Secretome Analysis of Thermothelomyces thermophilus LMBC 162 Cultivated with Tamarindus indica Seeds Reveals CAZymes for Degradation of Lignocellulosic Biomass
Abstract
1. Introduction
2. Methods and Materials
2.1. Maintenance of the Fungus and Culture Medium
2.2. Submerged Cultivation of T. thermophilus LMBC 162 for Protein Secretion Induction
2.3. Protein Quantification
2.4. Sample Processing
2.5. Characterization of the T. thermophilus LMBC 162 Using Liquid Chromatography-Tandem Mass Spectrometry (LC-MS/MS)
3. Results and Discussion
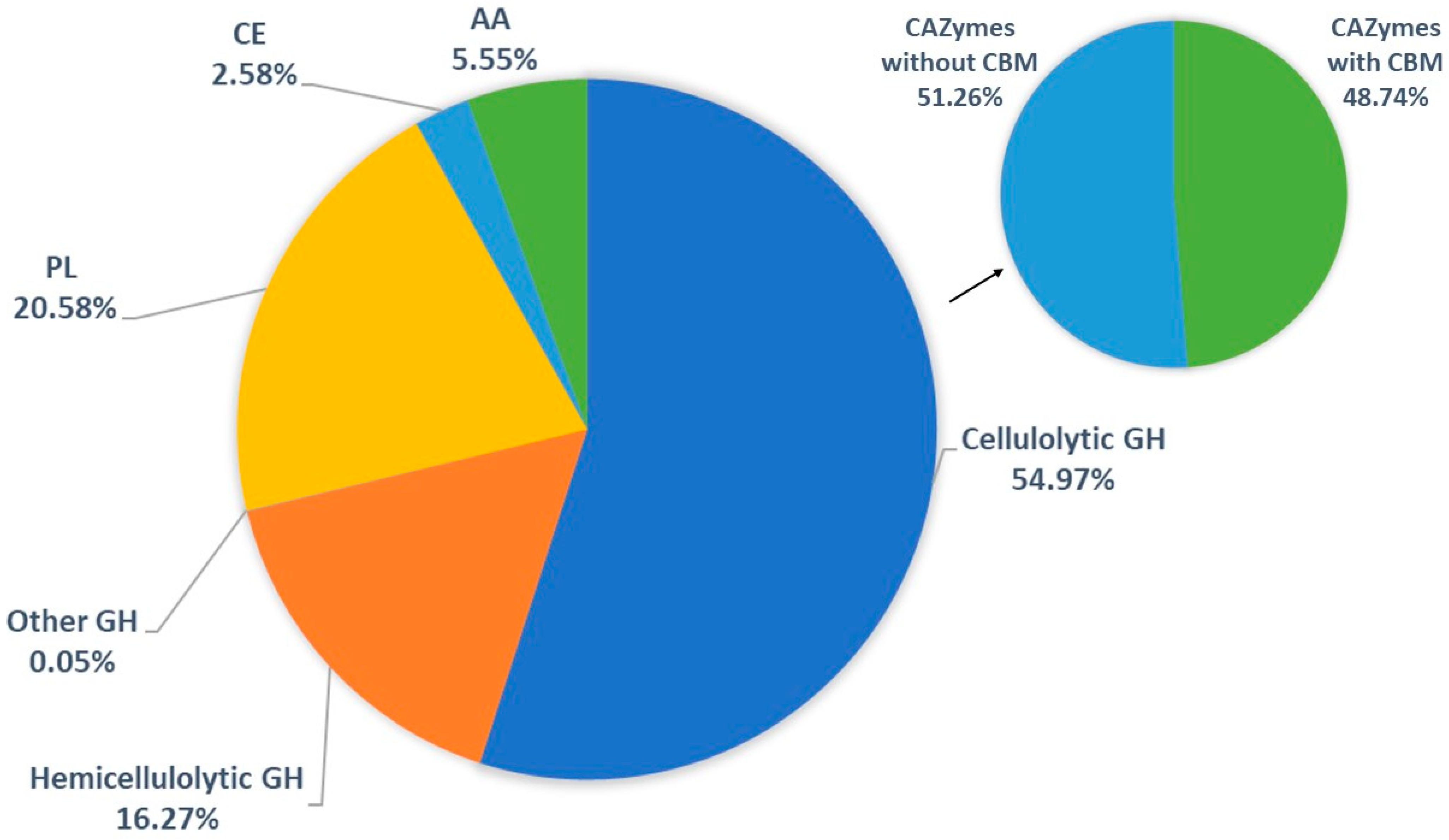
Analysis of Secretome Protein Composition
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yamaguchi, A.; Mimura, N.; Shirai, M.; Sato, O. Cascade utilization of biomass: Strategy for conversion of cellulose, hemicellulose, and lignin into useful chemicals. ACS Sustain. Chem. Eng. 2019, 7, 10445–10451. [Google Scholar] [CrossRef]
- Ravindra, K.; Singh, T.; Mor, S. Emissions of air pollutants from primary crop residue burning in India and their mitigation strategies for cleaner emissions. J. Clean. Prod. 2019, 208, 261–273. [Google Scholar] [CrossRef]
- Yadav, M.; Paritosh, K.; Pareek, N.; Vivekanand, V. Coupled treatment of lignocellulosic agricultural residues for augmented biomethanation. J. Clean. Prod. 2019, 213, 75–88. [Google Scholar] [CrossRef]
- Singh, T.A.; Sharma, M.; Sharma, M.; Sharma, G.D.; Passari, A.K.; Bhasin, S. Valorization of agro-industrial residues for production of commercial biorefinery products. Fuel 2022, 322, 124284. [Google Scholar] [CrossRef]
- Ortega, F.; Versino, F.; López, O.V.; García, M.A. Biobased composites from agro-industrial wastes and by-products. Emergent Mater. 2022, 5, 873–921. [Google Scholar] [CrossRef]
- Bechara, R.; Gomez, A.; Saint-Antonin, V.; Schweitzer, J.M.; Maréchal, F.; Ensinas, A. Review of design works for the conversion of sugarcane to first and second-generation ethanol and electricity. Renew. Sust. Energy Rev. 2018, 91, 152–164. [Google Scholar] [CrossRef]
- Klein, B.C.; Sampaio, I.L.M.; Mantelatto, P.E.; Maciel Filho, R.; Bonomi, A. Beyond ethanol, sugar, and electricity: A critical review of product diversification in Brazilian sugarcane mills. Biofuel Bioprod. Biorefining 2019, 13, 809–821. [Google Scholar] [CrossRef]
- Magalhães, A.I., Jr.; de Carvalho, J.C.; Pereira, G.V.M.; Karp, S.G.; Câmara, M.C.; Medina, J.D.C.; Soccol, C.R. Lignocellulosic biomass from agro-industrial residues in South America: Current developments and perspectives. Biofuel Bioprod. Biorefining 2019, 13, 1505–1519. [Google Scholar] [CrossRef]
- Patel, A.; Shah, A.R. Integrated lignocellulosic biorefinery: Gateway for production of second generation ethanol and value added products. J. Bioresour. Bioprod. 2021, 6, 108–128. [Google Scholar] [CrossRef]
- Chukwuma, O.B.; Rafatullah, M.; Tajarudin, H.A.; Ismail, N. Lignocellulolytic enzymes in biotechnological and industrial processes: A review. Sustainability 2020, 12, 7282. [Google Scholar] [CrossRef]
- dos Santos, J.R.; Moreira, L.R.S.; Ferreira Filho, E.X. Cellulose-degrading enzymes: Key players in biorefinery development. Biologia 2023, 78, 1759–1772. [Google Scholar] [CrossRef]
- Borin, G.P.; Sanchez, C.C.; de Souza, A.P.; de Santana, E.S.; de Souza, A.T.; Leme, A.F.P.; Squina, F.M.; Buckeridge, M.S.; Goldman, G.H.; Oliveira, J.V.D.C. Comparative secretome analysis of Trichoderma reesei and Aspergillus niger during growth on sugarcane biomass. PLoS ONE 2015, 10, e0129275. [Google Scholar] [CrossRef]
- Basotra, N.; Kaur, B.; Di Falco, M.; Tsang, A.; Chadha, B.S. Mycothermus thermophilus (Syn. Scytalidium thermophilum): Repertoire of a diverse array of efficient cellulases and hemicellulases in the secretome revealed. Bioresour. Technol. 2016, 222, 413–421. [Google Scholar] [CrossRef] [PubMed]
- Raheja, Y.; Kaur, B.; di Falco, M.; Tsang, A.; Chadha, B.S. Secretome analysis of Talaromyces emersonii reveals distinct CAZymes profile and enhanced cellulase production through response surface methodology. Ind. Crop. Prod. 2020, 152, 112554. [Google Scholar] [CrossRef]
- Wijayawardene, N.N.; Boonyuen, N.; Ranaweera, C.B.; de Zoysa, H.K.; Padmathilake, R.E.; Nifla, F.; Dai, D.Q.; Liu, X.; Suwannarachi, N.; Kumla, J.; et al. OMICS and other advanced technologies in mycological applications. J. Fungi 2023, 9, 688. [Google Scholar] [CrossRef]
- Barrett, K.; Jensen, K.; Meyer, A.S.; Frisvad, J.C.; Lange, L. Fungal secretome profile categorization of CAZymes by function and family corresponds to fungal phylogeny and taxonomy: Example Aspergillus and Penicillium. Sci. Rep. 2020, 10, 5158. [Google Scholar] [CrossRef] [PubMed]
- Nin-Hill, A.; Piniello, B.; Rovira, C. In silico modelling of the function of disease-related CAZymes. Essays Biochem. 2023, 67, 355–372. [Google Scholar] [CrossRef] [PubMed]
- Salazar-Cerezo, S.; de Vries, R.P.; Garrigues, S. Strategies for the development of industrial fungal producing strains. J. Fungi 2023, 9, 834. [Google Scholar] [CrossRef]
- Perez, C.L.; Casciatori, F.P.; Thomeo, J.C. Strategies for scaling-up packed-bed bioreactors for solid-state fermentation: The case of cellulolytic enzymes production by a thermophilic fungus. Chem. Eng. J. 2019, 361, 1142–1151. [Google Scholar] [CrossRef]
- Liu, E.; Segato, F.; Wilkins, M.R. Fed-batch production of Thermothelomyces thermophilus lignin peroxidase using a recombinant Aspergillus nidulans strain in stirred-tank bioreactor. Bioresour. Technol. 2021, 325, 124700. [Google Scholar] [CrossRef]
- Yang, J.; Yue, H.R.; Pan, L.Y.; Feng, J.X.; Zhao, S.; Suwannarangsee, S.; Champreda, V.; Liu, C.G.; Zhao, X.Q. Fungal strain improvement for efficient cellulase production and lignocellulosic biorefinery: Current status and future prospects. Bioresour. Technol. 2023, 385, 129449. [Google Scholar] [CrossRef] [PubMed]
- Berka, R.M.; Grigoriev, I.V.; Otillar, R.; Salamov, A.; Grimwood, J.; Reid, I.; Ishmael, N.; John, T.; Darmond, C.; Moisan, M.C.; et al. Comparative genomic analysis of the thermophilic biomass-degrading fungi Myceliophthora thermophila and Thielavia terrestris. Nat. Biotechnol. 2011, 29, 922–927. [Google Scholar] [CrossRef] [PubMed]
- Marin-Felix, Y.; Stchigel, A.M.; Miller, A.N.; Guarro, J.; Cano-Lira, J.F. A re-evaluation of the genus Myceliophthora (Sordariales, Ascomycota): Its segregation into four genera and description of Corynascus fumimontanus sp. nov. Mycologia 2015, 107, 619–632. [Google Scholar] [CrossRef] [PubMed]
- dos Santos, H.B.; Bezerra, T.M.S.; Pradella, J.C.G.; Delabona, P.; Lima, D.; Gomes, E.; Hartson, S.D.; Rogers, J.; Couger, B.; Prade, R. Myceliophthora thermophila M77 utilizes hydrolytic and oxidative mechanisms to deconstruct biomass. AMB Express 2016, 6, 103. [Google Scholar] [CrossRef] [PubMed]
- Chorozian, K.; Karnaouri, A.; Karantonis, A.; Souli, M.; Topakas, E. Characterization of a dual cellulolytic/xylanolytic AA9 lytic polysaccharide monooxygenase from Thermothelomyces thermophilus and its utilization toward nanocellulose production in a multi-step bioprocess. ACS Sustain. Chem. Eng. 2022, 10, 8919–8929. [Google Scholar] [CrossRef]
- Balabanova, L.; Seitkalieva, A.; Yugay, Y.; Rusapetova, T.; Slepchenko, L.; Podvolotskaya, A.; Yatsunskaya, M.; Vasyutkina, E.; Son, O.; Tekutyeva, L.; et al. Engineered fungus Thermothelomyces thermophilus producing plant storage proteins. J. Fungi 2022, 8, 119. [Google Scholar] [CrossRef]
- Contato, A.G.; Nogueira, K.M.V.; Buckeridge, M.S.; Silva, R.N.; Polizeli, M.L.T.M. Trichoderma longibrachiatum and Thermothelomyces thermophilus co-culture: Improvement the saccharification profile of different sugarcane bagasse varieties. Biotechnol. Lett. 2023, 45, 1093–1102. [Google Scholar] [CrossRef]
- Ramesh, T.; Rajalaksmi, N.; Dhatathrevan, K.S. Activated carbons derived from tamarind seeds for hydrogen storage. J. Energy Storage 2015, 4, 89–95. [Google Scholar] [CrossRef]
- Reis, P.M.C.L.; Dariva, C.; Vieira, G.A.B.; Hense, H. Extraction and evaluation of antioxidant potential of the extracts obtained from tamarind seeds (Tamarindus indica), sweet variety. J. Food Eng. 2016, 173, 116–123. [Google Scholar] [CrossRef]
- Gonçalves, G.R.; Gandolfi, O.R.; Bonomo, R.C.F.; Fontan, R.D.C.I.; Veloso, C.M. Synthesis of activated carbon from hydrothermally carbonized tamarind seeds for lipase immobilization: Characterization and application in aroma ester synthesis. J. Chem. Technol. Biotechnol. 2021, 96, 3316–3329. [Google Scholar] [CrossRef]
- Contato, A.G.; de Oliveira, T.B.; Aranha, G.M.; de Freitas, E.N.; Vici, A.C.; Nogueira, K.M.V.; de Lucas, R.C.; Scarcella, A.S.A.; Buckeridge, M.S.; Silva, R.N.; et al. Prospection of fungal lignocellulolytic enzymes produced from jatoba (Hymenaea courbaril) and tamarind (Tamarindus indica) seeds: Scaling for bioreactor and saccharification profile of sugarcane bagasse. Microorganisms 2021, 9, 533. [Google Scholar] [CrossRef] [PubMed]
- Calderaro, F.; Keser, M.; Akeroyd, M.; Bevers, L.E.; Eijsink, V.G.H.; Várnai, A.; van den Berg, M.A. Characterization of an AA9 LPMO from Thielavia australiensis, TausLPMO9B, under industrially relevant lignocellulose saccharification conditions. Biotechnol. Biofuels 2020, 13, 1–17. [Google Scholar] [CrossRef]
- Buckeridge, M.S.; Dietrich, S.M.C. Galactomannan from Brazilian legume seeds. Rev. Bras. Bot. 1990, 13, 109–112. [Google Scholar]
- Khanna, P.; Sundari, S.S.; Kumar, N.J. Production, isolation, and partial purification of xylanases from an Aspergillus sp. World J. Microbiol. Biotechnol. 1995, 11, 242–243. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef] [PubMed]
- Weber, K.; Osborn, M. The reliability of molecular weight determinations by dodecyl sulfate-polyacrylamide gel electrophoresis. J. Biol. Chem. 1969, 244, 4406–4412. [Google Scholar] [CrossRef]
- Voruganti, S.; Kline, J.T.; Balch, M.J.; Rogers, J.; Matts, R.L.; Hartson, S.D. Proteomic profiling of Hsp90 inhibitors. In Chaperones; Humana Press: New York, NY, USA, 2018; pp. 139–162. [Google Scholar]
- Cox, J.; Mann, M. MaxQuant enables high peptide identification rates, individualized ppb-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008, 26, 1367–1372. [Google Scholar] [CrossRef]
- Machado, A.S.; Valadares, F.; Silva, T.F.; Milagres, A.M.; Segato, F.; Ferraz, A. The secretome of Phanerochaete chrysosporium and Trametes versicolor grown in microcrystalline cellulose and use of the enzymes for hydrolysis of lignocellulosic materials. Front. Bioeng. Biotechnol. 2020, 8, 826. [Google Scholar] [CrossRef]
- Levasseur, A.; Drula, E.; Lombard, V.; Coutinho, P.M.; Henrissat, B. Expansion of the enzymatic repertoire of the CAZy database to integrate auxiliary redox enzymes. Biotechnol. Biofuels 2013, 6, 41. [Google Scholar] [CrossRef]
- Biely, P. Microbial carbohydrate esterases deacetylating plant polysaccharides. Biotechnol. Adv. 2012, 30, 1575–1588. [Google Scholar] [CrossRef]
- Rocha, V.A.L.; Maeda, R.N.; Pereira, N., Jr.; Kern, M.F.; Elias, L.; Simister, R.; Steele-King, C.; Gómez, L.D.; McQueen-Mason, S.J. Characterization of the cellulolytic secretome of Trichoderma harzianum during growth on sugarcane bagasse and analysis of the activity boosting effects of swollenin. Biotechnol. Progress 2016, 32, 327–336. [Google Scholar] [CrossRef] [PubMed]
- Kanelli, M.; Topakas, E. Acylation of soluble polysaccharides in a biphasic system catalyzed by a CE2 acetylesterase. Carbohydr. Polym. 2017, 163, 208–215. [Google Scholar] [CrossRef] [PubMed]
- Satapathy, S.; Rout, J.R.; Kerry, R.G.; Thatoi, H.; Sahoo, S.L. Biochemical prospects of various microbial pectinase and pectin: An approachable concept in pharmaceutical bioprocessing. Front. Nutr. 2020, 7, 117. [Google Scholar] [CrossRef] [PubMed]
- Garron, M.L.; Cygler, M. Uronic polysaccharide degrading enzymes. Curr. Opin. Struct. Biol. 2014, 28, 87–95. [Google Scholar] [CrossRef]
- Verma, S.; Gazara, R.K.; Nizam, S.; Parween, S.; Chattopadhyay, D.; Verma, P.K. Draft genome sequencing and secretome analysis of fungal phytopathogen Ascochyta rabiei provides insight into the necrotrophic effector repertoire. Sci. Rep. 2016, 6, 24638. [Google Scholar] [CrossRef]
- Rubio, M.V.; Zubieta, M.P.; Cairo, J.P.L.F.; Calzado, F.; Leme, A.F.P.; Squina, F.M.; Prade, R.A.; Damásio, A.R.L. Mapping N-linked glycosylation of carbohydrate-active enzymes in the secretome of Aspergillus nidulans grown on lignocellulose. Biotechnol. Biofuels 2016, 9, 168. [Google Scholar] [CrossRef] [PubMed]
- Bonnin, E.; Pelloux, J. Pectin degrading enzymes. In Pectin: Technological and Physiological Properties; Kontogiorgos, V., Ed.; Springer International Publishing: Cham, Switzerland, 2020; pp. 37–60. [Google Scholar]
- Prieto, A.; de Eugenio, L.; Méndez-Líter, J.A.; Nieto-Domínguez, M.; Murgiondo, C.; Barriuso, J.; Bejarano-Muñoz, L.; Martínez, M.J. Fungal glycosyl hydrolases for sustainable plant biomass valorization: Talaromyces amestolkiae as a model fungus. Int. Microbiol. 2021, 24, 545–558. [Google Scholar] [CrossRef]
- Berezina, O.V.; Herlet, J.; Rykov, S.V.; Kornberger, P.; Zavyalov, A.; Kozlov, D.; Sakhibgaraeva, L.; Krestyanova, I.; Schwarz, W.H.; Zverlov, Z.Z.; et al. Thermostable multifunctional GH74 xyloglucanase from Myceliophthora thermophila: High-level expression in Pichia pastoris and characterization of the recombinant protein. Appl. Microbiol. Biotechnol. 2017, 101, 5653–5666. [Google Scholar] [CrossRef]
- Velasco, J.; Oliva, B.; Gonçalves, A.L.; Lima, A.S.; Ferreira, G.; França, B.A.; Mulinari, E.J.; Gonçalves, T.A.; Squina, F.M.; Kadowaki, M.A.S.; et al. Functional characterization of a novel thermophilic exo-arabinanase from Thermothielavioides terrestris. Appl. Microbiol. Biotechnol. 2020, 104, 8309–8326. [Google Scholar] [CrossRef]
- Lafond, M.; Navarro, D.; Haon, M.; Couturier, M.; Berrin, J.G. Characterization of a broad-specificity β-glucanase acting on β-(1,3)-, β-(1,4)-, and β-(1,6)-glucans that defines a new glycoside hydrolase family. Appl. Environ. Microbiol. 2012, 78, 8540–8546. [Google Scholar] [CrossRef]
- Bamford, N.C.; Snarr, B.D.; Gravelat, F.N.; Little, D.J.; Lee, M.J.; Zacharias, C.A.; Chabot, J.C.; Geller, A.M.; Baptista, S.D.; Baker, P.; et al. Sph3 is a glycoside hydrolase required for the biosynthesis of galactosaminogalactan in Aspergillus fumigatus. J. Biol. Chem. 2015, 290, 27438–27450. [Google Scholar] [CrossRef] [PubMed]
- Harvey, A.J.; Hrmova, M.; De Gori, R.; Varghese, J.N.; Fincher, G.B. Comparative modeling of the three-dimensional structures of family 3 glycoside hydrolases. Proteins 2000, 41, 257–269. [Google Scholar] [CrossRef] [PubMed]
- Velasco, J.; Oliva, B.; Mulinari, E.J.; Quintero, L.P.; Lima, A.S.; Gonçalves, A.L.; Gonçalves, T.A.; Damasio, A.; Squina, F.M.; Milagres, A.M.F.; et al. Heterologous expression and functional characterization of a GH10 endoxylanase from Aspergillus fumigatus var. niveus with potential biotechnological application. Biotechnol. Rep. 2019, 24, e00382. [Google Scholar] [CrossRef]
- Liberato, M.V.; Prates, E.T.; Gonçalves, T.A.; Bernardes, A.; Vilela, N.; Fattori, J.; Ematsu, G.C.; Chinaglia, M.; Gomes, E.R.M.; Figueira, A.C.M.; et al. Insights into the dual cleavage activity of the GH16 laminarinase enzyme class on β-1,3 and β-1,4 glycosidic bonds. J. Biol. Chem. 2021, 296, 100385. [Google Scholar] [CrossRef]
- Mewis, K.; Lenfant, N.; Lombard, V.; Henrissat, B. Dividing the large glycoside hydrolase family 43 into subfamilies: A motivation for detailed enzyme characterization. Appl. Environ. Microbiol. 2016, 82, 1686–1692. [Google Scholar] [CrossRef]
- Bianchetti, C.M.; Takasuka, T.E.; Deutsch, S.; Udell, H.S.; Yik, E.J.; Bergeman, L.F.; Fox, B.G. Active site and laminarin binding in glycoside hydrolase family 55. J. Biol. Chem. 2015, 290, 11819–11832. [Google Scholar] [CrossRef]
- Sakaguchi, M. Diverse and common features of trehalases and their contributions to microbial trehalose metabolism. Appl. Microbiol. Biotechnol. 2020, 104, 1837–1847. [Google Scholar] [CrossRef]
- Silchenko, A.S.; Rubtsov, N.K.; Zueva, A.O.; Kusaykin, M.I.; Rasin, A.B.; Ermakova, S.P. Fucoidan-active α-L-fucosidases of the GH29 and GH95 families from a fucoidan degrading cluster of the marine bacterium Wenyingzhuangia fucanilytica. Arch. Biochem. Biophys. 2022, 728, 109373. [Google Scholar] [CrossRef]
- Guillén, D.; Sánchez, S.; Rodríguez-Sanoja, R. Carbohydrate-binding domains: Multiplicity of biological roles. Appl. Microbiol. Biotechnol. 2010, 85, 1241–1249. [Google Scholar] [CrossRef]
- McDonald, W.H.; Yates, J.R., III. Shotgun proteomics and biomarker discovery. Dis. Markers 2002, 18, 99–105. [Google Scholar] [CrossRef]
| MS/MS View: Identified Proteins | Accession Number | Molecular Weight (kDa) a | CAZy Domain | BLAST E-Value b | iBAQ c | iBAQ/Total iBAQ (%) | |
|---|---|---|---|---|---|---|---|
| UniProt/Swiss-Prot Database | NCBI Database | ||||||
| glucose–methanol–choline GMC oxidoreductase | G2PZJ2 | XP_003660923.1 | 69 | AA3 | 9.5 × 10−153 | 11699000 | 0.35 |
| cellobiose dehydrogenase | G2QGP4 | XP_003663851.1 | 68 | AA3—CBM1 | 2.2 × 10−76 | 231140 | 0.73 |
| cellobiose dehydrogenase | G2QNS9 | XP_003666548.1 | 57 | AA3 | 7 × 10−197 | 210360 | 0.01 |
| glyoxal oxidase | G2Q335 | XP_003658743.1 | 105 | AA5 | 4.8 × 10−142 | 86049 | 0.01 |
| FAD linked oxidase | G2QG48 | XP_003663758.1 | 54 | AA7 | 5.9 × 10−92 | 818280 | 0.02 |
| FAD linked oxidase | G2Q654 | XP_003660778.1 | 69 | AA7 | 1.8 × 10−45 | 98150 | 0.01 |
| cellobiose dehydrogenase | G2QFY4 | XP_003664543.1 | 84 | AA8 | 6 × 10−65 | 984050 | 0.03 |
| cellobiose dehydrogenase | A9XK88 | XP_003663382.1 | 89 | AA8—CBM1 | 2.5 × 10−71 | 846980 | 0.02 |
| lytic polysaccharide monooxygenase G | G2Q4M0 | XP_003659754.1 | 32 | AA9—CBM1 | 1.7 × 10−67 | 65875000 | 1.93 |
| lytic polysaccharide monooxygenase B | G2QCJ3 | XP_003663414.1 | 32 | AA9—CBM1 | 1.4 × 10−74 | 46761000 | 1.36 |
| lytic polysaccharide monooxygenase H | G2Q9T3 | XP_003661787.1 | 35 | AA9—CBM1 | 4.3 × 10−71 | 25188000 | 0.74 |
| lytic polysaccharide monooxygenase J | G2Q7A5 | XP_003661261.1 | 26 | AA9 | 1.4 × 10−74 | 6202800 | 0.18 |
| lytic polysaccharide monooxygenase F | G2QK49 | XP_003665200.1 | 24 | AA9 | 2.7 × 10−69 | 3043500 | 0.09 |
| lytic polysaccharide monooxygenase I | G2Q9F7 | XP_003661661.1 | 31 | AA9—CBM1 | 2 × 10−71 | 445310 | 0.01 |
| lytic polysaccharide monooxygenase A | G2QI82 | XP_003665516.1 | 24 | AA9 | 3.8 × 10−72 | 153330 | 0.01 |
| pyrroloquinoline quinone-dependent oxidoreductase | G2QES6 | XP_003664200.1 | 54 | AA12 | 1 × 10−171 | 542100 | 0.02 |
| lytic polysaccharide monooxygenase | G2QH80 | XP_003663985.1 | 20 | AA13 | 2.5 × 10−65 | 1054800 | 0.03 |
| MS/MS View: Identified Proteins | Accession Number | Molecular Weight (kDa) a | CAZy Domain | BLAST E-Value b | iBAQ c | iBAQ/Total iBAQ (%) | |
|---|---|---|---|---|---|---|---|
| UniProt/Swiss-Prot Database | NCBI Database | ||||||
| acetylxylan esterase | G2QD29 | XP_003663492.1 | 34 | CE1 | 1.5 × 10−23 | 5647500 | 0.17 |
| lipase | G2QGB0 | XP_003664615.1 | 26 | CE3 | 4.5 × 10−54 | 135180 | 0.01 |
| acetylxylan esterase | G2QJ94 | XP_003665705.1 | 32 | CE5—CBM1 | 1.3 × 10−41 | 2033500 | 0.06 |
| pectinesterase | G2QLD0 | XP_003666007.1 | 36 | CE8 | 1.1 × 10−75 | 40224000 | 1.19 |
| pectinesterase | G2QMM2 | XP_003666447.1 | 35 | CE8 | 7 × 10−79 | 760070 | 0.02 |
| rhamnogalacturonan acetylesterase | G2QMH3 | XP_003666398.1 | 28 | CE12 | 6.2 × 10−45 | 1178900 | 0.03 |
| acetylesterase | G2QJ27 | XP_003664847.1 | 32 | CE16 | 6.1 × 10−101 | 37435000 | 1.10 |
| MS/MS View: Identified Proteins | Accession Number | Molecular Weight (kDa) a | CAZy Domain | BLAST E-Value b | iBAQ c | iBAQ/Total iBAQ (%) | |
|---|---|---|---|---|---|---|---|
| UniProt/Swiss-Prot Database | NCBI Database | ||||||
| pectate lyase | G2QH79 | XP_003663984.1 | 34 | PL1 | 8.3 × 10−82 | 676130000 | 19.95 |
| pectate lyase | G2Q1K5 | XP_003660241.1 | 35 | PL1 | 1.3 × 10−94 | 10096000 | 0.30 |
| pectate lyase | G2QMM3 | XP_003666448.1 | 33 | PL1 | 1.9 × 10−45 | 1454500 | 0.04 |
| pectate lyase | G2QG50 | XP_003663760.1 | 40 | PL1 | 6.5 × 10−86 | 346420 | 0.01 |
| pectate lyase | G2QG74 | XP_003664579.1 | 26 | PL3 | 2.3 × 10−82 | 2939800 | 0.09 |
| rhamnogalacturonase B | G2QFG7 | XP_003664441.1 | 58 | PL4 | 4.4 × 10−211 | 6525300 | 0.19 |
| MS/MS View: Identified Proteins | Accession Number | Molecular Weight (kDa) a | CAZy Domain | BLAST E-Value b | iBAQ c | iBAQ/Total iBAQ (%) | |
|---|---|---|---|---|---|---|---|
| UniProt/Swiss-Prot Database | NCBI Database | ||||||
| cellobiohydrolase | G2QA39 | XP_003661032.1 | 51 | GH6—CBM1 | 8.2 × 10−97 | 167100000 | 4.93 |
| cellobiohydrolase | G2QFW6 | XP_003664525.1 | 42 | GH6 | 2.7 × 10−91 | 5419800 | 0.16 |
| cellobiohydrolase | G2Q665 | XP_003660789.1 | 56 | GH7—CBM1 | 5.5 × 10−198 | 1066100000 | 31.43 |
| glycoside hydrolase | G2QNN8 | XP_003666507.1 | 49 | GH7 | 2 × 10−175 | 464770000 | 13.71 |
| endoglucanase | G2QCS4 | XP_003663441.1 | 49 | GH7—CBM1 | 1 × 10−138 | 142270000 | 4.20 |
| endoglucanase | G2QGA1 | XP_003664606.1 | 49 | GH7 | 3.2 × 10−153 | 9135400 | 0.27 |
| glycoside hydrolase | G2Q359 | XP_003658767.1 | 49 | GH7 | 1.1 × 10−190 | 137980 | 0.01 |
| glucan 1,4-α-glucosidase | G2QPS0 | XP_003666828.1 | 67 | GH15—CBM20 | 2.4 × 10−74 | 2191000 | 0.06 |
| glycoside hydrolase | G2QAE3 | XP_003661084.1 | 103 | GH31 | 1.2 × 10−148 | 28072 | 0.01 |
| endoglucanase | G2Q0Y0 | XP_003659323.1 | 24 | GH45 | 1.1 × 10−92 | 6393300 | 0.19 |
| MS/MS View: Identified Proteins | Accession Number | Molecular Weight (kDa) a | CAZy Domain | BLAST E-Value b | iBAQ c | iBAQ/Total iBAQ (%) | |
|---|---|---|---|---|---|---|---|
| UniProt/Swiss-Prot Database | NCBI Database | ||||||
| β-galactosidase | G2QGS8 | XP_003664680.1 | 96 | GH2 | 5.7 × 10−120 | 1017800 | 0.03 |
| β-glucosidase | G2QDN2 | XP_003663588.1 | 78 | GH3 | 7.1 × 10−61 | 1628000 | 0.05 |
| β-glucosidase | G2QCQ3 | XP_003663420.1 | 95 | GH3 | 2.8 × 10−61 | 880340 | 0.03 |
| xylan 1,4-β-xylosidase | G2QKP9 | XP_003665776.1 | 90 | GH3 | 2.4 × 10−59 | 365920 | 0.01 |
| xylanase | G2QJ91 | XP_003665702.1 | 45 | GH10—CBM1 | 1.1 × 10−99 | 71371000 | 2.11 |
| xylanase | G2QGN6 | XP_003663843.1 | 42 | GH10 | 1.3 × 10−91 | 1989200 | 0.06 |
| xylanase | G2QG07 | XP_003664565.1 | 36 | GH10 | 2.7 × 10−106 | 528610 | 0.02 |
| xylanase | G2Q4M3 | XP_003659757.1 | 24 | GH11 | 2 × 10−53 | 87638000 | 2.59 |
| glycoside hydrolase | G2QLD1 | XP_003666008.1 | 31 | GH16 | 2.1 × 10−53 | 8443400 | 0.25 |
| glycoside hydrolase | G2QHP5 | XP_003664150.1 | 41 | GH16 | 2.5 × 10−71 | 2031600 | 0.06 |
| glycoside hydrolase | G2Q2L1 | XP_003658675.1 | 44 | GH16 | 2.9 × 10−71 | 37364 | 0.01 |
| mannan endo-1,4-β-mannosidase A | G2Q4H7 | XP_003658915.1 | 53 | GH26—CBM35 | 2.3 × 10−35 | 8956300 | 0.26 |
| α-galactosidase | G2QNU8 | XP_003667369.1 | 45 | GH27 | 1.1 × 10−56 | 3528300 | 0.10 |
| α-L-arabinofuranosidase 1 | G2QFK1 | XP_003663668.1 | 35 | GH43 | 1.4 × 10−97 | 23454000 | 0.69 |
| arabinanase | G2QCC8 | XP_003662548.1 | 61 | GH43 | 2 × 10−132 | 18453000 | 0.54 |
| arabinan endo-1,5-α-L-arabinosidase | G2QFK0 | XP_003663667.1 | 35 | GH43 | 1.5 × 10−125 | 4569300 | 0.13 |
| arabinanase | G2QDD9 | XP_003663549.1 | 49 | GH43—CBM35 | 1.9 × 10−84 | 4257400 | 0.13 |
| arabinanase | G2QQ09 | XP_003666917.1 | 55 | GH43 | 1.9 × 10−115 | 2470100 | 0.07 |
| arabinanase | G2QHQ9 | XP_003664164.1 | 39 | GH43 | 5.4 × 10−100 | 1494900 | 0.04 |
| α-1,2-mannosidase | G2QHL4 | XP_003664119.1 | 58 | GH47 | 1.5 × 10−136 | 446310 | 0.01 |
| exo-β-1,3-glucanase | G2QCT8 | XP_003663454.1 | 84 | GH55 | 0 | 2534300 | 0.07 |
| exo-β-1,3-glucanase | G2PZK7 | XP_003660938.1 | 95 | GH55 | 1.6 × 10−259 | 1030200 | 0.03 |
| exo-β-1,3-glucanase | G2QF48 | XP_003664322.1 | 82 | GH55 | 6.9 × 10−292 | 241200 | 0.01 |
| exo-β-1,3-glucanase | G2QIM4 | XP_003665591.1 | 82 | GH55 | 4.7 × 10−217 | 27566 | 0.01 |
| α-L-arabinofuranosidase | G2QJQ6 | XP_003665058.1 | 35 | GH62 | 2.1 × 10−131 | 6301300 | 0.19 |
| α-L-arabinofuranosidase | G2QLV4 | XP_003666179.1 | 40 | GH62—CBM1 | 1.7 × 10−125 | 295260 | 0.01 |
| 1,3-β-glucanosyltransferase | G2QN92 | XP_003667210.1 | 52 | GH72 | 9.3 × 10−122 | 745680 | 0.02 |
| 1,3-β-glucanosyltransferase | G2QAD1 | XP_003661926.1 | 57 | GH72 | 1.8 × 10−128 | 603890 | 0.02 |
| xyloglucanase | G2QHR7 | XP_003664172.1 | 79 | GH74 | 1.2 × 10−22 | 183060000 | 5.39 |
| α-mannanase | G2QL30 | XP_003665907.1 | 46 | GH76 | 1.6 × 10−96 | 39672 | 0.01 |
| glycoside hydrolase | G2QJT2 | XP_003665083.1 | 51 | GH79 | 7.1 × 10−64 | 1411000 | 0.04 |
| α-mannosidase | G2Q1N1 | XP_003660267.1 | 90 | GH92 | 3.4 × 10−160 | 135570 | 0.01 |
| exo-α-L-1,5-arabinanase | G2Q5Q6 | XP_003660737.1 | 42 | GH93 | 2.8 × 10−108 | 78089000 | 2.30 |
| glycoside hydrolase | G2QGJ6 | XP_003664651.1 | 55 | GH125 | 3.4 × 10−145 | 106580 | 0.01 |
| glycoside hydrolase | G2QNK3 | XP_003667321.1 | 37 | GH131—CBM1 | 1.2 × 10−111 | 25649000 | 0.76 |
| uncharacterized protein MYCTH_2295704 | G2Q5V8 | XP_003659079.1 | 32 | GH131 | 3.6 × 10−76 | 3439200 | 0.10 |
| uncharacterized protein MYCTH_2301831 | G2QAC0 | XP_003661915.1 | 33 | GH135 | 1.4 × 10−43 | 3426300 | 0.10 |
| MS/MS View: Identified Proteins | Accession Number | Molecular Weight (kDa) a | CAZy Domain | BLAST E-Value b | iBAQ c | iBAQ/Total iBAQ (%) | |
|---|---|---|---|---|---|---|---|
| UniProt/Swiss-Prot Database | NCBI Database | ||||||
| trehalase | G2PZS2 | XP_003658392.1 | 78 | GH37 | 1.4 × 10−159 | 154860 | 0.01 |
| α-L-fucosidase | G2QDI5 | XP_003662742.1 | 91 | GH95 | 7.4 × 10−209 | 1873100 | 0.04 |
| MS/MS View: Identified Proteins | Accession Number | CAZy Domain | BLAST E-Value a | iBAQ b | iBAQ/Total iBAQ (%) | |
|---|---|---|---|---|---|---|
| UniProt/Swiss-Prot Database | NCBI Database | |||||
| cellobiose dehydrogenase | G2QGP4 | XP_003663851.1 | AA3—CBM1 | 2.2 × 10−76 | 231140 | 0.73 |
| cellobiose dehydrogenase | A9XK88 | XP_003663382.1 | AA8—CBM1 | 2.5 × 10−71 | 846980 | 0.02 |
| lytic polysaccharide monooxygenase G | G2Q4M0 | XP_003659754.1 | AA9—CBM1 | 1.7 × 10−67 | 65875000 | 1.93 |
| lytic polysaccharide monooxygenase B | G2QCJ3 | XP_003663414.1 | AA9—CBM1 | 1.4 × 10−74 | 46761000 | 1.36 |
| lytic polysaccharide monooxygenase H | G2Q9T3 | XP_003661787.1 | AA9—CBM1 | 4.3 × 10−71 | 25188000 | 0.74 |
| lytic polysaccharide monooxygenase I | G2Q9F7 | XP_003661661.1 | AA9—CBM1 | 2 × 10−71 | 445310 | 0.01 |
| acetylxylan esterase | G2QJ94 | XP_003665705.1 | CE5—CBM1 | 1.3 × 10−41 | 2033500 | 0.06 |
| cellobiohydrolase | G2QA39 | XP_003661032.1 | GH6—CBM1 | 8.2 × 10−97 | 167100000 | 4.93 |
| cellobiohydrolase | G2Q665 | XP_003660789.1 | GH7—CBM1 | 5.5 × 10−198 | 1066100000 | 31.43 |
| endoglucanase | G2QCS4 | XP_003663441.1 | GH7—CBM1 | 1 × 10−138 | 142270000 | 4.20 |
| glucan 1,4-α-glucosidase | G2QPS0 | XP_003666828.1 | GH15—CBM20 | 2.4 × 10−74 | 2191000 | 0.06 |
| xylanase | G2QJ91 | XP_003665702.1 | GH10—CBM1 | 1.1 × 10−99 | 71371000 | 2.11 |
| mannan endo-1,4-β-mannosidase A | G2Q4H7 | XP_003658915.1 | GH26—CBM35 | 2.3 × 10−35 | 8956300 | 0.26 |
| arabinanase | G2QDD9 | XP_003663549.1 | GH43—CBM35 | 1.9 × 10−84 | 4257400 | 0.13 |
| α-L-arabinofuranosidase | G2QLV4 | XP_003666179.1 | GH62—CBM1 | 1.7 × 10−125 | 295260 | 0.01 |
| glycoside hydrolase | G2QNK3 | XP_003667321.1 | GH131—CBM1 | 1.2 × 10−111 | 25649000 | 0.76 |
| MS/MS View: Identified Proteins | Accession Number | Molecular Weight (kDa) a | CAZy Domain | iBAQ/Total iBAQ (%) b | Degraded Biomass | |
|---|---|---|---|---|---|---|
| UniProt/Swiss-Prot Database | NCBI Database | |||||
| cellobiohydrolase | G2Q665 | XP_003660789.1 | 56 | GH7—CBM1 | 31.43 | cellulose |
| polysaccharide lyase | G2QH79 | XP_003663984.1 | 34 | PL1 | 19.95 | pectin |
| glycoside hydrolase | G2QNN8 | XP_003666507.1 | 49 | GH7 | 13.71 | cellulose |
| xyloglucanase | G2QHR7 | XP_003664172.1 | 79 | GH74 | 5.39 | hemicellulose |
| cellobiohydrolase | G2QA39 | XP_003661032.1 | 51 | GH6—CBM1 | 4.93 | cellulose |
| endoglucanase | G2QCS4 | XP_003663441.1 | 49 | GH7—CBM1 | 4.20 | cellulose |
| xylanase | G2Q4M3 | XP_003659757.1 | 24 | GH11 | 2.59 | hemicellulose |
| exo-α-L-1,5-arabinanase | G2Q5Q6 | XP_003660737.1 | 42 | GH93 | 2.30 | hemicellulose |
| xylanase | G2QJ91 | XP_003663843.1 | 45 | GH10—CBM1 | 2.11 | hemicellulose |
| lytic polysaccharide monooxygenase G | G2Q4M0 | XP_003659754.1 | 32 | AA9—CBM1 | 1.93 | cellulose |
| lytic polysaccharide monooxygenase B | G2QCJ3 | XP_003663414.1 | 32 | AA9—CBM1 | 1.36 | cellulose |
| pectinesterase | G2QLD0 | XP_003666007.1 | 36 | CE8 | 1.19 | pectin |
| acetylesterase | G2QJ27 | XP_003664847.1 | 32 | CE16 | 1.10 | pectin |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Contato, A.G.; Borelli, T.C.; Buckeridge, M.S.; Rogers, J.; Hartson, S.; Prade, R.A.; Polizeli, M.d.L.T.d.M. Secretome Analysis of Thermothelomyces thermophilus LMBC 162 Cultivated with Tamarindus indica Seeds Reveals CAZymes for Degradation of Lignocellulosic Biomass. J. Fungi 2024, 10, 121. https://doi.org/10.3390/jof10020121
Contato AG, Borelli TC, Buckeridge MS, Rogers J, Hartson S, Prade RA, Polizeli MdLTdM. Secretome Analysis of Thermothelomyces thermophilus LMBC 162 Cultivated with Tamarindus indica Seeds Reveals CAZymes for Degradation of Lignocellulosic Biomass. Journal of Fungi. 2024; 10(2):121. https://doi.org/10.3390/jof10020121
Chicago/Turabian StyleContato, Alex Graça, Tiago Cabral Borelli, Marcos Silveira Buckeridge, Janet Rogers, Steven Hartson, Rolf Alexander Prade, and Maria de Lourdes Teixeira de Moraes Polizeli. 2024. "Secretome Analysis of Thermothelomyces thermophilus LMBC 162 Cultivated with Tamarindus indica Seeds Reveals CAZymes for Degradation of Lignocellulosic Biomass" Journal of Fungi 10, no. 2: 121. https://doi.org/10.3390/jof10020121
APA StyleContato, A. G., Borelli, T. C., Buckeridge, M. S., Rogers, J., Hartson, S., Prade, R. A., & Polizeli, M. d. L. T. d. M. (2024). Secretome Analysis of Thermothelomyces thermophilus LMBC 162 Cultivated with Tamarindus indica Seeds Reveals CAZymes for Degradation of Lignocellulosic Biomass. Journal of Fungi, 10(2), 121. https://doi.org/10.3390/jof10020121






