Prognostic Value of Machine Learning in Patients with Acute Myocardial Infarction
Abstract
:1. Introduction
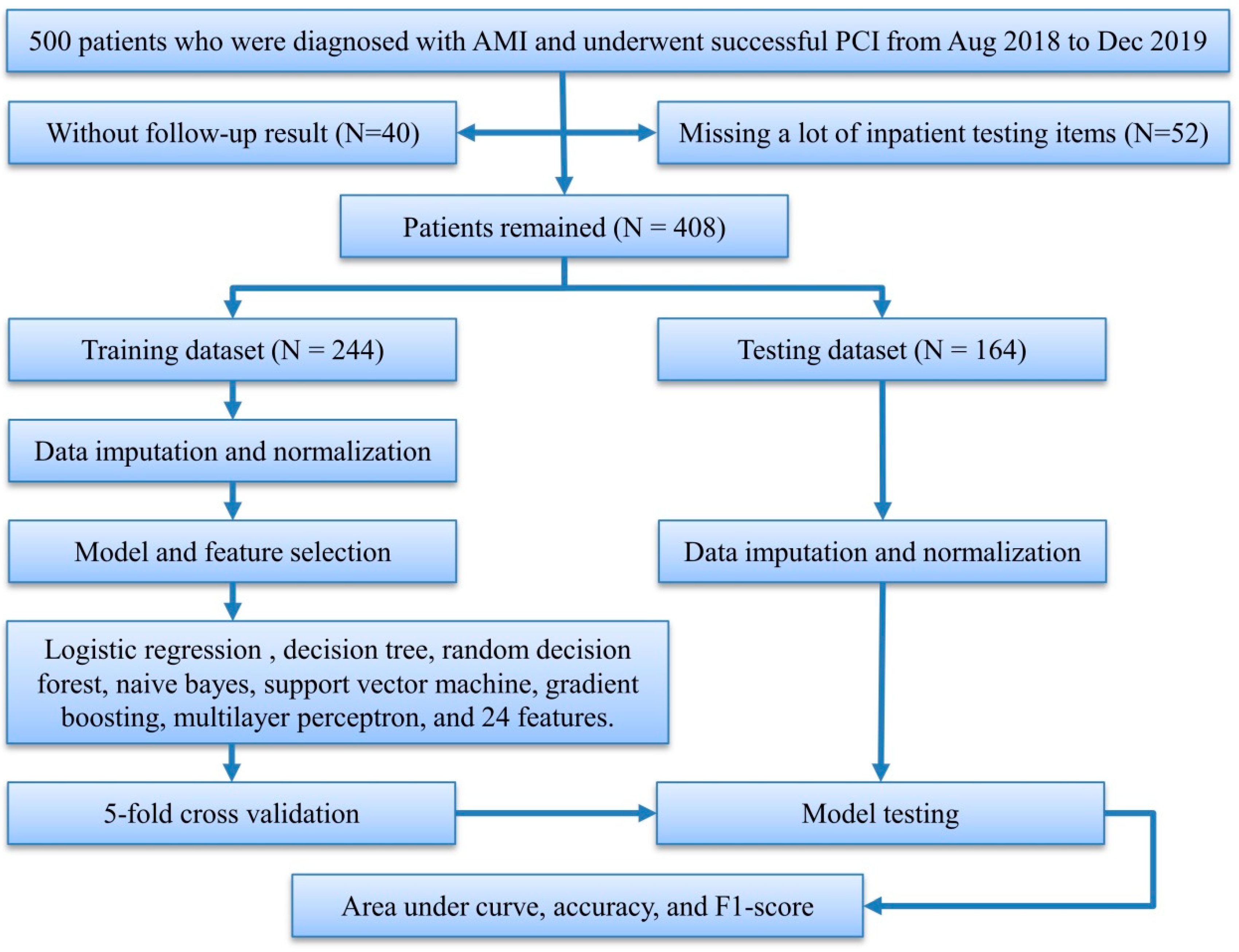
2. Materials and Methods
2.1. Study Population and Statistical Analysis
2.2. Preprocessing and Feature Selection
2.3. LR Analysis
2.4. Model Development
2.5. Model Testing
3. Results
3.1. Clinical Characteristics of the Selected Patients
3.2. The Selected and Independent Predictors
3.3. Comparative Performance between the ML and LR Models
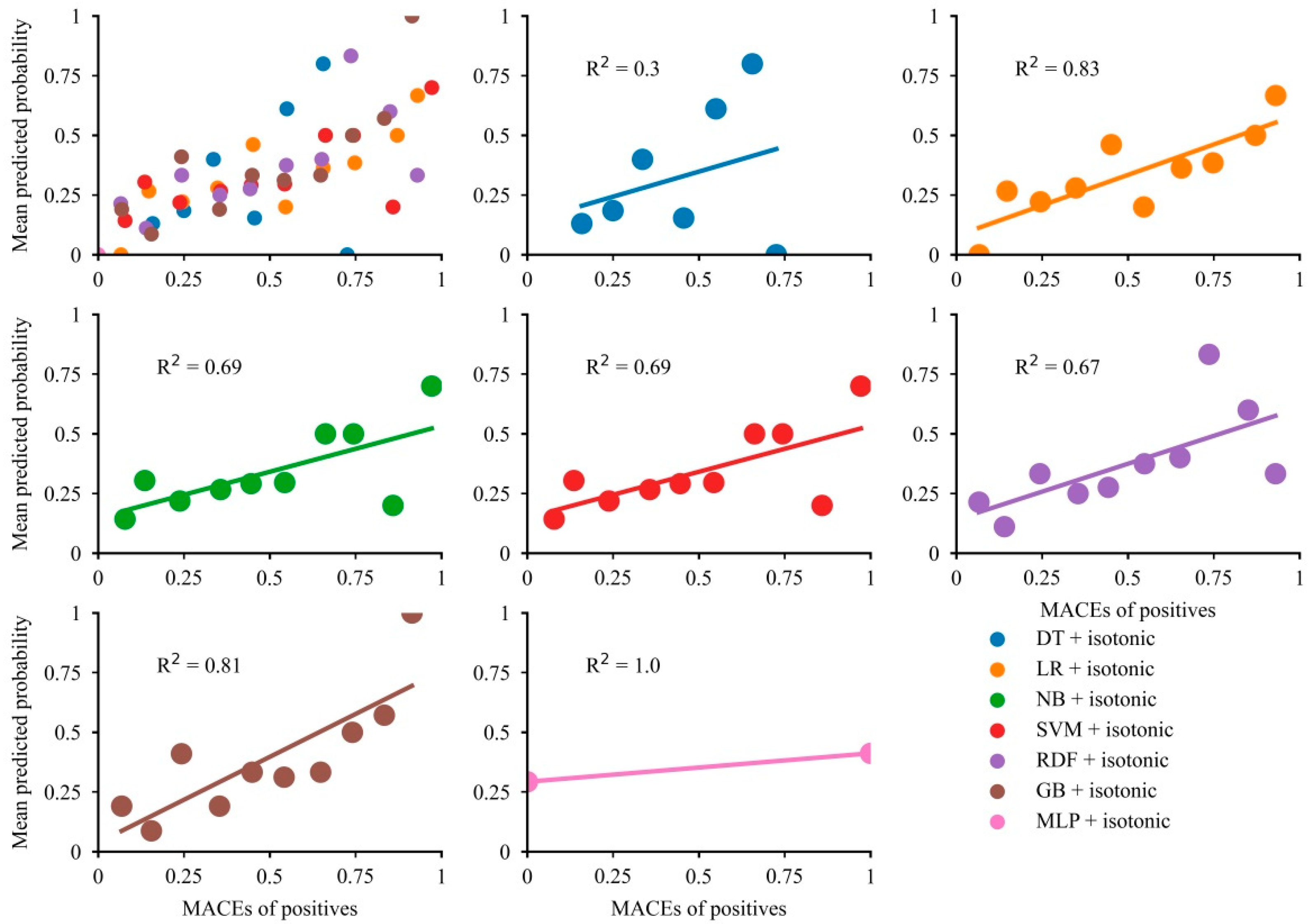
3.4. Calibration Plots for All Models
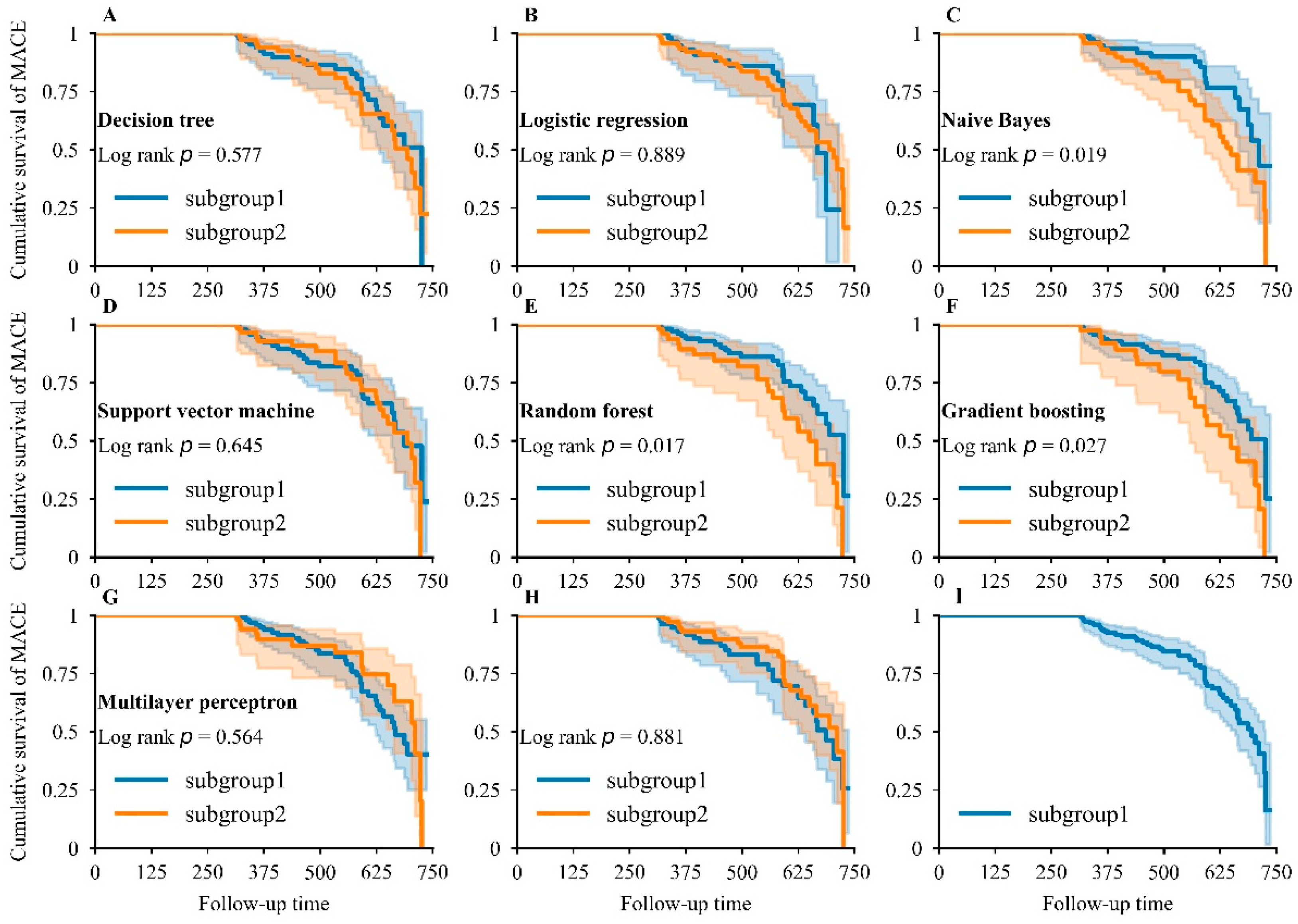
3.5. Kaplan–Meier Analysis for MACEs
4. Discussion
5. Limitations
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Virani, S.S.; Alonso, A.; Benjamin, E.J.; Bittencourt, M.S.; Callaway, C.W.; Carson, A.P.; Chamberlain, A.M.; Chang, A.R.; Cheng, S.; Delling, F.N.; et al. Heart disease and stroke statistics-2020 update: A report from the American Heart Association. Circulation 2020, 141, e139–e596. [Google Scholar] [CrossRef]
- Sanchez-Perez, I.; Abellan-Huerta, J.; Jurado-Roman, A.; Lopez-Lluva, M.T.; Pinilla-Echeverri, N.; Perez-Diaz, P.; Piqueras-Flores, J.; Lozano-Ruiz-Poveda, F. Long-term follow-up of percutaneous coronary intervention with paclitaxel-eluting balloon catheter. Angiology 2021, 72, 364–370. [Google Scholar] [CrossRef]
- Kim, J.M.; Stewart, R.; Lee, Y.S.; Lee, H.J.; Kim, M.C.; Kim, J.W.; Kang, H.J.; Bae, K.Y.; Kim, S.W.; Shin, I.S.; et al. Effect of Escitalopram vs. Placebo treatment for depression on long-term cardiac outcomes in patients with acute coronary syndrome A randomized clinical trial. JAMA 2018, 320, 350–357. [Google Scholar] [CrossRef] [Green Version]
- Benz, D.C.; Kaufmann, P.A.; von Felten, E.; Benetos, G.; Rampidis, G.; Messerli, M.; Giannopoulos, A.A.; Fuchs, T.A.; Grani, C.; Gebhard, C.; et al. Prognostic value of quantitative metrics from positron emission tomography in ischemic heart failure. Cardiovasc. Imaging 2021, 14, 454–464. [Google Scholar] [CrossRef]
- Guo, Y.Y.; Zhao, X.; Wang, L.; Zhao, X.Y. Related factors of left ventricular thrombus formation within two weeks in patients with acute ST-segment elevation myocardial infarction and left ventricular aneurysm. Zhonghua Xin Xue Guan Bing Za Zhi 2021, 49, 360–367. [Google Scholar]
- Sato, T.; Saito, Y.; Matsumoto, T.; Yamashita, D.; Saito, K.; Wakabayashi, S.; Kitahara, H.; Sano, K.; Kobayashi, Y. Impact of CADILLAC and GRACE risk scores on short- and long-term clinical outcomes in patients with acute myocardial infarction. J. Cardiol. 2021, 78, 201–205. [Google Scholar] [CrossRef]
- Maznyczka, A.M.; McCartney, P.J.; Oldroyd, K.G.; Lindsay, M.; McEntegart, M.; Eteiba, H.; Rocchiccioli, J.P.; Good, R.; Shaukat, A.; Robertson, K.; et al. Risk stratification guided by the index of microcirculatory resistance and left ventricular end-diastolic pressure in acute myocardial infarction. Cardiovasc. Interv. 2021, 14, e009529. [Google Scholar]
- Mitarai, T.; Tanabe, Y.; Akashi, Y.J.; Maeda, A.; Ako, J.; Ikari, Y.; Ebina, T.; Namiki, A.; Fukui, K.; Michishita, I.; et al. A novel risk stratification system “Angiographic GRACE Score” for predicting in-hospital mortality of patients with acute myocardial infarction: Data from the K-ACTIVE Registry. J. Cardiol. 2021, 77, 179–185. [Google Scholar] [CrossRef]
- Ito, H.; Masuda, J.; Kurita, T.; Ida, M.; Yamamoto, A.; Takasaki, A.; Takeuchi, T.; Sato, Y.; Omura, T.; Sawai, T.; et al. Effect of left ventricular ejection fraction on the prognostic impact of chronic total occlusion in a non-infarct-related artery in patients with acute myocardial infarction. Int. J. Cardiol. Heart Vasc. 2021, 33, 100738. [Google Scholar] [CrossRef]
- Li, X.; Wang, X.; Sun, T.; Ping, Y.; Dai, Y.; Liu, Z.; Wang, Y.; Wang, D.; Xia, X.; Shan, H.; et al. S100A1 is a sensitive and specific cardiac biomarker for early diagnosis and prognostic assessment of acute myocardial infarction measured by chemiluminescent immunoassay. Clin. Chim. Acta 2021, 516, 71–76. [Google Scholar] [CrossRef]
- Chen, Z.; Xiao, C.; Qiu, H.; Tan, X.; Jin, L.; He, Y.; Guo, Y.; He, N. Recent advances of artificial intelligence in cardiovascular disease. J. Biomed. Nanotechnol. 2020, 16, 1065–1081. [Google Scholar] [CrossRef]
- Jordan, M.I.; Mitchell, T.M. Machine learning: Trends, perspectives, and prospects. Science 2015, 349, 255–260. [Google Scholar] [CrossRef]
- Al’Aref, S.J.; Anchouche, K.; Singh, G.; Slomka, P.J.; Kolli, K.K.; Kumar, A.; Pandey, M.; Maliakal, G.; van Rosendael, A.R.; Beecy, A.N.; et al. Clinical applications of machine learning in cardiovascular disease and its relevance to cardiac imaging. Eur. Heart J. 2019, 40, 1975–1986. [Google Scholar] [CrossRef]
- Jamin, A.; Abraham, P.; Humeau-Heurtier, A. Machine learning for predictive data analytics in medicine: A review illustrated by cardiovascular and nuclear medicine examples. Clin. Physiol. Funct. Imaging 2021, 41, 113–127. [Google Scholar] [CrossRef]
- Gambella, C.; Ghaddar, B.; Naoum-Sawaya, J. Optimization problems for machine learning: A survey. Eur. J. Oper. Res. 2021, 290, 807–828. [Google Scholar] [CrossRef]
- Maheswari, S.; Pitchai, R. Heart disease prediction system using decision tree and naive bayes Algorithm. Curr. Med. Imaging 2019, 15, 712–717. [Google Scholar] [CrossRef]
- Jackins, V.; Vimal, S.; Kaliappan, M.; Lee, M.Y. AI-based smart prediction of clinical disease using random forest classifier and Naive Bayes. J. Supercomput. 2021, 77, 5198–5219. [Google Scholar] [CrossRef]
- Huang, S.; Cai, N.; Pacheco, P.P.; Narandes, S.; Wang, Y.; Xu, W. Applications of support vector machine (SVM) learning in cancer genomics. Cancer Genom. Proteom. 2018, 15, 41–51. [Google Scholar]
- Mohan, S.; Thirumalai, C.; Srivastava, G. Effective heart disease prediction using hybrid machine learning techniques. IEEE Access 2019, 7, 81542–81554. [Google Scholar] [CrossRef]
- Gumaei, A.; Al-Rakhami, M.; Al Rahhal, M.M.; Albogamy, F.R.H.; Al Maghayreh, E.; AlSalman, H. Prediction of COVID-19 confirmed cases using gradient boosting regression method. Comput. Mater. Contin. 2021, 66, 315–329. [Google Scholar] [CrossRef]
- Masih, N.; Naz, H.; Ahuja, S. Multilayer perceptron based deep neural network for early detection of coronary heart disease. Health Technol. 2021, 11, 127–138. [Google Scholar] [CrossRef]
- Liu, N.; Chee, M.L.; Koh, Z.X.; Leow, S.L.; Ho, A.F.W.; Guo, D.; Ong, M.E.H. Utilizing machine learning dimensionality reduction for risk stratification of chest pain patients in the emergency department. BMC Med. Res. Methodol. 2021, 21, 74. [Google Scholar] [CrossRef] [PubMed]
- Khera, R.; Haimovich, J.; Hurley, N.C.; McNamara, R.; Spertus, J.A.; Desai, N.; Rumsfeld, J.S.; Masoudi, F.A.; Huang, C.; Normand, S.-L.; et al. Use of machine learning models to predict death after acute myocardial infarction. JAMA Cardiol. 2021, 6, 633–641. [Google Scholar] [CrossRef]
- Lee, H.C.; Park, J.S.; Choe, J.C.; Ahn, J.H.; Lee, H.W.; Oh, J.H.; Choi, J.H.; Cha, K.S.; Hong, T.J.; Jeong, M.H.; et al. Prediction of 1-year mortality from acute myocardial infarction using machine learning. Am. J. Cardiol. 2020, 133, 23–31. [Google Scholar] [CrossRef]
- Tokodi, M.; Schwertner, W.R.; Kovacs, A.; Toser, Z.; Staub, L.; Sarkany, A.; Lakatos, B.K.; Behon, A.; Boros, A.M.; Perge, P.; et al. Machine learning-based mortality prediction of patients undergoing cardiac resynchronization therapy: The SEMMELWEIS-CRT score. Eur. Heart J. 2020, 41, 1747–1756. [Google Scholar] [CrossRef]
- Liu, W.C.; Lin, C.S.; Tsai, C.S.; Tsao, T.P.; Cheng, C.C.; Liou, J.T.; Lin, W.S.; Cheng, S.M.; Lou, Y.S.; Lee, C.C.; et al. A deep learning algorithm for detecting acute myocardial infarction. EuroIntervention 2021, 17, 765–773. [Google Scholar] [CrossRef]
- Kuderer, N.M.; Choueiri, T.K.; Shah, D.P.; Shyr, Y.; Rubinstein, S.M.; Rivera, D.R.; Shete, S.; Hsu, C.Y.; Desai, A.; de Lima Lopes, G., Jr.; et al. Clinical impact of COVID-19 on patients with cancer (CCC19): A cohort study. Lancet 2020, 395, 1907–1918. [Google Scholar] [CrossRef]
- Topal, D.G.; Aleksov Ahtarovski, K.; Lonborg, J.; Hofsten, D.; Nepper-Christensen, L.; Kyhl, K.; Schoos, M.; Ghotbi, A.A.; Goransson, C.; Bertelsen, L.; et al. Impact of age on reperfusion success and long-term prognosis in ST-segment elevation myocardial infarction—A cardiac magnetic resonance imaging study. Int. J. Cardiol. Heart Vasc. 2021, 33, 100731. [Google Scholar]
- Wang, S.J.; Cheng, Z.X.; Fan, X.T.; Lian, Y.G. Development of an optimized risk score to predict short-term death among acute myocardial infarction patients in rural China. Clin. Cardiol. 2021, 44, 699–707. [Google Scholar] [CrossRef]
- Kang, Y.; Fang, X.-Y.; Wang, D.; Wang, X.-J. Factors associated with acute myocardial infarction in older patients after hospitalization with community-acquired pneumonia: A cross-sectional study. BMC Geriatr. 2021, 21, 113. [Google Scholar] [CrossRef]
- Shivakumar, B.G.; Shivakumar, N.; Gosavi, S.; Shastry, S. The importance of serum uric acid levels and Killip classification in predicting prognosis of acute myocardial infarction. J. Evol. Med. Dent. Sci. 2021, 10, 409–413. [Google Scholar]
- Fox, K.A.A.; Goldberg, R.J.; Pieper, K.; Cannon, C.P.; Van de Werfs, F.; Goodman, S.G.; Dabbous, O.; Granger, C.B.; Investigators, G. Predictors of in-hospital mortality in the global registry of acute coronary events (GRACE). Eur. Heart J. 2002, 23, 626. [Google Scholar]
- Li, Z.; Hui, Z.; Zheng, Y.; Yu, J.; Zhang, J. Comparative study on the effect of phase II remote home-based rehabilitation and traditional outpatient rehabilitation in patients with acute myocardial infarction after PCI. Basic Clin. Pharmacol. Toxicol. 2020, 127, 107. [Google Scholar]
- Sax, D.R.; Mark, D.G.; Huang, J.; Sofrygin, O.; Rana, J.S.; Collins, S.P.; Storrow, A.B.; Liu, D.; Reed, M.E. Use of machine learning to develop a risk-stratification tool for emergency department patients with acute heart failure. Ann. Emerg. Med. 2021, 77, 237–248. [Google Scholar] [CrossRef]
- Gupta, S.; Ko, D.T.; Azizi, P.; Bouadjenek, M.R.; Koh, M.; Chong, A.; Austin, P.C.; Sanner, S. Evaluation of machine learning algorithms for predicting readmission after acute myocardial infarction using routinely collected clinical data. Can. J. Cardiol. 2020, 36, 878–885. [Google Scholar] [CrossRef]
- Zhang, P.I.; Hsu, C.C.; Kao, Y.; Chen, C.J.; Kuo, Y.W.; Hsu, S.L.; Liu, T.L.; Lin, H.J.; Wang, J.J.; Liu, C.F.; et al. Real-time AI prediction for major adverse cardiac events in emergency department patients with chest pain. Scand. J. Trauma Resusc. Emerg. Med. 2020, 28, 93. [Google Scholar] [CrossRef]
- Zhang, Z.; Qiu, H.; Li, W.; Chen, Y. A stacking-based model for predicting 30-day all-cause hospital readmissions of patients with acute myocardial infarction. BMC Med. Inform. Decis. Mak. 2020, 20, 335. [Google Scholar] [CrossRef]
- Yeung, W.; Sia, C.-H.; Pollard, T.; Leow, A.S.-T.; Tan, B.Y.-Q.; Kaur, R.; Yeo, T.-C.; Tay, E.L.-W.; Yeo, L.L.-L.; Chan, M.Y.-Y.; et al. Predicting mortality, thrombus recurrence and persistence in patients with post-acute myocardial infarction left ventricular thrombus. J. Thromb. Thrombolysis 2021, 52, 654–661. [Google Scholar] [CrossRef]
- Ozer, I.; Cetin, O.; Gorur, K.; Temurtas, F. Improved machine learning performances with transfer learning to predicting need for hospitalization in arboviral infections against the small dataset. Neural Comput. Appl. 2021, 33, 14975–14989. [Google Scholar] [CrossRef]
- Wong, N.C.; Lam, C.; Patterson, L.; Shayegan, B. Use of machine learning to predict early biochemical recurrence after robot-assisted prostatectomy. BJU Int. 2019, 123, 51–57. [Google Scholar] [CrossRef] [Green Version]
- Liang, X.W.; Jiang, A.P.; Li, T.; Xue, Y.Y.; Wang, G.T. LR-SMOTE—An improved unbalanced data set oversampling based on K-means and SVM. Knowl.-Based Syst. 2020, 196, 105845. [Google Scholar] [CrossRef]
- Pandey, S.K.; Mishra, R.B.; Tripathi, A.K. Machine learning based methods for software fault prediction: A survey. Expert Syst. Appl. 2021, 172, 114595. [Google Scholar] [CrossRef]
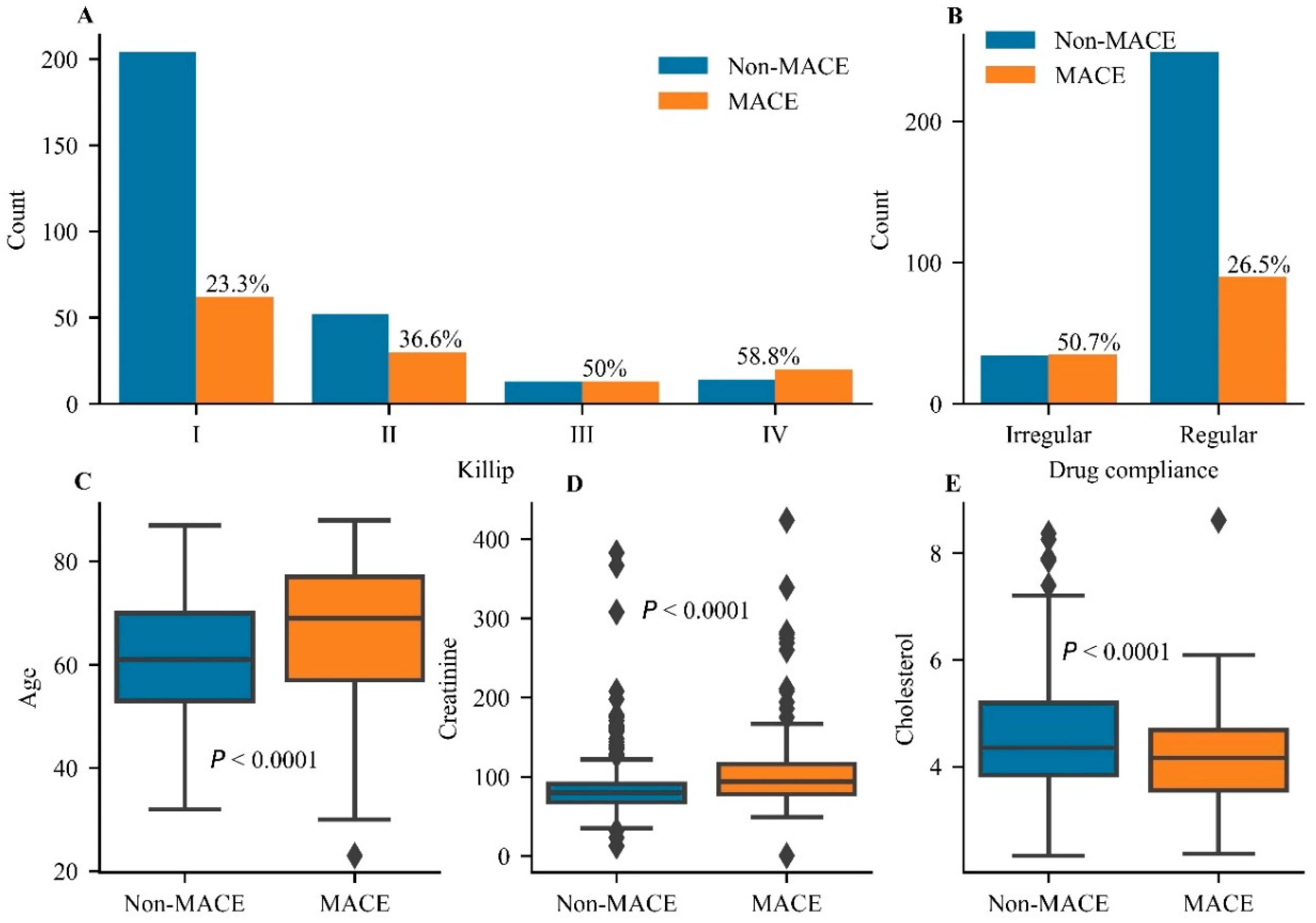

| Characteristic | All (N = 408) | Non-MACEs (N = 283) | MACEs (N = 125) | p-Value |
|---|---|---|---|---|
| Age (y) | 62.95 ± 12.98 | 61.32 ± 12.32 | 66.65 ± 13.71 | <0.0001 |
| Male (%) | 317 (77.7%) | 223 (78.8%) | 94 (75.2%) | 0.421 |
| Follow-up time (d) | 516.45 ± 137.82 | 509.88 ± 139.37 | 531.30 ± 133.61 | 0.148 |
| Coronary lesion vessels (num) | 2.14 ± 1.00 | 2.08 ± 1.00 | 2.27 ± 0.98 | 0.081 |
| Number of stents implanted | 1.79 ± 1.06 | 1.74 ± 1.08 | 1.90 ± 1.00 | 0.145 |
| Electrocardiogram (%) (ST segment elevation) | 258 (63.2%) | 179 (63.3%) | 79 (63.2%) | 0.992 |
| Killip classification (I:II:III:IV) | <0.0001 | |||
| I | 266 (65.2%) | 204 (72.1%) | 62 (49.6%) | |
| II | 82 (20.1%) | 52 (18.4%) | 30 (24.0%) | |
| III | 26 (6.4%) | 13 (4.6%) | 13 (10.4%) | |
| IV | 34 (8.3%) | 14 (4.9%) | 20 (16%) | |
| Cholesterol (mmol/L) | 4.44 ± 1.04 | 4.56 ± 1.07 | 4.16 ± 0.93 | <0.0001 |
| Low density lipoprotein (mmol/L) | 2.68 ± 0.87 | 2.76 ± 0.87 | 2.49 ± 0.83 | 0.004 |
| C-reactive protein (mg/L) | 18.14 ± 19.97 | 17.02 ± 19.49 | 20.70 ± 20.89 | 0.086 |
| Left ventricular diameter (mm) | 47.18 ± 5.85 | 46.80 ± 5.39 | 48.04 ± 6.73 | 0.049 |
| Left ventricular ejection fraction (%) | 0.52 ± 0.10 | 0.53 ± 0.09 | 0.50 ± 0.11 | 0.002 |
| Creatinine (µmol/L) | 92.59 ± 46.70 | 85.63 ± 38.40 | 108.35 ± 58.68 | <0.0001 |
| Uric acid (µmol/L) | 349.94 ± 101.27 | 337.86 ± 98.94 | 377.29 ± 101.54 | 2.64 × 10−4 |
| Glucose (mmol/L) | 8.78 ± 5.14 | 8.53 ± 5.13 | 9.36 ± 5.13 | 0.131 |
| White blood cells (109/L) | 10.46 ± 4.29 | 10.21 ± 4.00 | 11.02 ± 4.85 | 0.103 |
| Neutrophils (109/L) | 8.07 ± 4.11 | 7.89 ± 4.03 | 8.49 ± 4.28 | 0.168 |
| Hypertension (%) | 228 (55.9%) | 153 (54.1%) | 75 (60%) | 0.206 |
| Cigarettes (%) | 199 (48.8%) | 145 (51.2%) | 54 (43.2%) | 0.134 |
| Past medical history (%) | 132 (32.4%) | 83 (29.3%) | 49 (39.2%) | 0.049 |
| Diabetes (%) | 101 (24.8%) | 66 (23.3%) | 35 (28%) | 0.313 |
| Drug compliance (%) | 339 (83.4%) | 249 (88.0%) | 90 (72%) | <0.0001 |
| Revascularization time (min) | 4885 ± 11,126 | 4736 ± 10,264 | 5224 ± 12,917 | 0.683 |
| Number of diseased vessels | 2.14 ± 1.00 | 2.09 ± 1.01 | 2.26 ± 0.98 | 0.100 |
| Characteristic | OR | 95% Confidence Interval | p-Value |
|---|---|---|---|
| Killip classification III vs. I IV vs. I | 0.001 | ||
| 2.849 | (1.181–6.873) | 0.02 | |
| 4.386 | (1.943–9.904) | <0.0001 | |
| Drug compliance (irregular vs. regular) | 3.06 | (1.721–5.438) | <0.0001 |
| Age (per year) | 1.025 | (1.006–1.044) | 0.01 |
| Creatinine (1 µmol/L) | 1.007 | (1.002, 1.012) | 0.004 |
| Cholesterol (1 mmol/L) | 0.708 | (0.556–0.903) | 0.005 |
| Classifier | AUC, Mean (95%CI) | Accuracy, Mean (95%CI) | F1-Score, Mean (95%CI) |
|---|---|---|---|
| Logistic regression | 0.717 (0.692–0.743) | 0.721 (0.648–0.795) | 0.565 (0.483–0.646) |
| Decision tree | 0.664 (0.488–0.840) | 0.644 (0.463–0.825) | 0.532 (0.374–0.690) |
| Naive Bayes | 0.733 (0.650–0.718) | 0.742 (0.681–0.803) | 0.503 (0.358–0.648) |
| Support vector machine | 0.717 (0.687–0.746) | 0.725 (0.639–0.811) | 0.570 (0.483–0.656) |
| Random forest | 0. 749 (0.644–0.853) | 0.734 (0.647–0.820) | 0.480 (0.358–0.602) |
| Gradient boosting | 0.737 (0.637–0.838) | 0.729 (0.628–0.831) | 0.453 (0.206–0.701) |
| Multilayer perceptron | 0.663 (0.532–0.794) | 0.697 (0.599–0.795) | 0.103 (−0.162–0.368) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xiao, C.; Guo, Y.; Zhao, K.; Liu, S.; He, N.; He, Y.; Guo, S.; Chen, Z. Prognostic Value of Machine Learning in Patients with Acute Myocardial Infarction. J. Cardiovasc. Dev. Dis. 2022, 9, 56. https://doi.org/10.3390/jcdd9020056
Xiao C, Guo Y, Zhao K, Liu S, He N, He Y, Guo S, Chen Z. Prognostic Value of Machine Learning in Patients with Acute Myocardial Infarction. Journal of Cardiovascular Development and Disease. 2022; 9(2):56. https://doi.org/10.3390/jcdd9020056
Chicago/Turabian StyleXiao, Changhu, Yuan Guo, Kaixuan Zhao, Sha Liu, Nongyue He, Yi He, Shuhong Guo, and Zhu Chen. 2022. "Prognostic Value of Machine Learning in Patients with Acute Myocardial Infarction" Journal of Cardiovascular Development and Disease 9, no. 2: 56. https://doi.org/10.3390/jcdd9020056
APA StyleXiao, C., Guo, Y., Zhao, K., Liu, S., He, N., He, Y., Guo, S., & Chen, Z. (2022). Prognostic Value of Machine Learning in Patients with Acute Myocardial Infarction. Journal of Cardiovascular Development and Disease, 9(2), 56. https://doi.org/10.3390/jcdd9020056








