Ergosterol Is Critical for Sporogenesis in Cryptococcus neoformans
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains and Growth Conditions
2.2. Mating
2.3. Growth Assays
2.4. Gene Manipulation
2.5. RNA Extraction and Real-Time PCR
2.6. Microscopy
2.7. Filipin Staining
3. Results
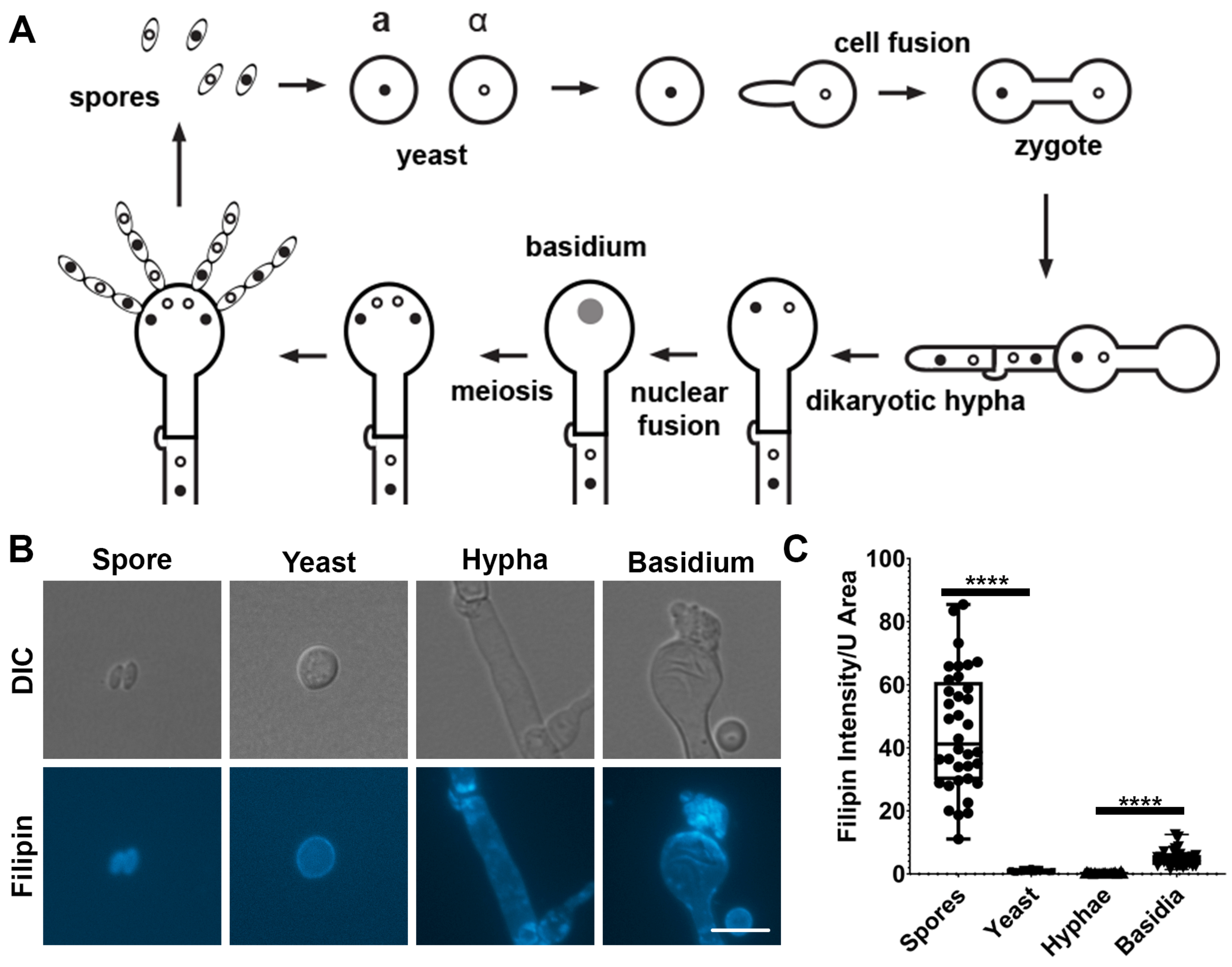
3.1. Ergosterol Is Enriched in Basidia and Spores Based on Filipin Staining
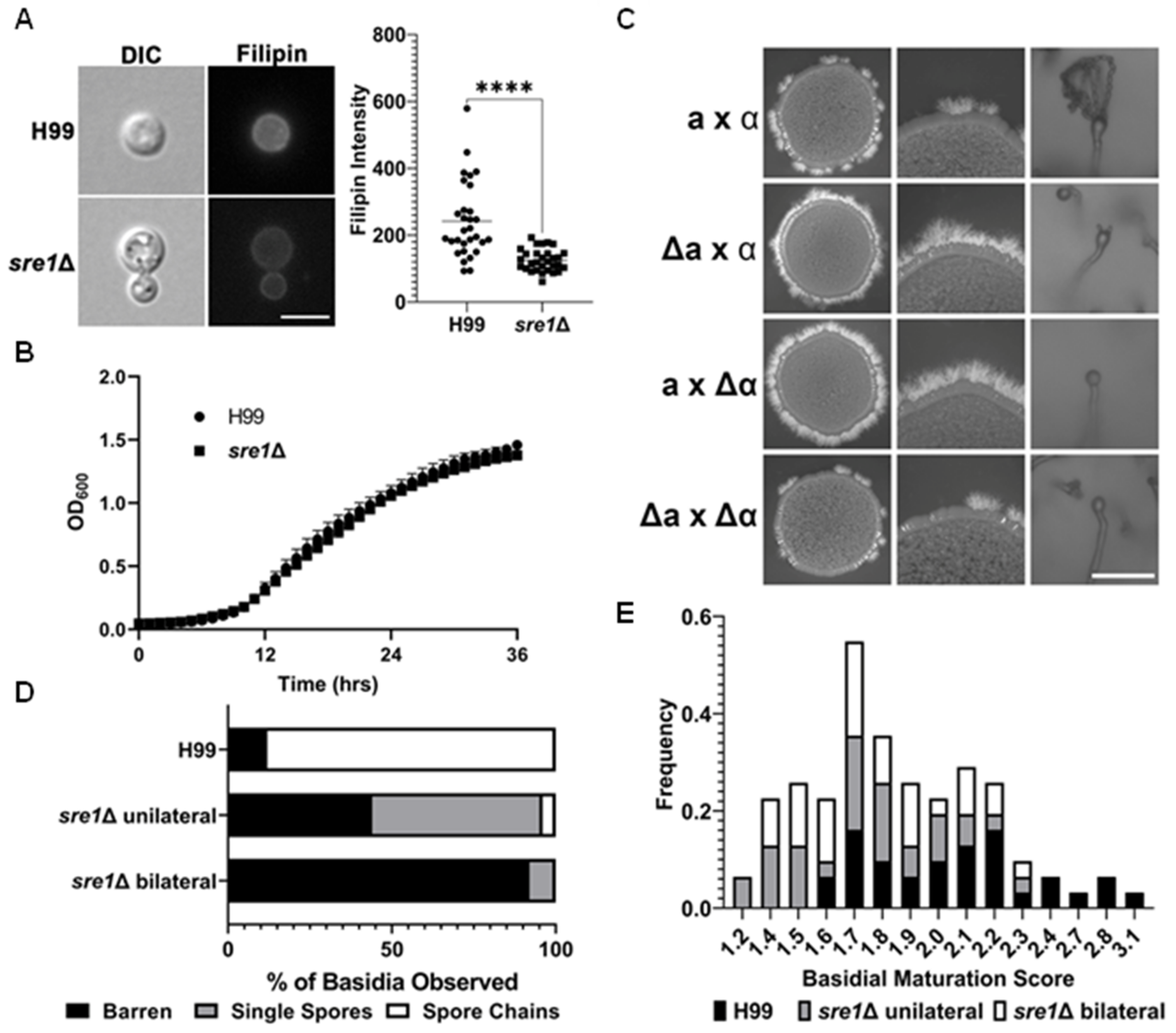
3.2. Crosses Involving sre1Δ Produce Abundant Hyphae but Are Defective at Sporulation
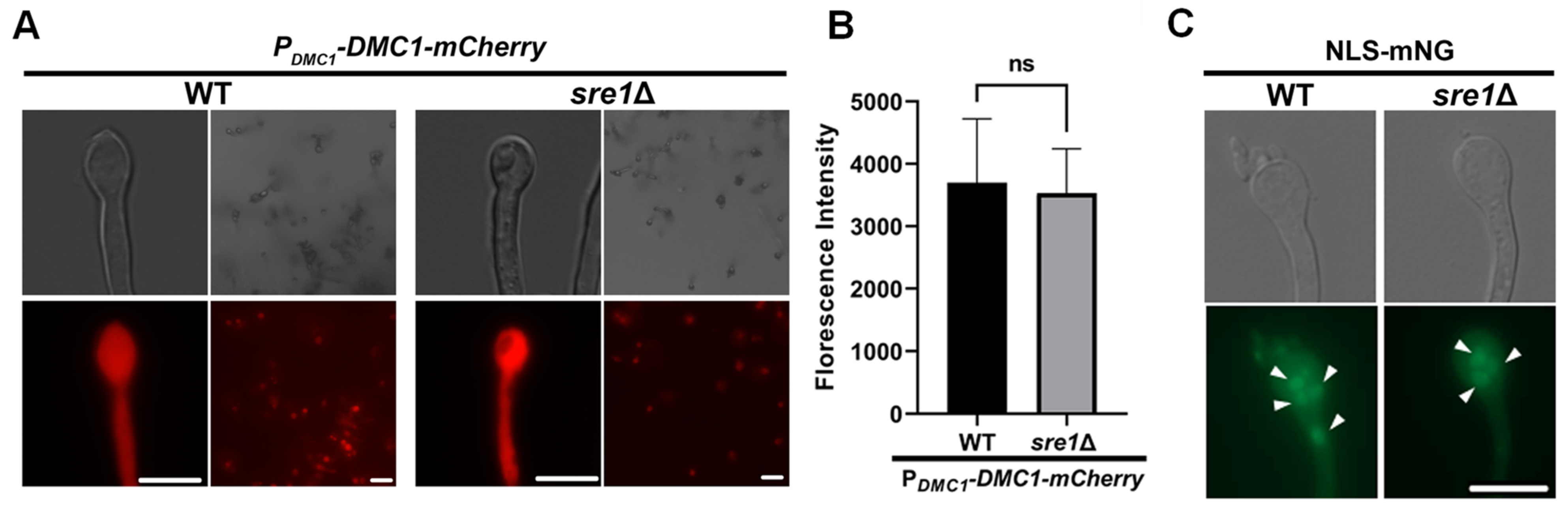
3.3. sre1Δ Sporulation Defect Is Not Due to a Defect in Meiosis Gene Activation
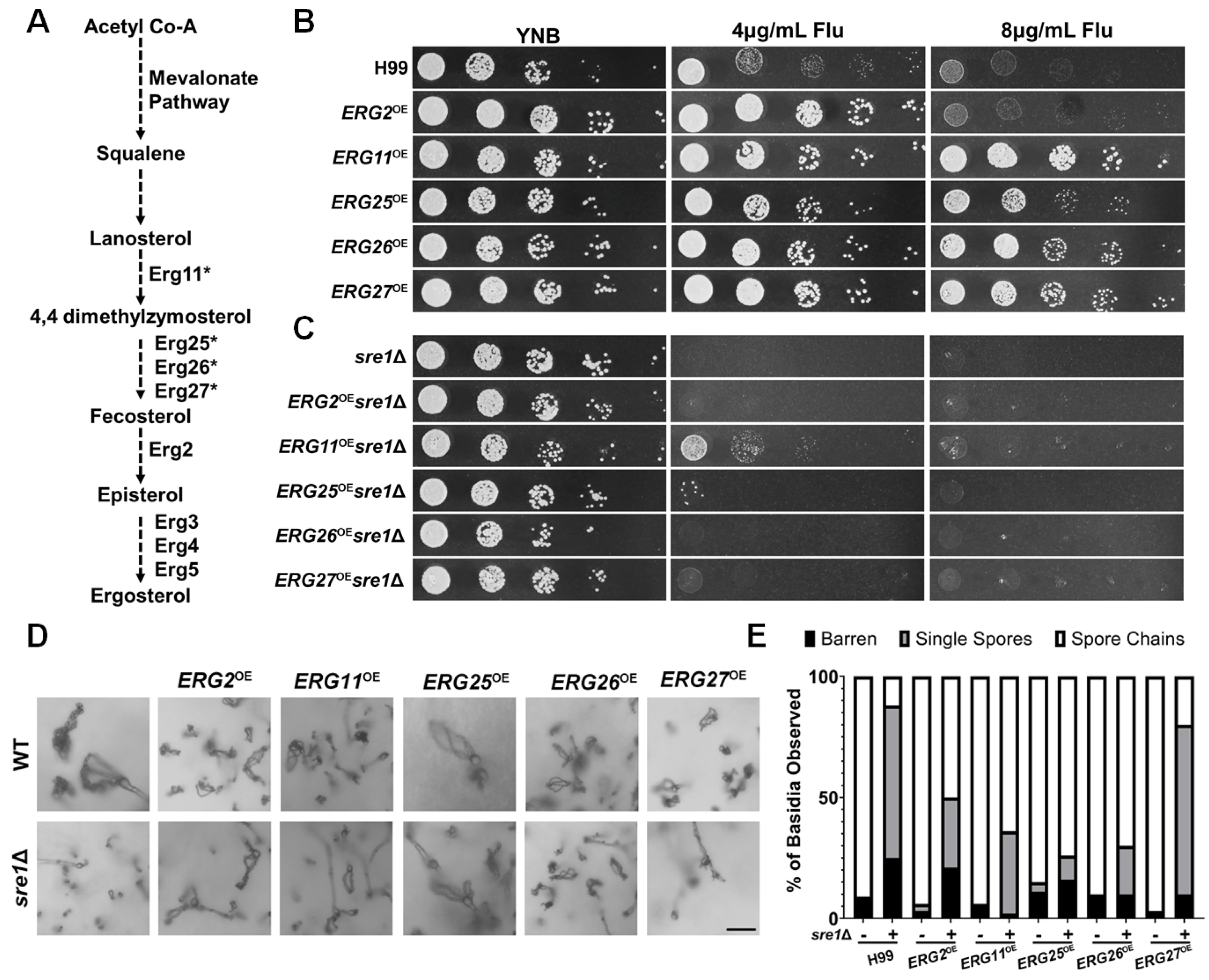
3.4. Defective EBP Pathway Leads to Sporulation Defect
3.5. Overexpression of Multiple Individual EBP Genes Partially Restore sre1Δ’s Sporulation Defect
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Errington, J. Regulation of endospore formation in Bacillus subtilis. Nat. Rev. Microbiol. 2003, 1, 117–126. [Google Scholar] [CrossRef] [PubMed]
- Setlow, P. Spores of Bacillus subtilis: Their resistance to and killing by radiation, heat and chemicals. J. Appl. Microbiol. 2006, 101, 514–525. [Google Scholar] [CrossRef]
- Busse, O. Uber parasitare Zelleinschlusse und ihre Zuchtung. Zentralbl Bakteriol. 1894, 16, 175–180. [Google Scholar]
- Zhao, Y.; Lin, J.; Fan, Y.; Lin, X. Life Cycle of Cryptococcus neoformans. Annu. Rev. Microbiol. 2019, 73, 17–42. [Google Scholar] [CrossRef]
- Kwon-Chung, K. A new genus, Filobasidiella, the perfect state of Cryptococcus neoformans. Mycologia 1975, 67, 1197–1200. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Idnurm, A.; Lin, X. Morphology and its underlying genetic regulation impact the interaction between Cryptococcus neoformans and its hosts. Med. Mycol. 2015, 53, 493–504. [Google Scholar] [CrossRef]
- Fu, M.S.; Liporagi-Lopes, L.C.; dos Santos, S.R.; Tenor, J.L.; Perfect, J.R.; Cuomo, C.A.; Casadevall, A. Amoeba predation of Cryptococcus neoformans results in pleiotropic changes to traits associated with virulence. MBio 2021, 12, 10–1128. [Google Scholar] [CrossRef]
- Velagapudi, R.; Hsueh, Y.-P.; Geunes-Boyer, S.; Wright, J.R.; Heitman, J. Spores as infectious propagules of Cryptococcus neoformans. Infect. Immun. 2009, 77, 4345–4355. [Google Scholar] [CrossRef]
- Giles, S.S.; Dagenais, T.R.; Botts, M.R.; Keller, N.P.; Hull, C.M. Elucidating the pathogenesis of spores from the human fungal pathogen Cryptococcus neoformans. Infect. Immun. 2009, 77, 3491–3500. [Google Scholar] [CrossRef]
- Zhao, Y.; Lin, X. Cryptococcus neoformans: Sex, morphogenesis, and virulence. Infect. Genet. Evol. 2021, 89, 104731. [Google Scholar] [CrossRef]
- Kwon-Chung, K. Morphogenesis of Filobasidiella neoformans, the sexual state of Cryptococcus neoformans. Mycologia 1976, 68, 821–833. [Google Scholar] [CrossRef] [PubMed]
- Hull, C.M.; Davidson, R.C.; Heitman, J. Cell identity and sexual development in Cryptococcus neoformans are controlled by the mating-type-specific homeodomain protein Sxi1α. Genes. Dev. 2002, 16, 3046–3060. [Google Scholar] [CrossRef] [PubMed]
- McClelland, C.M.; Chang, Y.C.; Varma, A.; Kwon-Chung, K. Uniqueness of the mating system in Cryptococcus neoformans. Trends Microbiol. 2004, 12, 208–212. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Hull, C.M.; Heitman, J. Sexual reproduction between partners of the same mating type in Cryptococcus neoformans. Nature 2005, 434, 1017–1021. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Jackson, J.C.; Feretzaki, M.; Xue, C.; Heitman, J. Transcription factors Mat2 and Znf2 operate cellular circuits orchestrating opposite-and same-sex mating in Cryptococcus neoformans. PLoS Genet. 2010, 6, e1000953. [Google Scholar] [CrossRef]
- Sun, S.; Priest, S.J.; Heitman, J. Cryptococcus neoformans mating and genetic crosses. Curr. Protoc. Microbiol. 2019, 53, e75. [Google Scholar] [CrossRef]
- Matha, A.R.; Lin, X. Current perspectives on uniparental mitochondrial inheritance in Cryptococcus neoformans. Pathogens 2020, 9, 743. [Google Scholar] [CrossRef]
- Huang, M.; Hebert, A.S.; Coon, J.J.; Hull, C.M. Protein composition of infectious spores reveals novel sexual development and germination factors in Cryptococcus. PLoS Genet. 2015, 11, e1005490. [Google Scholar] [CrossRef]
- Wyatt, T.T.; Wösten, H.A.; Dijksterhuis, J. Fungal spores for dispersion in space and time. Adv. Appl. Microbiol. 2013, 85, 43–91. [Google Scholar]
- Mukhopadhyay, K.; Prasad, T.; Saini, P.; Pucadyil, T.J.; Chattopadhyay, A.; Prasad, R. Membrane sphingolipid-ergosterol interactions are important determinants of multidrug resistance in Candida albicans. Antimicrob. Agents Chemother. 2004, 48, 1778–1787. [Google Scholar] [CrossRef]
- Abe, F.; Hiraki, T. Mechanistic role of ergosterol in membrane rigidity and cycloheximide resistance in Saccharomyces cerevisiae. Biochim. Biophys. Acta (BBA)-Biomembr. 2009, 1788, 743–752. [Google Scholar] [CrossRef] [PubMed]
- Ortiz, S.C.; Huang, M.; Hull, C.M. Spore germination as a target for antifungal therapeutics. Antimicrob. Agents Chemother. 2019, 63, 10–1128. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.C.; Bien, C.M.; Lee, H.; Espenshade, P.J.; Kwon-Chung, K.J. Sre1p, a regulator of oxygen sensing and sterol homeostasis, is required for virulence in Cryptococcus neoformans. Mol. Microbiol. 2007, 64, 614–629. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Bien, C.M.; Hughes, A.L.; Espenshade, P.J.; Kwon-Chung, K.J.; Chang, Y.C. Cobalt chloride, a hypoxia-mimicking agent, targets sterol synthesis in the pathogenic fungus Cryptococcus neoformans. Mol. Microbiol. 2007, 65, 1018–1033. [Google Scholar] [CrossRef] [PubMed]
- Byskov, A.G.; Andersen, C.Y.; Nordholm, L.; Thogersen, H.; Guoliang, X.; Wassmann, O.; Andersen, J.V.; Guddal, E.; Roed, T. Chemical structure of sterols that activate oocyte meiosis. Nature 1995, 374, 559–562. [Google Scholar] [CrossRef] [PubMed]
- Byskov, A.G.; Andersen, C.Y.; Leonardsen, L. Role of meiosis activating sterols, MAS, in induced oocyte maturation. Mol. Cell. Endocrinol. 2002, 187, 189–196. [Google Scholar] [CrossRef] [PubMed]
- Grøndahl, C.; Breinholt, J.; Wahl, P.; Murray, A.; Hansen, T.H.; Faerge, I.; Stidsen, C.E.; Raun, K.; Hegele-Hartung, C. Physiology of meiosis-activating sterol: Endogenous formation and mode of action. Hum. Reprod. 2003, 18, 122–129. [Google Scholar] [CrossRef][Green Version]
- Tian, X.; He, G.-J.; Hu, P.; Chen, L.; Tao, C.; Cui, Y.-L.; Shen, L.; Ke, W.; Xu, H.; Zhao, Y.; et al. Cryptococcus neoformans sexual reproduction is controlled by a quorum sensing peptide. Nat. Microbiol. 2018, 3, 698–707. [Google Scholar] [CrossRef]
- Liu, L.; He, G.-J.; Chen, L.; Zheng, J.; Chen, Y.; Shen, L.; Tian, X.; Li, E.; Yang, E.; Liao, G.; et al. Genetic basis for coordination of meiosis and sexual structure maturation in Cryptococcus neoformans. eLife 2018, 7, e38683. [Google Scholar] [CrossRef]
- Jung, K.-W.; Yang, D.-H.; Maeng, S.; Lee, K.-T.; So, Y.-S.; Hong, J.; Choi, J.; Byun, H.-J.; Kim, H.; Bang, S.; et al. Systematic functional profiling of transcription factor networks in Cryptococcus neoformans. Nat. Commun. 2015, 6, 6757. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Lin, X. Multiple applications of a transient CRISPR-Cas9 coupled with electroporation (TRACE) system in the Cryptococcus neoformans species complex. Genetics 2018, 208, 1357–1372. [Google Scholar] [CrossRef]
- Lin, J.; Fan, Y.; Lin, X. Transformation of Cryptococcus neoformans by electroporation using a transient CRISPR-Cas9 expression (TRACE) system. Fungal Genet. Biol. 2020, 138, 103364. [Google Scholar] [CrossRef]
- Upadhya, R.; Lam, W.C.; Maybruck, B.T.; Donlin, M.J.; Chang, A.L.; Kayode, S.; Ormerod, K.L.; Fraser, J.A.; Doering, T.L.; Lodge, J.K. A fluorogenic C. neoformans reporter strain with a robust expression of m-cherry expressed from a safe haven site in the genome. Fungal Genet. Biol. 2017, 108, 13–25. [Google Scholar] [CrossRef] [PubMed]
- Baek, S.; Utomo, J.C.; Lee, J.Y.; Dalal, K.; Yoon, Y.J.; Ro, D.K. The yeast platform engineered for synthetic gRNA-landing pads enables multiple gene integrations by a single gRNA/Cas9 system. Metab. Eng. 2021, 64, 111–121. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhai, B.; Lin, X. The link between morphotype transition and virulence in Cryptococcus neoformans. PLoS Pathog. 2012, 8, e1002765. [Google Scholar] [CrossRef] [PubMed]
- Sephton-Clark, P.C.; Voelz, K. Spore germination of pathogenic filamentous fungi. In Advances in Applied Microbiology; Elsevier: Amsterdam, The Netherlands, 2018; pp. 117–157. [Google Scholar]
- Van Leeuwen, M.; Smant, W.; De Boer, W.; Dijksterhuis, J. Filipin is a reliable in situ marker of ergosterol in the plasma membrane of germinating conidia (spores) of Penicillium discolor and stains intensively at the site of germ tube formation. J. Microbiol. Methods 2008, 74, 64–73. [Google Scholar] [CrossRef] [PubMed]
- Eberlé, D.; Hegarty, B.; Bossard, P.; Ferré, P.; Foufelle, F. SREBP transcription factors: Master regulators of lipid homeostasis. Biochimie 2004, 86, 839–848. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Tian, X.; Gyawali, R.; Upadhyay, S.; Foyle, D.; Wang, G.; Cai, J.J.; Lin, X. Morphotype transition and sexual reproduction are genetically associated in a ubiquitous environmental pathogen. PLoS Pathog. 2014, 10, e1004185. [Google Scholar] [CrossRef]
- Shi, R.; Lin, X. lluminating Cryptococcus neoformans: Unveiling Intracellular Structures with Fluorescent-Protein-Based Markers. Genetics 2024, submitted.
- Bhattacharya, S.; Esquivel, B.D.; White, T.C. Overexpression or deletion of ergosterol biosynthesis genes alters doubling time, response to stress agents, and drug susceptibility in Saccharomyces cerevisiae. MBio 2018, 9, e01291-18. [Google Scholar] [CrossRef] [PubMed]
- Mo, C.; Valachovic, M.; Randall, S.; Nickels, J.; Bard, M. Protein–protein interactions among C-4 demethylation enzymes involved in yeast sterol biosynthesis. Proc. Natl. Acad. Sci. USA 2002, 99, 9739–9744. [Google Scholar] [CrossRef]
- Jordá, T.; Puig, S. Regulation of ergosterol biosynthesis in Saccharomyces cerevisiae. Genes 2020, 11, 795. [Google Scholar] [CrossRef]
- Shim, Y.H.; Chun, J.H.; Lee, E.Y.; Paik, Y.K. Role of cholesterol in germ-line development of Caenorhabditis elegans. Mol. Reprod. Dev. 2002, 61, 358–366. [Google Scholar] [CrossRef]
- Long, N.; Xu, X.; Zeng, Q.; Sang, H.; Lu, L. Erg4A and Erg4B are required for conidiation and azole resistance via regulation of ergosterol biosynthesis in Aspergillus fumigatus. Appl. Environ. Microbiol. 2017, 83, e02924-16. [Google Scholar] [CrossRef] [PubMed]
- Long, N.; Zhong, G. The C-22 sterol desaturase Erg5 is responsible for ergosterol biosynthesis and conidiation in Aspergillus fumigatus. J. Microbiol. 2022, 60, 620–626. [Google Scholar] [CrossRef] [PubMed]
- Alcazar-Fuoli, L.; Mellado, E.; Garcia-Effron, G.; Buitrago, M.J.; Lopez, J.F.; Grimalt, J.O.; Cuenca-Estrella, J.M.; Rodriguez-Tudela, J.L. Aspergillus fumigatus C-5 sterol desaturases Erg3A and Erg3B: Role in sterol biosynthesis and antifungal drug susceptibility. Antimicrob. Agents Chemother. 2006, 50, 453–460. [Google Scholar] [CrossRef]
- Han, S.; Sheng, B.; Zhu, D.; Chen, J.; Cai, H.; Zhang, S.; Guo, C. Role of FoERG3 in Ergosterol Biosynthesis by Fusarium oxysporum and the Associated Regulation by Bacillus subtilis HSY21. Plant Dis. 2023, 107, 1565–1575. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Niu, Y.; Huang, H.; He, B.; Ma, L.; Tu, Y.; Tran, V.-T.; Zeng, B.; Hu, Z. Mevalonate diphosphate decarboxylase MVD/Erg19 is required for ergosterol biosynthesis, growth, sporulation and stress tolerance in Aspergillus oryzae. Front. Microbiol. 2019, 10, 1074. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Niu, Y.; Jin, Q.; Qin, K.; Wang, L.; Shang, Y.; Zeng, B.; Hu, Z. Identification of six thiolases and their effects on fatty acid and ergosterol biosynthesis in Aspergillus oryzae. Appl. Environ. Microbiol. 2022, 88, e02372-21. [Google Scholar] [CrossRef] [PubMed]
- Jin, Q.; Li, G.; Qin, K.; Shang, Y.; Yan, H.; Liu, H.; Zeng, B.; Hu, Z. The expression pattern, subcellular localization and function of three sterol 14α-demethylases in Aspergillus oryzae. Front. Genet. 2023, 14, 1009746. [Google Scholar] [CrossRef]
- Willger, S.D.; Puttikamonkul, S.; Kim, K.-H.; Burritt, J.B.; Grahl, N.; Metzler, L.J.; Barbuch, R.; Bard, M.; Lawrence, C.B.; Cramer, R.A. A sterol-regulatory element binding protein is required for cell polarity, hypoxia adaptation, azole drug resistance, and virulence in Aspergillus fumigatus. PLoS Pathog. 2008, 4, e1000200. [Google Scholar] [CrossRef]
- Chung, D.; Barker, B.M.; Carey, C.C.; Merriman, B.; Werner, E.R.; Lechner, B.E.; Dhingra, S.; Cheng, C.; Xu, W.; Blosser, S.J.; et al. ChIP-seq and in vivo transcriptome analyses of the Aspergillus fumigatus SREBP SrbA reveals a new regulator of the fungal hypoxia response and virulence. PLoS Pathog. 2014, 10, e1004487. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Matha, A.R.; Xie, X.; Lin, X. Ergosterol Is Critical for Sporogenesis in Cryptococcus neoformans. J. Fungi 2024, 10, 106. https://doi.org/10.3390/jof10020106
Matha AR, Xie X, Lin X. Ergosterol Is Critical for Sporogenesis in Cryptococcus neoformans. Journal of Fungi. 2024; 10(2):106. https://doi.org/10.3390/jof10020106
Chicago/Turabian StyleMatha, Amber R., Xiaofeng Xie, and Xiaorong Lin. 2024. "Ergosterol Is Critical for Sporogenesis in Cryptococcus neoformans" Journal of Fungi 10, no. 2: 106. https://doi.org/10.3390/jof10020106
APA StyleMatha, A. R., Xie, X., & Lin, X. (2024). Ergosterol Is Critical for Sporogenesis in Cryptococcus neoformans. Journal of Fungi, 10(2), 106. https://doi.org/10.3390/jof10020106





