 × 105 for the Cadap1/Cadap1 mutant (n = 2), and 4.42 × 106 for the RS (n = 2). The microscopic examination of kidney tissues revealed a similar pattern to those infected with the WT RM1000 and RS, with heavy infiltration by hyphal filaments. In contrast, those infected with the Cadap1/Cadap1 mutant exhibited fewer filaments, albeit in a similar pattern, along with yeast-like fungal cells. Those injected with saline buffer (CK) exhibited no Ca cells (Figure 5B). Collectively, these findings suggest that CaDAP1 deletion markedly impacts the virulence of Ca cells.
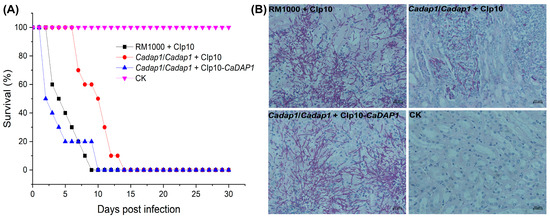
Figure 5.
Assessment of virulence. (A) The survival rates of mice (n = 10) were evaluated after infection with WT RM1000, homozygous Cadap1/Cadap1 mutant, and RS. Daily monitoring for morbidity and survival tracking was conducted over a 30-day period. (B) Kidney tissues from mice infected with WT strain, Cadap1/Cadap1 homozygous mutant, and RS were subjected to histopathological examination. Saline buffer injection (CK) served as a negative control. Sections from infected kidney tissues were stained with periodic acid–Schiff’s reagent. Representative images of 5 kidney cross-sections (n = 2 mice/strain) were captured at 40× magnification.
4. Discussion
In S. cerevisiae, ScDap1 is essential for ergosterol biosynthesis and confers resistance to azoles [14,15,16,17,18]. In the fungal pathogen C. glabrata, CgDap1 is involved in mediating azole tolerance [38]. In the human pathogen Aspergillus fumigatus, AfDapA is necessary for ergosterol biosynthesis and azole resistance [20]. The present investigation has identified and characterized the CaDAP1 gene. Similar to the orthologs in C. glabrata and S. cerevisiae, CaDap1 contains a cytochrome b5-like heme-binding domain (Figure S1, Supplementary Materials). Our experiments demonstrated that CaDAP1 deletion renders Ca cells susceptible to terbinafine and resistant to fluconazole (Figure 2). This outcome implies that the response mechanism of Cadap1 to fluconazole stress is different from those of Scdap1, Cgdap1, and AfdapA. Additionally, we found that the deletion of CaDAP1 increased the sensitivity of Ca to SDS-induced cell membrane stress (Figure 3A), indicating a potential alteration in the composition of the cell membrane due to CaDAP1 deletion.
Chitin is a critical component of fungal cell walls and septa, playing pivotal roles in maintaining fungal pathogenicity and adaption to stress [39,40,41]. A previous study showed that a Ca mutant with low chitin levels exhibited increased resistance to CFW and elevated sensitivity to SDS [42]. Our study reveals that the absence of CaDAP1 also leads to cell resistance to CFW and sensitivity to SDS. In addition, the chitin content was reduced in the Cadap1/Cadap1 mutant cell walls. In Ca, chitin synthesis is facilitated by four chitin synthase enzymes encoded by CaCHS1, CaCHS2, CaCHS3, and CaCHS8, with CaChs3 primarily responsible for chitin synthesis [40,43,44]. Compared to the WT strain, the expression level of CaCHS3 was remarkably decreased in the Cadap1/Cadap1 mutant. Munro et al. demonstrated an obvious reduction in chitin content in the Camkc1/Camkc1 mutant, implying the post-transcriptional regulation of CaChs3 through PKC signaling [36,37]. Han et al. reported that the blocking of β-1,6-glucan synthesis triggers the phosphorylation of CaMkc1, leading to the activation of chitin synthase CaChs3 via a post-transcriptional mechanism, thereby maintaining cell wall chitin levels to ensure cell viability [39]. The knockout of CaDAP1 decreases the CaMkc1 phosphorylation level, which may result in a reduced expression level of CaCHS3 and/or the post-transcriptional regulation of CaChs3. Alternatively, CaDAP1 may serve as a transcriptional activator of CaCHS3 gene expression, and its deletion may affect the transcriptional activation of CaCHS3, leading to decreased chitin levels in the Cadap1/Cadap1 mutant. Previous studies have indicated that elevating chitin content in the Ca cell wall can enhance cell resistance to echinocandins [31,45,46]. The Cadap1/Cadap1 deletion strain is sensitive to caspofungin, probably due to reduced chitin content in the cell wall.
Rowbottom and co-workers reported that CaBNI4 mutants exhibit low cell-wall chitin levels and display smooth colony morphology on serum agar [42]. Compared to the wild-type strain, the Cadap1/Cadap1 mutant displayed smoother colonies on a serum-containing solid medium, possibly due to a reduced chitin level in the cell walls. A previous study has highlighted the necessity of normal chitin content in cell walls for the virulence of Ca [40]. Decreased chitin levels in the Cadap1/Cadap1 mutant may potentially decrease the virulence of Ca. In conclusion, our research indicates that CaDap1 is indispensable for hyphal development, antifungal drug resistance, the maintenance of chitin content in the cell wall, and virulence. Thus, this investigation offers novel insights into the functional role of CaDAP1 in Ca.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/jof10050316/s1, Figure S1: Predicted yeast Dap1 ortholog, CaDap1 (orf19.1034) in Candida albicans; Figure S2: The Cadap1/Cadap1 mutant exhibited insensitivity to fluconazole; Figure S3: Deletion of CaDAP1 leads to unchanged β-1,3-glucan content in cell walls.
Author Contributions
D.X. and F.L. designed the research. D.X. and M.W. conducted the research. X.Z. (Xing Zhang), H.M. and H.X. contributed to the experiments. B.Z. and X.Z. (Xin Zeng) contributed to the analysis of the data. D.X. and F.L. analyzed the data and wrote the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Anhui Provincial Natural Science Foundation (Grant 1808085MC74, supported by the Department of Science and Technology of Anhui Province), the University Natural Science Research Key Projects in Anhui Province (Grant KJ2018A0676, supported by the Department of Education of Anhui Province), the Anhui Province University Outstanding Talents Cultivation Funding Project (Grant gxgnfx2021118, supported by the Department of Education of Anhui Province), and the Anhui Province University Innovation Team Project (Grant 2023AH010045, supported by the Department of Education of Anhui Province).
Institutional Review Board Statement
This study was approved by the Ethics Committee of Huaibei Normal University (permission ECHN191016).
Informed Consent Statement
Not applicable.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author (xudayonghello@163.com or rx2500@163.com) upon reasonable request.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Gow, N.A.R.; van de Veerdonk, F.L.; Brown, A.J.P.; Netea, M.G. Candida albicans morphogenesis and host defence: Discriminating invasion from colonization. Nat. Rev. Microbiol. 2012, 10, 112–122. [Google Scholar] [CrossRef] [PubMed]
- Talapko, J.; Juzbasic, M.; Matijevic, T.; Pustijanac, E.; Bekic, S.; Kotris, I.; Skrlec, I. Candida albicans—The Virulence Factors and Clinical Manifestations of Infection. J. Fungi 2021, 7, 79. [Google Scholar] [CrossRef] [PubMed]
- Calderone, R.A.; Fonzi, W.A. Virulence factors of Candida albicans. Trends Microbiol. 2001, 9, 327–335. [Google Scholar] [CrossRef] [PubMed]
- Richardson, M.D. Changing patterns and trends in systemic fungal infections. J. Antimicrob. Chemother. 2005, 56 (Suppl. S1), i5–i11. [Google Scholar] [CrossRef] [PubMed]
- Mayer, F.L.; Wilson, D.; Hube, B. Candida albicans pathogenicity mechanisms. Virulence 2013, 4, 119–128. [Google Scholar] [CrossRef] [PubMed]
- Malinovská, Z.; Conková, E.; Váczi, P. Biofilm Formation in Medically Important Candida Species. J. Fungi 2023, 9, 955. [Google Scholar] [CrossRef] [PubMed]
- Gow Neil, A.R.; Latge, J.-P.; Munro Carol, A. The Fungal Cell Wall: Structure, Biosynthesis, and Function. Microbiol. Spectr. 2017, 5, 10–1128. [Google Scholar] [CrossRef]
- Lenardon, M.D.; Sood, P.; Dorfmueller, H.C.; Brown, A.J.P.; Gow, N.A.R. Scalar nanostructure of the Candida albicans cell wall; a molecular, cellular and ultrastructural analysis and interpretation. Cell Surf. 2020, 6, 100047. [Google Scholar] [CrossRef]
- Denning, D.W. Echinocandins: A new class of antifungal. J. Antimicrob. Chemother. 2002, 49, 889–891. [Google Scholar] [CrossRef] [PubMed]
- Hopke, A.; Brown, A.J.P.; Hall, R.A.; Wheeler, R.T. Dynamic Fungal Cell Wall Architecture in Stress Adaptation and Immune Evasion. Trends Microbiol. 2018, 26, 284–295. [Google Scholar] [CrossRef]
- Alves, R.; Barata-Antunes, C.; Casal, M.; Brown, A.J.P.; Van Dijck, P.; Paiva, S. Adapting to survive: How Candida overcomes host-imposed constraints during human colonization. PLoS Pathog. 2020, 16, e1008478. [Google Scholar] [CrossRef] [PubMed]
- Ibe, C.; Munro, C.A. Fungal Cell Wall Proteins and Signaling Pathways Form a Cytoprotective Network to Combat Stresses. J. Fungi 2021, 7, 739. [Google Scholar] [CrossRef] [PubMed]
- Lenardon, M.D.; Munro, C.A.; Gow, N.A. Chitin synthesis and fungal pathogenesis. Curr. Opin. Microbiol. 2010, 13, 416–423. [Google Scholar] [CrossRef]
- Mifsud, W.; Bateman, A. Membrane-bound progesterone receptors contain a cytochrome b5-like ligand-binding domain. Genome Biol. 2002, 3, RESEARCH0068. [Google Scholar] [CrossRef]
- Hand, R.A.; Jia, N.; Bard, M.; Craven, R.J. Saccharomyces cerevisiae Dap1p, a novel DNA damage response protein related to the mammalian membrane-associated progesterone receptor. Eukaryot. Cell 2003, 2, 306–317. [Google Scholar] [CrossRef] [PubMed]
- Mallory, J.C.; Crudden, G.; Johnson, B.L.; Mo, C.; Pierson, C.A.; Bard, M.; Craven, R.J. Dap1p, a heme-binding protein that regulates the cytochrome P450 protein Erg11p/Cyp51p in Saccharomyces cerevisiae. Mol. Cell. Biol. 2005, 25, 1669–1679. [Google Scholar] [CrossRef] [PubMed]
- Craven, R.J.; Mallory, J.C.; Hand, R.A. Regulation of iron homeostasis mediated by the heme-binding protein Dap1 (damage resistance protein 1) via the P450 protein Erg11/Cyp51. J. Biol. Chem. 2007, 282, 36543–36551. [Google Scholar] [CrossRef] [PubMed]
- Jo, W.J.; Kim, J.H.; Oh, E.; Jaramillo, D.; Holman, P.; Loguinov, A.V.; Arkin, A.P.; Nislow, C.; Giaever, G.; Vulpe, C.D. Novel insights into iron metabolism by integrating deletome and transcriptome analysis in an iron deficiency model of the yeast Saccharomyces cerevisiae. BMC Genom. 2009, 10, 130. [Google Scholar] [CrossRef] [PubMed]
- Hughes, A.L.; Powell, D.W.; Bard, M.; Eckstein, J.; Barbuch, R.; Link, A.J.; Espenshade, P.J. Dap1/PGRMC1 binds and regulates cytochrome P450 enzymes. Cell Metab. 2007, 5, 143–149. [Google Scholar] [CrossRef]
- Song, J.X.; Zhai, P.F.; Zhang, Y.W.; Zhang, C.Y.; Sang, H.; Han, G.Z.; Keller, N.P.; Lu, L. The Aspergillus fumigatus Damage Resistance Protein Family Coordinately Regulates Ergosterol Biosynthesis and Azole Susceptibility. mBio 2016, 7, 13. [Google Scholar] [CrossRef]
- Lee, C.-M.; Nantel, A.; Jiang, L.; Whiteway, M.; Shen, S.-H. The serine/threonine protein phosphatase SIT4 modulates yeast-to-hypha morphogenesis and virulence in Candida albicans. Mol. Microbiol. 2004, 51, 691–709. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.Y.; Cheng, J.Q.; Cao, C.L.; Wang, L.T.; Jiang, L.H. Genetic interactions between Rch1 and the high-affinity calcium influx system Cch1/Mid1/Ecm7 in the regulation of calcium homeostasis, drug tolerance, hyphal development and virulence in Candida albicans. FEMS Yeast Res. 2015, 15, 9. [Google Scholar] [CrossRef]
- Xu, D.Y.; Zhang, X.; Zhang, B.; Zeng, X.; Mao, H.C.; Xu, H.T.; Jiang, L.H.; Li, F. The lipid flippase subunit Cdc50 is required for antifungal drug resistance, endocytosis, hyphal development and virulence in Candida albicans. FEMS Yeast Res. 2019, 19, 11. [Google Scholar] [CrossRef] [PubMed]
- Negredo, A.; Monteoliva, L.; Gil, C.; Pla, J.; Nombela, C. Cloning, analysis and one-step disruption of the ARG5,6 gene of Candida albicans. Microbiology 1997, 143 Pt 2, 297–302. [Google Scholar] [CrossRef]
- Reuss, O.; Vik, A.; Kolter, R.; Morschhauser, J. The SAT1 flipper, an optimized tool for gene disruption in Candida albicans. Gene 2004, 341, 119–127. [Google Scholar] [CrossRef] [PubMed]
- Wilson, R.B.; Davis, D.; Mitchell, A.P. Rapid hypothesis testing with Candida albicans through gene disruption with short homology regions. J. Bacteriol. 1999, 181, 1868–1874. [Google Scholar] [CrossRef] [PubMed]
- Murad, A.M.; Lee, P.R.; Broadbent, I.D.; Barelle, C.J.; Brown, A.J. CIp10, an efficient and convenient integrating vector for Candida albicans. Yeast 2000, 16, 325–327. [Google Scholar] [CrossRef]
- Jia, C.; Zhang, K.; Zhang, D.; Yu, Q.L.; Zhao, Q.; Xiao, C.P.; Dong, Y.J.; Chu, M.P.; Li, M.C. Roles of VPH2 and VMA6 in localization of V-ATPase subunits, cell wall functions and filamentous development in Candida albicans. Fungal Genet. Biol. 2018, 114, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, D.; Abe, M.; Ohya, Y. Yeast Lrg1p acts as a specialized RhoGAP regulating 1,3-beta-glucan synthesis. Yeast 2001, 18, 943–951. [Google Scholar] [CrossRef] [PubMed]
- Juchimiuk, M.; Orlowski, J.; Gawarecka, K.; Swiezewska, E.; Ernst, J.F.; Palamarczyk, G. Candida albicans cis-prenyltransferase Rer2 is required for protein glycosylation, cell wall integrity and hypha formation. Fungal Genet. Biol. 2014, 69, 1–12. [Google Scholar] [CrossRef]
- Walker, L.A.; Munro, C.A.; de Bruijn, I.; Lenardon, M.D.; McKinnon, A.; Gow, N.A.R. Stimulation of chitin synthesis rescues Candida albicans from echinocandins. PLoS Pathog. 2008, 4, e1000040. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.H.; Chen, X.Q.; He, Y.M.; Yu, X.Y.; Li, S.S.; Gao, N.; Niu, L.D.; Mao, Y.H.; Wang, Y.Y.; Wu, X.W.; et al. Mitochondrial complex I bridges a connection between regulation of carbon flexibility and gastrointestinal commensalism in the human fungal pathogen Candida albicans. PLoS Pathog. 2017, 13, 35. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.B.; Pande, K.; French, S.D.; Tuch, B.B.; Noble, S.M. An Iron Homeostasis Regulatory Circuit with Reciprocal Roles in Candida albicans Commensalism and Pathogenesis. Cell Host Microbe 2011, 10, 118–135. [Google Scholar] [CrossRef] [PubMed]
- Denning, D.W. Echinocandin antifungal drugs. Lancet 2003, 362, 1142–1151. [Google Scholar] [CrossRef] [PubMed]
- Perlin, D.S. Current perspectives on echinocandin class drugs. Future Microbiol. 2011, 6, 441–457. [Google Scholar] [CrossRef] [PubMed]
- Munro, C.A.; Selvaggini, S.; de Bruijn, I.; Walker, L.; Lenardon, M.D.; Gerssen, B.; Milne, S.; Brown, A.J.P.; Gow, N.A.R. The PKC, HOG and Ca2+ signalling pathways co-ordinately regulate chitin synthesis in Candida albicans. Mol. Microbiol. 2007, 63, 1399–1413. [Google Scholar] [CrossRef] [PubMed]
- Lenardon, M.D.; Milne, S.A.; Mora-Montes, H.M.; Kaffarnik, F.A.R.; Peck, S.C.; Brown, A.J.P.; Munro, C.A.; Gow, N.A.R. Phosphorylation regulates polarisation of chitin synthesis in Candida albicans. J. Cell Sci. 2010, 123, 2199–2206. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hosogaya, N.; Miyazaki, T.; Nagi, M.; Tanabe, K.; Minematsu, A.; Nagayoshi, Y.; Yamauchi, S.; Nakamura, S.; Imamura, Y.; Izumikawa, K.; et al. The heme-binding protein Dap1 links iron homeostasis to azole resistance via the P450 protein Erg11 in Candida glabrata. FEMS Yeast Res. 2013, 13, 411–421. [Google Scholar] [CrossRef] [PubMed]
- Han, Q.; Wang, N.; Pan, C.Y.; Wang, Y.; Sang, J.L. Elevation of cell wall chitin via Ca2+-calcineurin-mediated PKC signaling pathway maintains the viability of Candida albicans in the absence of β-1,6-glucan synthesis. Mol. Microbiol. 2019, 112, 960–972. [Google Scholar] [CrossRef]
- Bulawa, C.E.; Miller, D.W.; Henry, L.K.; Becker, J.M. Attenuated virulence of chitin-deficient mutants of Candida albicans. Proc. Natl. Acad. Sci. USA 1995, 92, 10570–10574. [Google Scholar] [CrossRef] [PubMed]
- Wagener, J.; MacCallum, D.M.; Brown, G.D.; Gow, N.A.R. Candida albicans Chitin Increases Arginase-1 Activity in Human Macrophages, with an Impact on Macrophage Antimicrobial Functions. mBio 2017, 8, 14. [Google Scholar] [CrossRef] [PubMed]
- Rowbottom, L.; Munro, C.A.; Gow, N.A.R. Candida albicans mutants in the BNI4 gene have reduced cell-wall chitin and alterations in morphogenesis. Microbiology 2004, 150, 3243–3252. [Google Scholar] [CrossRef] [PubMed]
- Mio, T.; Yabe, T.; Sudoh, M.; Satoh, Y.; Nakajima, T.; Arisawa, M.; Yamada-Okabe, H. Role of three chitin synthase genes in the growth of Candida albicans. J. Bacteriol. 1996, 178, 2416–2419. [Google Scholar] [CrossRef] [PubMed]
- Knafler, H.C.; Smaczynska-de Rooij, I.I.; Walker, L.A.; Lee, K.K.; Gow, N.A.R.; Ayscough, K.R. AP-2-Dependent Endocytic Recycling of the Chitin Synthase Chs3 Regulates Polarized Growth in Candida albicans. mBio 2019, 10, 20. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.K.; MacCallum, D.M.; Jacobsen, M.D.; Walker, L.A.; Odds, F.C.; Gow, N.A.R.; Munro, C.A. Elevated Cell Wall Chitin in Candida albicans Confers Echinocandin Resistance In Vivo. Antimicrob. Agents Chemother. 2012, 56, 208–217. [Google Scholar] [CrossRef] [PubMed]
- Walker, L.A.; Gow, N.A.R.; Munro, C.A. Elevated Chitin Content Reduces the Susceptibility of Candida Species to Caspofungin. Antimicrob. Agents Chemother. 2013, 57, 146–154. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).