Characterizing Neonatal Heart Maturation, Regeneration, and Scar Resolution Using Spatial Transcriptomics
Abstract
:1. Introduction
2. Materials and Methods
2.1. Mice
2.2. Apical Resection Surgery
2.3. Histology and Immunohistochemistry
2.4. Bulk RNA Sequencing
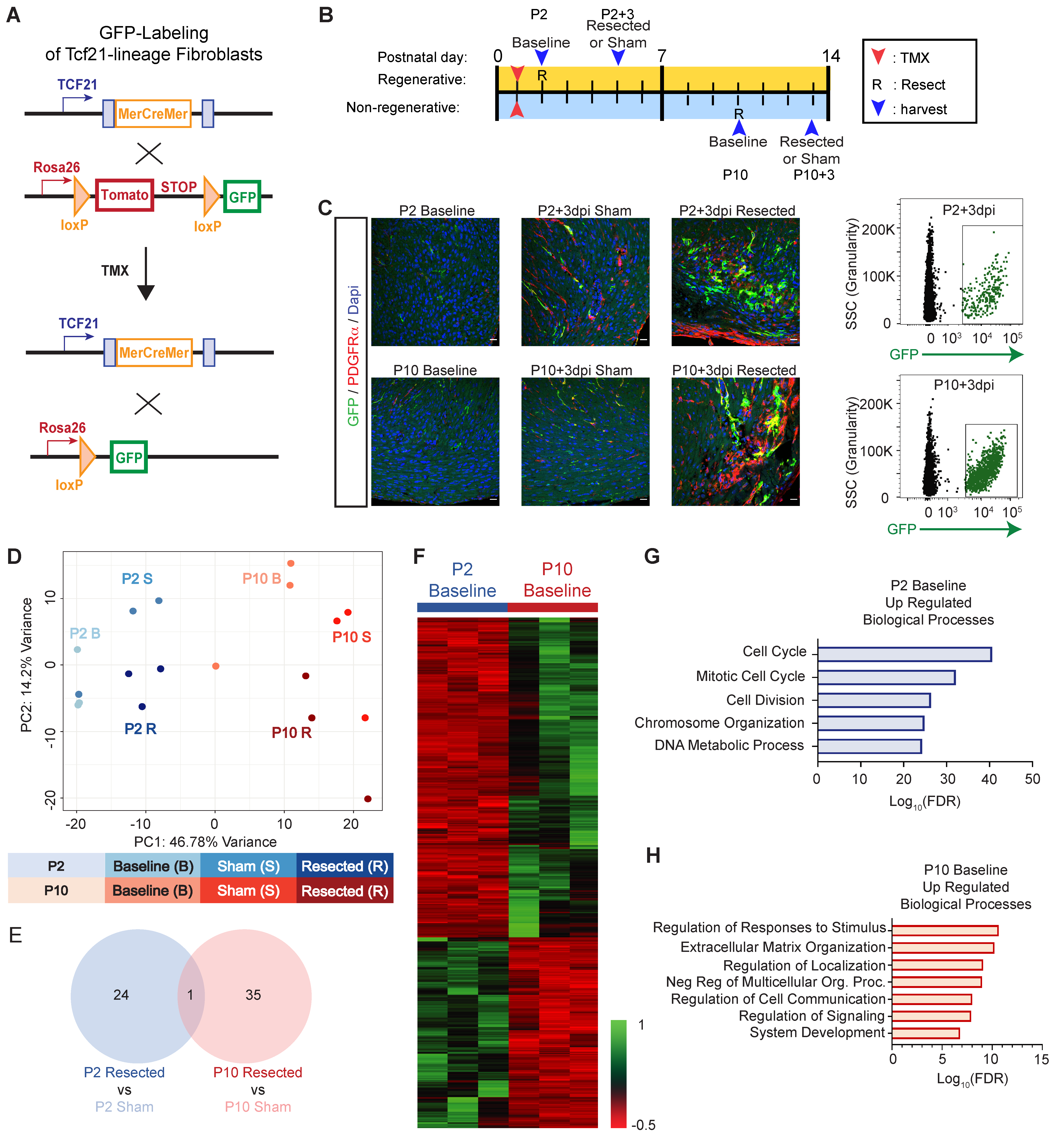
2.4.1. Flow Cytometry
2.4.2. Low-Input Library Prep
2.4.3. Analysis of RNA Sequencing and Statistical Analysis
2.5. Spatial Transcriptomics
2.5.1. Sample Preparation
2.5.2. mRNA Capture and Library Generation
2.5.3. Statistical Analysis of RNA Sequencing Data
3. Results
3.1. Acquisition of the Secretory Cardiac Fibroblast Phenotype Corresponds with the Loss of Regenerative Potential
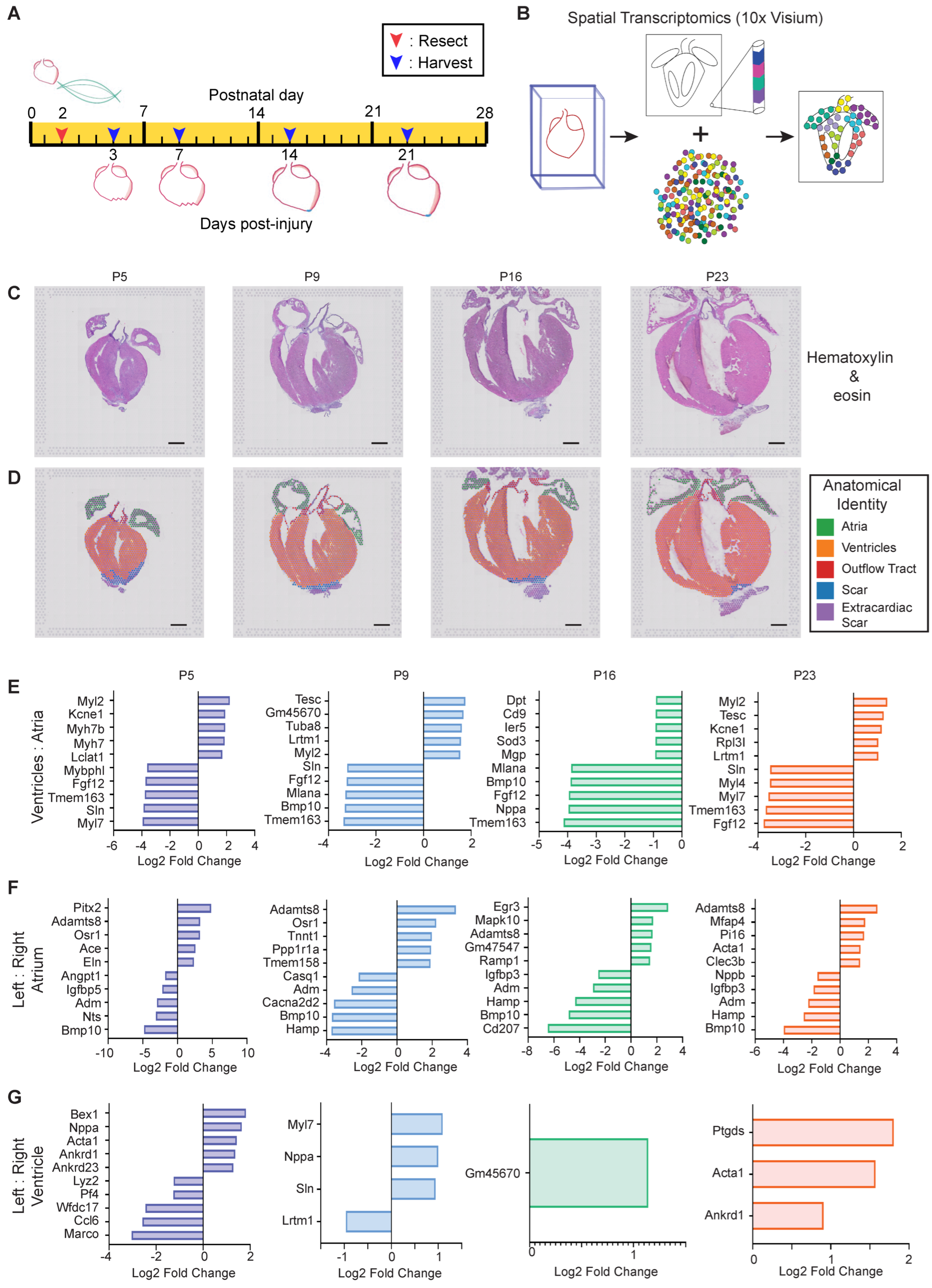
3.2. Interrogating Postnatal Heart Maturation and Regeneration Using Spatial Transcriptomics
3.2.1. Verifying Efficacy of Spatial Transcriptomics
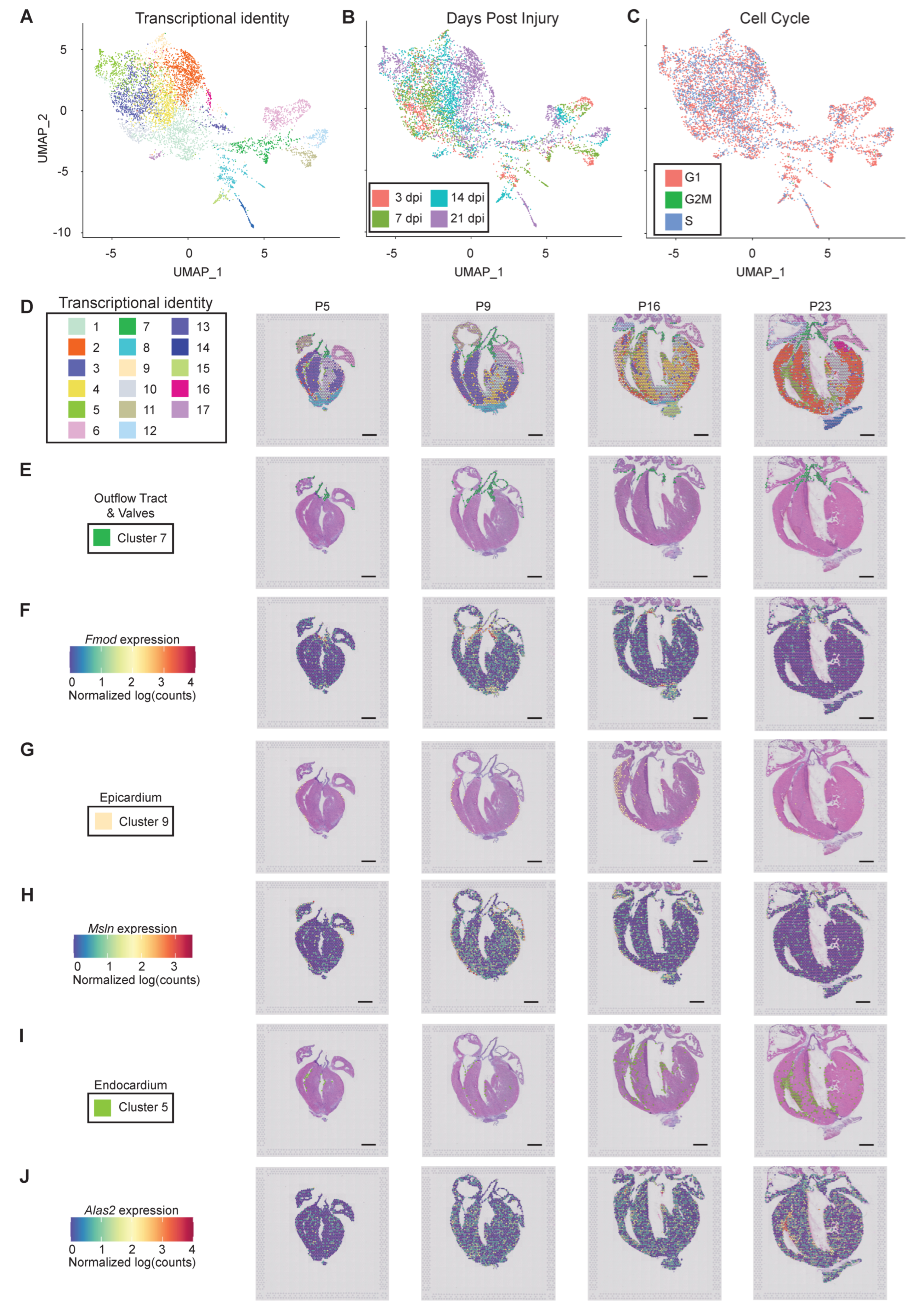
3.2.2. Cellular Resolution of Unbiased Spatial Transcriptomics
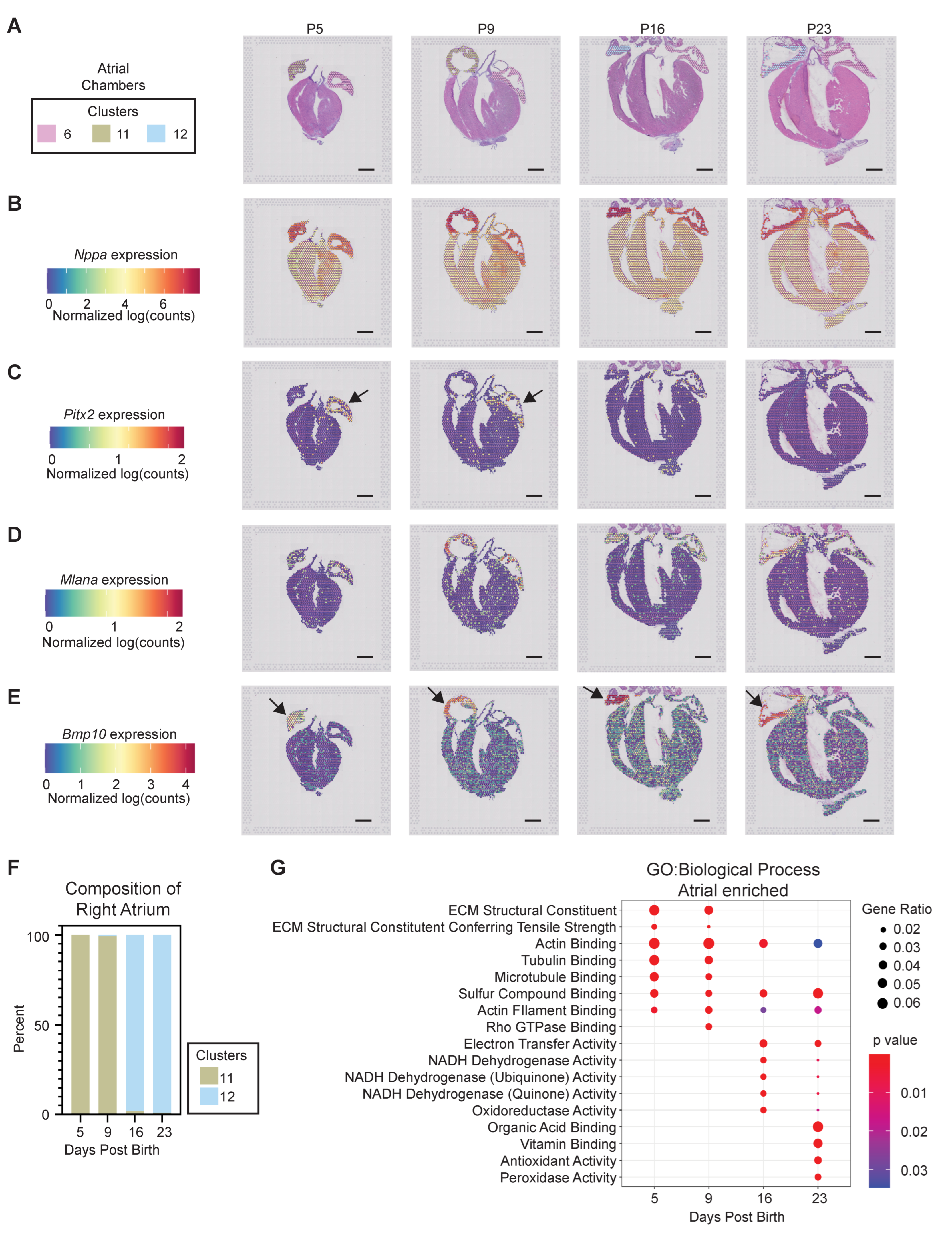
3.2.3. Atrial Chamber Maturation
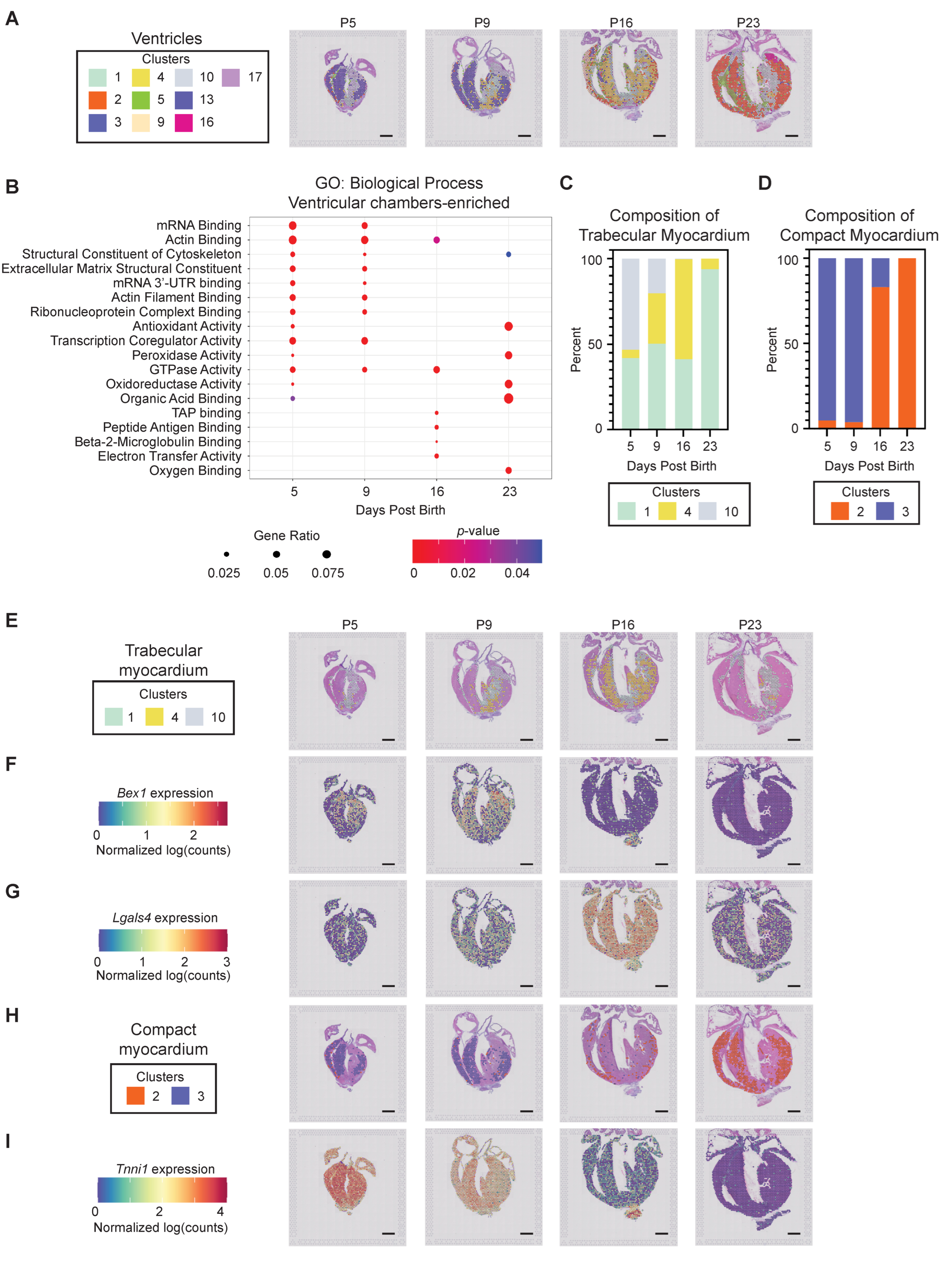
3.2.4. Ventricular Chamber Maturation
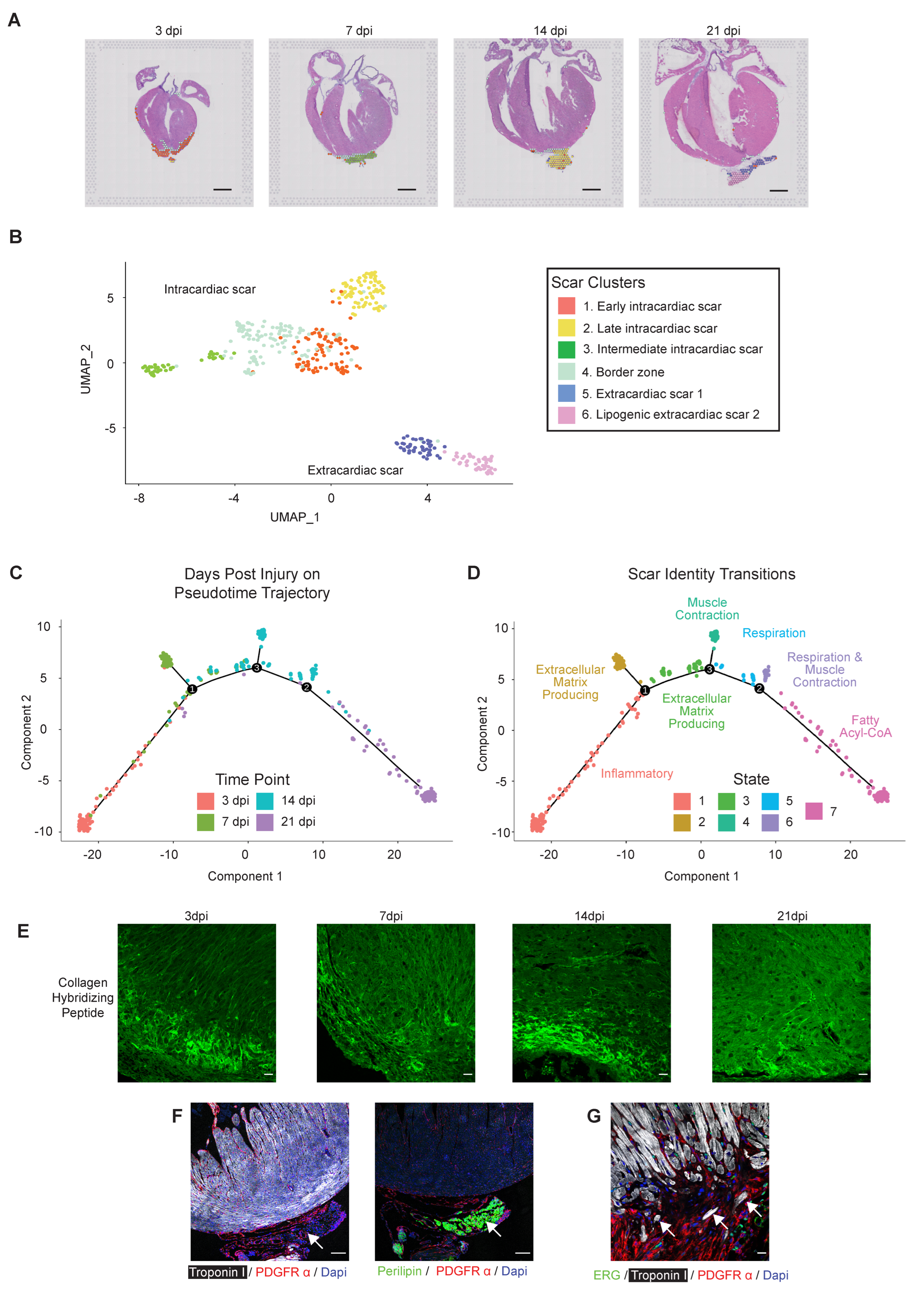
3.3. Interrogating Regenerative Scar Resolution Using Spatial Transcriptomics
3.3.1. Bi-Phasic Deposition of ECM
3.3.2. Extracardiac Cardiomyocytes and Lipogenic Tissue
3.3.3. Defining the Regenerative Border Zone
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Frangogiannis, N.G. The Extracellular Matrix in Ischemic and Nonischemic Heart Failure. Circ. Res. 2019, 125, 117–146. [Google Scholar] [CrossRef]
- Virani, S.S.; Alonso, A.; Aparicio, H.J.; Benjamin, E.J.; Bittencourt, M.S.; Callaway, C.W.; Carson, A.P.; Chamberlain, A.M.; Cheng, S.; Delling, F.N.; et al. Heart Disease and Stroke Statistics-2021 Update: A Report From the American Heart Association. Circulation 2021, 143, e254–e743. [Google Scholar] [CrossRef] [PubMed]
- Mahmoud, A.I.; Porrello, E.; Kimura, W.; Olson, E.N.; Sadek, H.A. Surgical models for cardiac regeneration in neonatal mice. Nat. Protoc. 2014, 9, 305–311. [Google Scholar] [CrossRef]
- Polizzotti, B.D.; Ganapathy, B.; Haubner, B.J.; Penninger, J.; Kühn, B.D.P.B.G.B. A cryoinjury model in neonatal mice for cardiac translational and regeneration research. Nat. Protoc. 2016, 11, 542–552. [Google Scholar] [CrossRef]
- Porrello, E.R.; Mahmoud, A.I.; Simpson, E.; Hill, J.A.; Richardson, J.A.; Olson, E.N.; Sadek, H. Transient Regenerative Potential of the Neonatal Mouse Heart. Science 2011, 331, 1078–1080. [Google Scholar] [CrossRef] [Green Version]
- Foglia, M.; Poss, K.D. Building and re-building the heart by cardiomyocyte proliferation. Development 2016, 143, 729–740. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, F.; Wang, X.; Bunger, P.C.; Gerdes, A. Formation of Binucleated Cardiac Myocytes in Rat Heart: I. Role of Actin–myosin Contractile Ring. J. Mol. Cell. Cardiol. 1997, 29, 1541–1551. [Google Scholar] [CrossRef]
- Walsh, S.; Pontén, A.; Fleischmann, B.K.; Jovinge, S. Cardiomyocyte cell cycle control and growth estimation in vivo—an analysis based on cardiomyocyte nuclei. Cardiovasc. Res. 2010, 86, 365–373. [Google Scholar] [CrossRef] [PubMed]
- Windmueller, R.; Leach, J.P.; Babu, A.; Zhou, S.; Morley, M.P.; Wakabayashi, A.; Petrenko, N.B.; Viatour, P.; Morrisey, E.E. Direct Comparison of Mononucleated and Binucleated Cardiomyocytes Reveals Molecular Mechanisms Underlying Distinct Proliferative Competencies. Cell Rep. 2020, 30, 3105–3116. [Google Scholar] [CrossRef] [Green Version]
- D’Uva, G.; Aharonov, A.; Lauriola, M.; Kain, D.; Yahalom-Ronen, Y.; Carvalho, S.; Weisinger, K.; Bassat, E.; Rajchman, D.; Yifa, O.; et al. ERBB2 triggers mammalian heart regeneration by promoting cardiomyocyte dedifferentiation and proliferation. Nat. Cell Biol. 2015, 17, 627–638. [Google Scholar] [CrossRef]
- Honkoop, H.; de Bakker, D.E.; Aharonov, A.; Kruse, F.; Shakked, A.; Nguyen, P.D.; de Heus, C.; Garric, L.; Muraro, M.J.; Shoffner, A.; et al. Single-cell analysis uncovers that metabolic reprogramming by ErbB2 signaling is essential for cardiomyocyte proliferation in the regenerating heart. Elife 2019, 8, e50163. [Google Scholar] [CrossRef] [PubMed]
- Mohamed, T.M.; Ang, Y.-S.; Radzinsky, E.; Zhou, P.; Huang, Y.; Elfenbein, A.; Foley, A.; Magnitsky, S.; Srivastava, D. Regulation of Cell Cycle to Stimulate Adult Cardiomyocyte Proliferation and Cardiac Regeneration. Cell 2018, 173, 104–116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Monroe, T.; Hill, M.; Morikawa, Y.; Leach, J.; Heallen, T.; Cao, S.; Krijger, P.; de Laat, W.; Wehrens, X.; Rodney, G.; et al. YAP Partially Reprograms Chromatin Accessibility to Directly Induce Adult Cardiogenesis In Vivo. Dev. Cell 2019, 48, 765–779. [Google Scholar] [CrossRef] [Green Version]
- Patterson, M.; Barske, L.; Van Handel, B.; Rau, C.D.; Gan, P.; Sharma, A.; Parikh, S.; Denholtz, M.; Huang, Y.; Yamaguchi, Y.; et al. Frequency of mononuclear diploid cardiomyocytes underlies natural variation in heart regeneration. Nat. Genet. 2017, 49, 1346–1353. [Google Scholar] [CrossRef] [PubMed]
- Huang, G.N.; Thatcher, J.E.; McAnally, J.; Kong, Y.; Qi, X.; Tan, W.; DiMaio, J.M.; Amatruda, J.F.; Gerard, R.D.; Hill, J.A.; et al. C/EBP Transcription Factors Mediate Epicardial Activation During Heart Development and Injury. Science 2012, 338, 1599–1603. [Google Scholar] [CrossRef] [Green Version]
- Aurora, A.B.; Porrello, E.; Tan, W.; Mahmoud, A.I.; Hill, J.A.; Bassel-Duby, R.; Sadek, H.A.; Olson, E.N. Macrophages are required for neonatal heart regeneration. J. Clin. Investig. 2014, 124, 1382–1392. [Google Scholar] [CrossRef] [Green Version]
- Simões, F.C.; Cahill, T.J.; Kenyon, A.; Gavriouchkina, D.; Vieira, J.M.; Sun, X.; Pezzolla, D.; Ravaud, C.; Masmanian, E.; Weinberger, M.; et al. Macrophages directly contribute collagen to scar formation during zebrafish heart regeneration and mouse heart repair. Nat. Commun. 2020, 11, 1–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- González-Rosa, J.M.; Martín, V.; Peralta, M.; Torres, M.; Mercader, N. Extensive scar formation and regression during heart regeneration after cryoinjury in zebrafish. Development 2011, 138, 1663–1674. [Google Scholar] [CrossRef] [Green Version]
- Sánchez-Iranzo, H.; Galardi-Castilla, M.; Sanz-Morejón, A.; González-Rosa, J.M.; Costa, R.; Ernst, A.; de Aja, J.S.; Langa, X.; Mercader, N. Transient fibrosis resolves via fibroblast inactivation in the regenerating zebrafish heart. Proc. Natl. Acad. Sci. USA 2018, 115, 4188–4193. [Google Scholar] [CrossRef] [Green Version]
- Bassat, E.; Mutlak, Y.E.; Genzelinakh, A.; Shadrin, I.; Umansky, K.B.; Yifa, O.; Kain, D.; Rajchman, D.; Leach, J.; Bassat, D.R.; et al. The extracellular matrix protein agrin promotes heart regeneration in mice. Nat. Cell Biol. 2017, 547, 179–184. [Google Scholar] [CrossRef] [PubMed]
- Marín-Juez, R.; El-Sammak, H.; Helker, C.S.M.; Kamezaki, A.; Mullapuli, S.T.; Bibli, S.-I.; Foglia, M.J.; Fleming, I.; Poss, K.D.; Stainier, D.Y.R. Coronary Revascularization During Heart Regeneration Is Regulated by Epicardial and Endocardial Cues and Forms a Scaffold for Cardiomyocyte Repopulation. Dev. Cell 2019, 51, 503–515. [Google Scholar] [CrossRef]
- Mewhort, H.E.; Svystonyuk, D.A.; Turnbull, J.D.; Teng, G.; Belke, D.D.; Guzzardi, D.G.; Park, D.S.; Kang, S.; Hollenberg, M.D.; Fedak, P.W. Bioactive Extracellular Matrix Scaffold Promotes Adaptive Cardiac Remodeling and Repair. JACC Basic Transl. Sci. 2017, 2, 450–464. [Google Scholar] [CrossRef] [PubMed]
- DeLaughter, D.M.; Bick, A.; Wakimoto, H.; McKean, D.; Gorham, J.M.; Kathiriya, I.S.; Hinson, J.T.; Homsy, J.; Gray, J.; Pu, W.; et al. Single-Cell Resolution of Temporal Gene Expression during Heart Development. Dev. Cell 2016, 39, 480–490. [Google Scholar] [CrossRef] [Green Version]
- Farbehi, N.; Patrick, R.; Dorison, A.; Xaymardan, M.; Janbandhu, V.; Wystub-Lis, K.; Ho, J.W.; Nordon, R.E.; Harvey, R.P. Single-cell expression profiling reveals dynamic flux of cardiac stromal, vascular and immune cells in health and injury. eLife 2019, 8, e43882. [Google Scholar] [CrossRef]
- Wang, Z.; Cui, M.; Shah, A.M.; Tan, W.; Liu, N.; Bassel-Duby, R.; Olson, E.N. Cell-Type-Specific Gene Regulatory Networks Underlying Murine Neonatal Heart Regeneration at Single-Cell Resolution. Cell Rep. 2020, 33, 108472. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Xu, A.; Sim, S.; Priest, J.R.; Tian, X.; Khan, T.; Quertermous, T.; Zhou, B.; Tsao, P.S.; Quake, S.R.; et al. Transcriptomic Profiling Maps Anatomically Patterned Subpopulations among Single Embryonic Cardiac Cells. Dev. Cell 2016, 39, 491–507. [Google Scholar] [CrossRef] [Green Version]
- van Duijvenboden, K.; De Bakker, D.E.; Man, J.; Janssen, R.; Günthel, M.; Hill, M.; Hooijkaas, I.B.; Van Der Made, I.; Van Der Kraak, P.H.; Vink, A.; et al. Conserved NPPB + Border Zone Switches From MEF2- to AP-1–Driven Gene Program. Circulation 2019, 140, 864–879. [Google Scholar] [CrossRef] [PubMed]
- Lacraz, G.P.; Junker, J.P.; Gladka, M.M.; Molenaar, B.; Scholman, K.T.; Vigil-Garcia, M.; Versteeg, D.; de Ruiter, H.; Vermunt, M.W.; Creyghton, M.P.; et al. Tomo-Seq Identifies SOX9 as a Key Regulator of Cardiac Fibrosis During Ischemic Injury. Circ. 2017, 136, 1396–1409. [Google Scholar] [CrossRef] [Green Version]
- Wu, C.-C.; Kruse, F.; Vasudevarao, M.D.; Junker, J.P.; Zebrowski, D.C.; Fischer, K.; Noël, E.S.; Grün, D.; Berezikov, E.; Engel, F.B.; et al. Spatially Resolved Genome-wide Transcriptional Profiling Identifies BMP Signaling as Essential Regulator of Zebrafish Cardiomyocyte Regeneration. Dev. Cell 2016, 36, 36–49. [Google Scholar] [CrossRef] [Green Version]
- Asp, M.; Giacomello, S.; Larsson, L.; Wu, C.; Fürth, D.; Qian, X.; Wärdell, E.; Custodio, J.; Reimegård, J.; Salmén, F.; et al. A Spatiotemporal Organ-Wide Gene Expression and Cell Atlas of the Developing Human Heart. Cell 2019, 179, 1647–1660. [Google Scholar] [CrossRef]
- Mantri, M.; Scuderi, G.J.; Abedini-Nassab, R.; Wang, M.F.Z.; McKellar, D.; Shi, H.; Grodner, B.; Butcher, J.T.; De Vlaminck, I. Spatiotemporal single-cell RNA sequencing of developing chicken hearts identifies interplay between cellular differentiation and morphogenesis. Nat. Commun. 2021, 12, 1–13. [Google Scholar] [CrossRef]
- Srivatsan, S.R.; Regier, M.C.; Barkan, E.; Franks, J.M.; Packer, J.S.; Grosjean, P.; Duran, M.; Saxton, S.; Ladd, J.J.; Spielmann, M.; et al. Embryo-scale, single-cell spatial transcriptomics. Science 2021, 373, 111–117. [Google Scholar] [CrossRef] [PubMed]
- Ståhl, P.L.; Salmén, F.; Vickovic, S.; Lundmark, A.; Navarro, J.F.; Magnusson, J.; Giacomello, S.; Asp, M.; Westholm, J.O.; Huss, M.; et al. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science 2016, 353, 78–82. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Das, S.; Goldstone, A.B.; Wang, H.; Farry, J.; D’Amato, G.; Paulsen, M.J.; Eskandari, A.; Hironaka, C.E.; Phansalkar, R.; Sharma, B.; et al. A Unique Collateral Artery Development Program Promotes Neonatal Heart Regeneration. Cell 2019, 176, 1128–1142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lizen, B.; Claus, M.; Jeannotte, L.; Rijli, F.M.; Gofflot, F. Perinatal induction of Cre recombination with tamoxifen. Transgenic Res. 2015, 24, 1065–1077. [Google Scholar] [CrossRef] [PubMed]
- Pinto, A.R.; Ilinykh, A.; Ivey, M.J.; Kuwabara, J.T.; D’Antoni, M.L.; Debuque, R.; Chandran, A.; Wang, L.; Arora, K.; Rosenthal, N.; et al. Revisiting Cardiac Cellular Composition. Circ. Res. 2016, 118, 400–409. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quijada, P.; Misra, A.; Velasquez, L.S.; Burke, R.M.; Lighthouse, J.K.; Mickelsen, D.M.; Dirkx, R.A.; Small, E.M. Pre-existing fibroblasts of epicardial origin are the primary source of pathological fibrosis in cardiac ischemia and aging. J. Mol. Cell. Cardiol. 2019, 129, 92–104. [Google Scholar] [CrossRef]
- Poss, K.D.; Wilson, L.G.; Keating, M.T. Heart Regeneration in Zebrafish. Science 2002, 298, 2188–2190. [Google Scholar] [CrossRef]
- Acharya, A.; Baek, S.T.; Huang, G.; Eskiocak, B.; Goetsch, S.; Sung, C.Y.; Banfi, S.; Sauer, M.F.; Olsen, G.S.; Duffield, J.S.; et al. The bHLH transcription factor Tcf21 is required for lineage-specific EMT of cardiac fibroblast progenitors. Development 2012, 139, 2139–2149. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanisicak, O.; Khalil, H.; Ivey, M.; Karch, J.; Maliken, B.D.; Correll, R.N.; Brody, M.; Lin, S.-C.J.; Aronow, B.J.; Tallquist, M.D.; et al. Genetic lineage tracing defines myofibroblast origin and function in the injured heart. Nat. Commun. 2016, 7, 12260. [Google Scholar] [CrossRef] [Green Version]
- Kuga, K.; Kusakari, Y.; Uesugi, K.; Semba, K.; Urashima, T.; Akaike, T.; Minamisawa, S. Fibrosis growth factor 23 is a promoting factor for cardiac fibrosis in the presence of transforming growth factor-beta1. PLoS ONE 2020, 15, e0231905. [Google Scholar] [CrossRef] [Green Version]
- Sheikh, F.; Lyon, R.C.; Chen, J. Functions of myosin light chain-2 (MYL2) in cardiac muscle and disease. Gene 2015, 569, 14–20. [Google Scholar] [CrossRef] [Green Version]
- Zhen, Y. Reactivation of Atrium Genes Is a Primer for Heart Infarction or Regeneration. Front. Cardiovasc. Med. 2020, 7, 575275. [Google Scholar] [CrossRef]
- Chen, H.; Shi, S.; Acosta, L.; Li, W.; Lu, J.; Bao, S.; Chen, Z.; Yang, Z.; Schneider, M.D.; Chien, K.R.; et al. BMP10 is essential for maintaining cardiac growth during murine cardiogenesis. Development 2004, 131, 2219–2231. [Google Scholar] [CrossRef] [Green Version]
- Jougasaki, M.; Burnett, J.C. Adrenomedullin: Potential in physiology and pathophysiology. Life Sci. 2000, 66, 855–872. [Google Scholar] [CrossRef]
- Lakhal-Littleton, S.; Wolna, M.; Chung, Y.J.; Christian, H.; Heather, L.C.; Brescia, M.D.; Ball, V.; Diaz, R.; Santos, A.; Biggs, D.; et al. An essential cell-autonomous role for hepcidin in cardiac iron homeostasis. eLife 2016, 5, e19804. [Google Scholar] [CrossRef]
- Trincot, C.E.; Xu, W.; Zhang, H.; Kulikauskas, M.R.; Caranasos, T.G.; Jensen, B.C.; Sabine, A.; Petrova, T.V.; Caron, K.M. Adrenomedullin Induces Cardiac Lymphangiogenesis After Myocardial Infarction and Regulates Cardiac Edema Via Connexin 43. Circ. Res. 2019, 124, 101–113. [Google Scholar] [CrossRef]
- Santamaria, S.; de Groot, R. ADAMTS proteases in cardiovascular physiology and disease. Open Biol. 2020, 10, 200333. [Google Scholar] [CrossRef]
- Liu, C.; Liu, W.; Lu, M.-F.; Brown, N.A.; Martin, J.F. Regulation of left-right asymmetry by thresholds of Pitx2c activity. Development 2001, 128, 2039–2048. [Google Scholar] [CrossRef]
- Andenæs, K.; Lunde, I.G.; Mohammadzadeh, N.; Dahl, C.P.; Aronsen, J.M.; Strand, M.E.; Palmero, S.; Sjaastad, I.; Christensen, G.; Engebretsen, K.V.T.; et al. The extracellular matrix proteoglycan fibromodulin is upregulated in clinical and experimental heart failure and affects cardiac remodeling. PLoS ONE 2018, 13, e0201422. [Google Scholar] [CrossRef]
- Delfín, D.A.; Deaguero, J.L.; McKown, E.N. The Extracellular Matrix Protein ABI3BP in Cardiovascular Health and Disease. Front. Cardiovasc. Med. 2019, 6, 23. [Google Scholar] [CrossRef]
- Fan, D.; Kassiri, Z. Biology of Tissue Inhibitor of Metalloproteinase 3 (TIMP3), and Its Therapeutic Implications in Cardiovascular Pathology. Front. Physiol. 2020, 11, 661. [Google Scholar] [CrossRef] [PubMed]
- Harikrishnan, K.; Cooley, M.A.; Sugi, Y.; Barth, J.L.; Rasmussen, L.M.; Kern, C.B.; Argraves, K.M.; Argraves, W.S. Fibulin-1 suppresses endothelial to mesenchymal transition in the proximal outflow tract. Mech. Dev. 2015, 136, 123–132. [Google Scholar] [CrossRef]
- Wang, X.; Chen, W.; Zhang, J.; Khan, A.; Li, L.; Huang, F.; Qiu, Z.; Wang, L.; Chen, X. Critical Role of ADAMTS2 (A Disintegrin and Metalloproteinase with Thrombospondin Motifs 2) in Cardiac Hypertrophy Induced by Pressure Overload. Hypertension 2017, 69, 1060–1069. [Google Scholar] [CrossRef]
- Zhou, Y.; Cashman, T.J.; Nevis, K.R.; Obregon, P.; Carney, S.A.; Liu, Y.; Gu, A.; Mosimann, C.; Sondalle, S.; Peterson, R.E.; et al. Latent TGF-beta binding protein 3 identifies a second heart field in zebrafish. Nature 474, 645–648. [CrossRef] [PubMed] [Green Version]
- Bruneau, B.G.; Logan, M.; Davis, N.; Levi, T.; Tabin, C.J.; Seidman, J.G.; Seidman, C.E. Chamber-Specific Cardiac Expression of Tbx5 and Heart Defects in Holt–Oram Syndrome. Dev. Biol. 1999, 211, 100–108. [Google Scholar] [CrossRef] [Green Version]
- Kahr, P.C.; Piccini, I.; Fabritz, L.; Greber, B.; Schöler, H.; Scheld, H.H.; Hoffmeier, A.; Brown, N.A.; Kirchhof, P. Systematic Analysis of Gene Expression Differences between Left and Right Atria in Different Mouse Strains and in Human Atrial Tissue. PLoS ONE 2011, 6, e26389. [Google Scholar] [CrossRef]
- Cuenca-Zamora, E.J.; Ferrer-Marín, F.; Rivera, J.; Teruel-Montoya, R. Tubulin in Platelets: When the Shape Matters. Int. J. Mol. Sci. 2019, 20, 3484. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, H.-F.; Yang, J.-F.; Wang, Q.; Li, R.-G.; Xu, Y.-J.; Qu, X.-K.; Fang, W.-Y.; Liu, X.; Yang, Y.-Q. Prevalence and spectrum of GJA5 mutations associated with lone atrial fibrillation. Mol. Med. Rep. 2012, 7, 767–774. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Woulfe, K.C.; Sucharov, C.C. Midkine’s Role in Cardiac Pathology. J. Cardiovasc. Dev. Dis. 2017, 4, 13. [Google Scholar] [CrossRef] [Green Version]
- Huang, Y.-J.; Zhou, Z.-W.; Xu, M.; Ma, Q.-W.; Yan, J.-B.; Wang, J.-Y.; Zhang, Q.-Q.; Huang, M.; Bao, L. Alteration of Gene Expression Profiling Including GPR174 and GNG2 is Associated with Vasovagal Syncope. Pediatr. Cardiol. 2014, 36, 475–480. [Google Scholar] [CrossRef]
- Uosaki, H.; Cahan, P.; Lee, D.I.; Wang, S.; Miyamoto, M.; Fernandez, L.; Kass, D.A.; Kwon, C. Transcriptional Landscape of Cardiomyocyte Maturation. Cell Rep. 2015, 13, 1705–1716. [Google Scholar] [CrossRef] [Green Version]
- Accornero, F.; Schips, T.G.; Petrosino, J.M.; Gu, S.-Q.; Kanisicak, O.; Van Berlo, J.H.; Molkentin, J.D. BEX1 is an RNA-dependent mediator of cardiomyopathy. Nat. Commun. 2017, 8, 1875. [Google Scholar] [CrossRef] [Green Version]
- Bunnell, T.M.; Ervasti, J.M. Delayed embryonic development and impaired cell growth and survival in Actg1 null mice. Cytoskeleton 2010, 67, 564–572. [Google Scholar] [CrossRef] [Green Version]
- Roselló-Lletí, E.; Carnicer, R.; Tarazón, E.; Ortega, A.; Gil-Cayuela, C.; Lago, F.; Juanatey, J.R.G.; Portoles, M.; Rivera, M. Human Ischemic Cardiomyopathy Shows Cardiac Nos1 Translocation and its Increased Levels are Related to Left Ventricular Performance. Sci. Rep. 2016, 6, 24060. [Google Scholar] [CrossRef] [Green Version]
- Fu, X.; Khalil, H.; Kanisicak, O.; Boyer, J.G.; Vagnozzi, R.J.; Maliken, B.D.; Sargent, M.A.; Prasad, V.; Valiente-Alandi, I.; Blaxall, B.C.; et al. Specialized fibroblast differentiated states underlie scar formation in the infarcted mouse heart. J. Clin. Investig. 2018, 128, 2127–2143. [Google Scholar] [CrossRef] [Green Version]
- Quijada, P.; Trembley, M.A.; Small, E.M. The Role of the Epicardium During Heart Development and Repair. Circ. Res. 2020, 126, 377–394. [Google Scholar] [CrossRef]
- Zhou, B.; Honor, L.B.; He, H.; Ma, Q.; Oh, J.-H.; Butterfield, C.; Lin, R.-Z.; Melero-Martin, J.M.; Dolmatova, E.; Duffy, H.S.; et al. Adult mouse epicardium modulates myocardial injury by secreting paracrine factors. J. Clin. Investig. 2011, 121, 1894–1904. [Google Scholar] [CrossRef] [Green Version]
- Lange, S.; Gehmlich, K.; Lun, A.S.; Blondelle, J.; Hooper, C.; Dalton, N.D.; Alvarez, E.A.; Zhang, X.; Bang, M.L.; Abassi, Y.A.; et al. MLP and CARP are linked to chronic PKCalpha signalling in dilated cardiomyopathy. Nat. Commun. 2016, 7, 12120. [Google Scholar] [CrossRef]
- Miller, M.K.; Bang, M.L.; Witt, C.C.; Labeit, D.; Trombitas, C.; Watanabe, K.; Granzier, H.; McElhinny, A.S.; Gregorio, C.C.; Labeit, S. The muscle ankyrin repeat proteins: CARP, ankrd2/Arpp and DARP as a family of titin filament-based stress response molecules. J. Mol. Biol. 2003, 333, 951–964. [Google Scholar] [CrossRef]
- Murphy, N.P.; Lubbers, E.R.; Mohler, P.J. Advancing our understanding of AnkRD1 in cardiac development and disease. Cardiovasc. Res. 2020, 116, 1402–1404. [Google Scholar] [CrossRef]
- Pradervand, S.; Yasukawa, H.; Muller, O.G.; Kjekshus, H.; Nakamura, T.; Amand, T.R.S.; Yajima, T.; Matsumura, K.; Duplain, H.; Iwatate, M.; et al. Small proline-rich protein 1A is a gp130 pathway- and stress-inducible cardioprotective protein. EMBO J. 2004, 23, 4517–4525. [Google Scholar] [CrossRef] [Green Version]
- Hortells, L.; Valiente-Alandi, I.; Thomas, Z.; Agnew, E.J.; Schnell, D.J.; York, A.J.; Vagnozzi, R.J.; Meyer, E.C.; Molkentin, J.D.; Yutzey, K.E. A specialized population of Periostin-expressing cardiac fibroblasts contributes to postnatal cardiomyocyte maturation and innervation. Proc. Natl. Acad. Sci. USA 2020, 117, 21469–21479. [Google Scholar] [CrossRef]
- Ivey, M.; Kuwabara, J.T.; Pai, J.T.; Moore, R.E.; Sun, Z.; Tallquist, M.D. Resident fibroblast expansion during cardiac growth and remodeling. J. Mol. Cell. Cardiol. 2018, 114, 161–174. [Google Scholar] [CrossRef]
- Ieda, M.; Tsuchihashi, T.; Ivey, K.N.; Ross, R.S.; Hong, T.T.; Shaw, R.M.; Srivastava, D. Cardiac fibroblasts regulate myocardial proliferation through beta1 integrin signaling. Dev. Cell 2009, 16, 233–244. [Google Scholar] [CrossRef] [Green Version]
- Houweling, A.C.; van Borren, M.M.; Moorman, A.F.; Christoffels, V.M. Expression and regulation of the atrial natriuretic factor encoding gene Nppa during development and disease. Cardiovasc. Res. 2005, 67, 583–593. [Google Scholar] [CrossRef] [Green Version]
- Sergeeva, I.A.; Hooijkaas, I.B.; Ruijter, J.M.; van der Made, I.; de Groot, N.E.; van de Werken, H.J.G.; Creemers, E.; Christoffels, V.M. Identification of a regulatory domain controlling the Nppa-Nppb gene cluster during heart development and stress. Development 2016, 143, 2135–2146. [Google Scholar] [CrossRef] [Green Version]
- Cao, J.; Poss, K.D. The epicardium as a hub for heart regeneration. Nat. Rev. Cardiol. 2018, 15, 631–647. [Google Scholar] [CrossRef]
- Kikuchi, K.; Holdway, J.E.; Major, R.J.; Blum, N.; Dahn, R.D.; Begemann, G.; Poss, K.D. Retinoic Acid Production by Endocardium and Epicardium Is an Injury Response Essential for Zebrafish Heart Regeneration. Dev. Cell 2011, 20, 397–404. [Google Scholar] [CrossRef] [Green Version]
- Lepilina, A.; Coon, A.N.; Kikuchi, K.; Holdway, J.E.; Roberts, R.W.; Burns, C.G.; Poss, K.D. A Dynamic Epicardial Injury Response Supports Progenitor Cell Activity during Zebrafish Heart Regeneration. Cell 2006, 127, 607–619. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Karra, R.; Dickson, A.L.; Poss, K.D. Fibronectin is deposited by injury-activated epicardial cells and is necessary for zebrafish heart regeneration. Dev. Biol. 2013, 382, 427–435. [Google Scholar] [CrossRef] [Green Version]
- Cavallero, S.; Shen, H.; Yi, C.; Lien, C.-L.; Kumar, S.R.; Sucov, H.M. CXCL12 Signaling Is Essential for Maturation of the Ventricular Coronary Endothelial Plexus and Establishment of Functional Coronary Circulation. Dev. Cell 2015, 33, 469–477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mellgren, A.M.; Smith, C.L.; Olsen, G.S.; Eskiocak, B.; Zhou, B.; Kazi, M.N.; Ruiz, F.R.; Pu, W.T.; Tallquist, M.D. Platelet-derived growth factor receptor beta signaling is required for efficient epicardial cell migration and development of two distinct coronary vascular smooth muscle cell populations. Circ. Res. 2008, 103, 1393–1401. [Google Scholar] [CrossRef]
- Quijada, P.; Trembley, M.A.; Misra, A.; Myers, J.A.; Baker, C.D.; Pérez-Hernández, M.; Myers, J.R.; Dirkx, R.A.; Cohen, E.D.; Delmar, M.; et al. Coordination of endothelial cell positioning and fate specification by the epicardium. Nat. Commun. 2021, 12, 1–18. [Google Scholar] [CrossRef]
- Trembley, M.A.; Velasquez, L.S.; Bentley, K.L.D.M.; Small, E.M. Myocardin-related transcription factors control the motility of epicardium-derived cells and the maturation of coronary vessels. Development 2015, 142, 21–30. [Google Scholar] [CrossRef] [Green Version]
- Grunewald, M.; Kumar, S.; Sharife, H.; Volinsky, E.; Gileles-Hillel, A.; Licht, T.; Permyakova, A.; Hinden, L.; Azar, S.; Friedmann, Y.; et al. Counteracting age-related VEGF signaling insufficiency promotes healthy aging and extends life span. Science 2021, 373, eabc8479. [Google Scholar] [CrossRef]
- Cao, J.; Wang, J.; Jackman, C.P.; Cox, A.H.; Trembley, M.A.; Balowski, J.J.; Cox, B.D.; De Simone, A.; Dickson, A.L.; Di Talia, S.; et al. Tension Creates an Endoreplication Wavefront that Leads Regeneration of Epicardial Tissue. Dev. Cell 2017, 42, 600–615. [Google Scholar] [CrossRef] [Green Version]

| Antyibody | Vendor/Cat No. | Concentration |
|---|---|---|
| Mouse anti-Cardiac Troponin I (C-4) | Santa Cruz sc133117 | 1/30 |
| Rabbit anti-ERG | Abcam ab92513 | 1/100 |
| Rabbit anti-GFP | Torrey Pines Biolabs TP401 | 1/200 |
| Goat anti-PDGFR-alpha | R&D Systems AF1062 | 1/200 |
| Rabbit anti-Perilipin | Abcam ab3526 | 1/100 |
| Rabbit anti-Sprr1a | Abcam ab125374 | 1/100 |
| Secondary & Tertiary Antibodies | ||
| Donkey anti-mouse 647 | Jackson ImmunoResearch Laboratories #715-605-150 | 1/100 |
| Donkey anti-rabbit Biotin | Jackson ImmunoResearch Laboratories #711-065-152 | 1/500 |
| Bovine anti-goat HRP | Jackson ImmunoResearch Laboratories #805-035-180 | 1/200 |
| Tyramide Cy3 | Perkin Elmer NEL744001KT | 1/200 |
| Streptavidin 488 | LifeTech S11223 | 1/100 |
| Streptavidin 555 | LifeTech S21381 | 1/100 |
| DAPI | Invitrogen D1306 | 1/10,000 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Misra, A.; Baker, C.D.; Pritchett, E.M.; Burgos Villar, K.N.; Ashton, J.M.; Small, E.M. Characterizing Neonatal Heart Maturation, Regeneration, and Scar Resolution Using Spatial Transcriptomics. J. Cardiovasc. Dev. Dis. 2022, 9, 1. https://doi.org/10.3390/jcdd9010001
Misra A, Baker CD, Pritchett EM, Burgos Villar KN, Ashton JM, Small EM. Characterizing Neonatal Heart Maturation, Regeneration, and Scar Resolution Using Spatial Transcriptomics. Journal of Cardiovascular Development and Disease. 2022; 9(1):1. https://doi.org/10.3390/jcdd9010001
Chicago/Turabian StyleMisra, Adwiteeya, Cameron D. Baker, Elizabeth M. Pritchett, Kimberly N. Burgos Villar, John M. Ashton, and Eric M. Small. 2022. "Characterizing Neonatal Heart Maturation, Regeneration, and Scar Resolution Using Spatial Transcriptomics" Journal of Cardiovascular Development and Disease 9, no. 1: 1. https://doi.org/10.3390/jcdd9010001
APA StyleMisra, A., Baker, C. D., Pritchett, E. M., Burgos Villar, K. N., Ashton, J. M., & Small, E. M. (2022). Characterizing Neonatal Heart Maturation, Regeneration, and Scar Resolution Using Spatial Transcriptomics. Journal of Cardiovascular Development and Disease, 9(1), 1. https://doi.org/10.3390/jcdd9010001






