Abstract
Hypertrophic cardiomyopathy (HCM) is a heterogeneous genetic disorder, most often caused by sarcomeric gene mutations, with a small proportion due to variants in non-sarcomeric loci. Phospholamban (PLN) is a phosphoprotein associated with the cardiac sarcoplasmic reticulum, a major determinant of cardiac contractility and relaxation. We conducted a retrospective study to determine the prevalence, phenotypical spectrum and clinical course of patients carrying the PLN p.Leu39* variant. A cohort including 11 PLN patients was identified among all patients with HCM (9/189, 4.8%) and DCM (2/62, 3.2%) who underwent genetic testing from two tertiary centers and five more were detected through cascade screening. Complete phenotyping was performed. PLN p.Leu39* variant-driven cardiomyopathy presented mostly as hypertrophic, with frequent progression to end-stage dilated HCM. We proceeded to compare these results to a similar analysis of a control cohort consisting of age-matched individuals that inherited pathogenic or likely pathogenic variants in common sarcomeric genes (MYBPC3/MYH7). Overall, the clinical characteristics and examination findings of patients carrying PLN p.Leu39* were not different from patients with cardiomyopathy related to sarcomeric mutations except for the presence of pathological Q waves and the incidence of non-sustained ventricular arrhythmias, which were higher in PLN patients than in those with MYBPC3/MYH7-related diseases.
1. Introduction
Hereditary cardiomyopathies are a heterogeneous group of primary myocardial diseases caused by genetic variants in which the heart muscle is structurally and functionally abnormal, resulting in various phenotypes. Hypertrophic cardiomyopathy is the most common subtype, representing a phenotype of left ventricular hypertrophy unexplained entirely by abnormal loading conditions. It is one of the leading causes of sudden cardiac death (SCD) in young people and athletes [1] and one of the most common inherited cardiovascular diseases, affecting 1 in 500 individuals. It is mostly caused by disease-causing variants in genes encoding sarcomeric proteins, which are identified in up to 60% of cases of the disease. Dilated cardiomyopathy (DCM) is also an important genetic heart disease defined by the presence of left ventricular dilation and systolic dysfunction, with a prevalence of 1:2500 in the general population [2]. DCM is one of the leading causes of heart failure and requires cardiac transplantation in severe cases.
Recent advances in genomics have shed more light on the molecular pathogenic mechanisms of cardiomyopathies, contributing to substantial advances in the diagnosis of the disease. Disease-causing variants in many genes are involved in cardiomyopathies, especially those in genes encoding cytoskeletal, sarcomere, and nuclear envelope proteins. Disease-causing variants in more 100 genes encoding proteins involved in many different subcellular systems have been identified to contribute to the genetic landscape of cardiomyopathies, indicating the diversity of pathways contributing to cardiac remodeling [3].
Phospholamban (PLN) is a 52-amino acid integral membrane protein that plays an essential role in regulating cardiac contractility via a reversible inhibitory association with the sarcoplasmic reticulum Ca2+ATPase (SERCA2a), the enzyme responsible for maintaining calcium homeostasis in the heart muscle. Pathogenic variants in the PLN gene may cause inherited cardiomyopathies due to a key role in the function of the sarcoplasmic reticulum (SR) which is the main dynamic Ca2+ storage compartment of the cell [4,5]. The Ca2+ is released from the SR into the cytosol, facilitating contraction of the myocytes and is transported back into the SR to initiate relaxation by the SERCA2a, the activity of which is regulated by PLN [6,7]. The PLN NM_002667.5, c.116T>G variant creates a premature translational stop signal (p.Leu39*) in the PLN gene, which is predicted to lead to a truncated protein, as the last 14 amino acids are lost. This alteration has been previously identified in multiple individuals with DCM [8], HCM [9,10,11,12,13] and unexplained cardiac arrest in the absence of a cardiomyopathy phenotype [14].
2. Materials and Methods
Patients with PLN p.Leu39* variants were identified at two tertiary referral centers that routinely evaluate patients with HCM/ DCM. Each center independently identified all genotype positive individuals among the consecutive HCM and DCM patients for whom clinical genetic testing had been performed for HCM or DCM. All probands and family members with confirmed PLN p.Leu39* variant were included to enable study of the variability in phenotypes of mutation carriers. Retrospective data were collected into a prespecified database.
Patients were given annotations based on their family status: the first digit stands for the family, and the second digit stands for the individual within the family (the proband has 1 as the second digit.
HCM or DCM was diagnosed by physical examination, electrocardiogram, echocardiogram and magnetic resonance imaging, according to the criteria of the ESC working group on cardiomyopathies [15]. Written informed consent was obtained from all patients before enrollment. Next-generation sequencing (NGS) was performed in 189 patients with HCM and 62 patients with DCM phenotype using cardiomyopathy dedicated gene panels: Invitae Dilated Cardiomyopathy and Left Ventricular Noncompaction (80 genes tested), Invitae Hypertrophic Cardiomyopathy Panel (44 genes tested)—INVITAE laboratory, United States, Hypertrophic Cardiomyopathy Panel (92 genes tested)—Blueprint Genetics laboratory, Finland or TruSightCardio Panel Illumina (174 genes tested)—Genomic Center of the University of Medicine and Pharmacy Victor Babeș Timișoara, depending on the patient and physician preferences, considering different turnaround time and reimbursement.
The sequencing was carried out by the sequencing instruments using the Illumina (San Diego, CA) sequencing-by-synthesis method. All sequence reads were mapped onto the human reference genome hg37. The American College of Medical Genetics and Genomics and the Association for Molecular Pathology (ACMG/AMP) was used for classification of genetic variants as benign (B), likely benign (LB), variant of uncertain significance (VUS), likely pathogenic (LP), and pathogenic (P) [16]. Results were categorized as disease causing/significant (if P or LP), negative (if B or LB), or uncertain (VUS), depending on the classification of the variant identified and the inheritance pattern of the associated condition.
Detailed clinical data were collected on all patients, at each tertiary care center, by reviewing the clinical records from the most recent visit of all participants and included pertinent personal (symptom onset and its aggravation) or family history (especially with regard to HCM or SCD with a three-generation family history) and treatment strategies. Biomarkers such as natriuretic peptides and creatine kinases were assessed. Echocardiographic parameters such as left ventricle (LV) ejection fraction (LVEF), LV end diastolic and systolic diameter (LVEDD, LVESD) and volume (LVEDV, LVESV), global longitudinal strain (GLS), wall thickness, LV outflow tract (LVOT) or mid-cavity pressure gradient, left atrium (LA) systolic volume and diastolic function parameters were determined. When available, cardiac magnetic resonance (CMR) findings were described: LVEF, LV end diastolic and systolic volumes, right ventricle ejection fraction (RVEF), maximum wall thickness, presence of late gadolinium enhancement (LGE). Resting electrocardiogram (ECG) and 24 h electrocardiographic Holter monitoring data were also analyzed. ECG changes were defined as described: ‘high QRS voltage’ is defined as sum of S in V1 and R in V5 exceeded 35 mm, ‘low QRS voltage is defined QRS by a amplitude of less than 5 mm in the limb leads and/or less than 10 mm in the precordial leads, ‘repolarization abnormalities’ are defined as presence of ST segment depression ≥1 mm or an inverted T wave opposite to the QRS axis in at least two contiguous leads, and ‘pathologic Q waves’ are defined as any Q wave with a width greater than 40 ms or a depth greater than one-third of the adjacent R wave. Survival status was identified using the Romanian National Health Insurance House to ensure that no deaths were missed.
Multiple clinical and outcome variables were compared between the phenotype-positive PLN group and a control group formed by age matching of phenotype-positive carriers of myosin-binding protein C (MYBPC3) or myosin heavy chain (MYH7) likely pathogenic or pathogenic variant, to define genotype-phenotype associations. Each PLN patients was age-matched with one MYBPC3 or MYH7 patient for a better description of the phenotypic evolution. Genotype-positive/phenotype-negative individuals were excluded from the analyses.
Statistical analyses were performed using IBM SPSS 26.0 (IBM Corp, Armonk, NY, USA). Univariable analysis was applied to both continuous and categorical variables. Normality was determined by the Shapiro–Wilk test. Continuous variables were reported as the mean ± standard deviation and/or as the median and interquartile range (IQR) when appropriate. Among-group comparisons were made using a non-parametric test (Mann–Whitney U test). Categorical variables were reported as counts and percentages. Among-group comparisons were made using a χ2 test. Statistics with a 2-sided p value < 0.05 indicating significant differences.
Due to the observational nature of this study, the use of anonymized data was based on the individual patient’s informed consent including acceptance of data utilization for research.
3. Results
3.1. PLN Cohort
Genetic investigations identified a pathogenic variant (NM_002667.5, c.116T>G or p.Leu39*, heterozygous) in the PLN gene in 11 probands (Pt 1.1, Pt 2.1, Pt 3.1, Pt 4.1, Pt 5.1, Pt 6.1, Pt 7.1, Pt 8.1, Pt 9.1, Pt 10.1, Pt 11.1) representing 9/189 (4.8%) patients tested for HCM and 2/62 (3.2%) patients tested for DCM. Five more cases (Pt 1.2, Pt 2.2, Pt 2.3, Pt 4.2, Pt 6.2) have been identified as a result of cascade screening in the affected families, including three asymptomatic carriers (Pt 2.2, Pt 2.3, Pt 6.2).
Multiple cardiomyopathy pathogenic variants were present in three patients from this cohort. One patient (Pt 10.1) carries a pathogenic variants in another HCM-related gene [(MYBPC3] NM_000256.3, c.1504C>T, p.Arg502Trp,) while two others (Pt 6.1 and Pt 11.1) carry a likely pathogenic variant ([MYH7] NM_000257.4, c.5134C>T, p.Arg1712Trp and titin [TTN] NM_001267550.2, c.94128del, p.Tyr31376*, respectively).
The clinical variables of patients enrolled in this study at the last visit are displayed in Table 1. Six patients (Pt 1.1, Pt 1.2, Pt 2.2, Pt 2.3, Pt 4.2 and Pt 6.2) had a family history of HCM, and another four (Pt 3.1, Pt 5.1, Pt 7.1, Pt 9.1) had a family history of SCD. At the initial presentation to our clinic, 4 (36%) out of 13 affected individuals were asymptomatic, but 9 of them had already developed heart failure symptoms (at least NYHA class II), while 5 patients complained of palpitations and 5 patients had angina. Syncope was recorded only for patient 7.1 and patient 11.1. The median age at the diagnosis was 46 years old, with a wide range reported (4 to 66). Pt 6.1 exhibited early-onset cardiomyopathy (during his first years of life) while his father (Pt 6.2, carrying the same PLN variant) is phenotype negative. The presence of an additionally HCM-related variant in the child (MYH7) could potentially explain the severity of the phenotype in this case. A total of 9 patients developed atrial fibrillation (AF), with a median age of onset of 54 (42.5–65) years old.

Table 1.
Baseline Characteristics of PLN patients.
The N-terminal pro B-type natriuretic peptide (NTproBNP) levels were significantly elevated (median value 4893.2 ± 6299.5 pg/dL). At 12-lead ECG examination (Figure 1), six patients (Pt 2.3, Pt 3.1, Pt 4.1, Pt 5.1, Pt 6.1, Pt 8.1) presented with high QRS voltage, and three patients had low QRS voltage (Pt 9.1, Pt 10.1, Pt 11.1). ECG of nine of the patients (Pt 1.1, Pt 1.2, Pt 2.1, Pt 3.1, Pt 4.1, Pt 5.1, Pt 6.1, Pt 6.2, Pt 8.1, Pt 11.1) showed inverted T waves. Pt 4.1, Pt 6.1, Pt 6.2, Pt 10.1 and Pt 11.1 developed pathological Q waves. Additionally, one patient (Pt 4.1) had a left bundle branch block morphology.
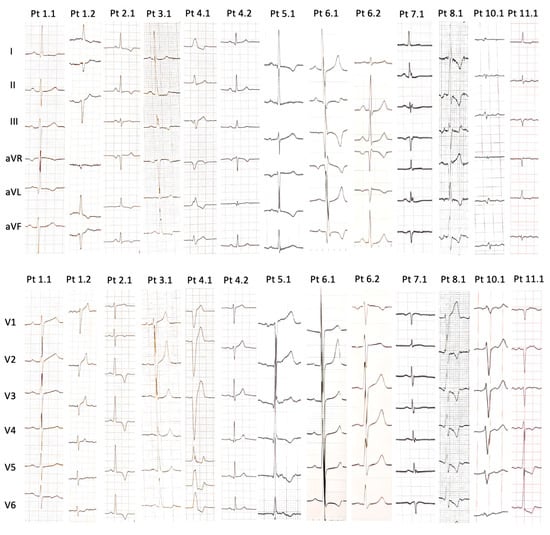
Figure 1.
Findings of 12-lead electrocardiograms of PLN patients (when available).
Echocardiographic and magnetic resonance imaging features are presented in Table 2. The majority of PLN subjects displayed a hypertrophied phenotype (8 individuals representing 57%) mostly with septal thickening, but also with concentric hypertrophy (Pt 1.2, Pt 5.1, Pt 7.1) or apical hypertrophy (Pt 2.1, Pt 11.1), but a dilated phenotype (2 patients) or a mixed one (3 patients) characterized by a dilated and hypertrophied LV were noted. The maximal LV walls thickness ranged from 9 mm to 30 mm (median 17 mm). Right ventricle (RV) involvement was noted in 10 patients.

Table 2.
Echocardiography and cardiac magnetic resonance characteristics of PLN patients.
Five patients (Pt 1.2, Pt 4.1, Pt 9.1, Pt 10.1, Pt 11.1) showed decreased LVEF; coronary artery disease could have been involved in the first case, but was absent for the other four individuals. LVOT dynamic obstruction was present in two cases (Pt 1.1, Pt 3.1). Another patient (Pt 2.1) experienced mid-cavity obstruction.
Only nine patients were examined with CMR (Figure 2). Therefore, eight (89%) of them exhibited LGE.
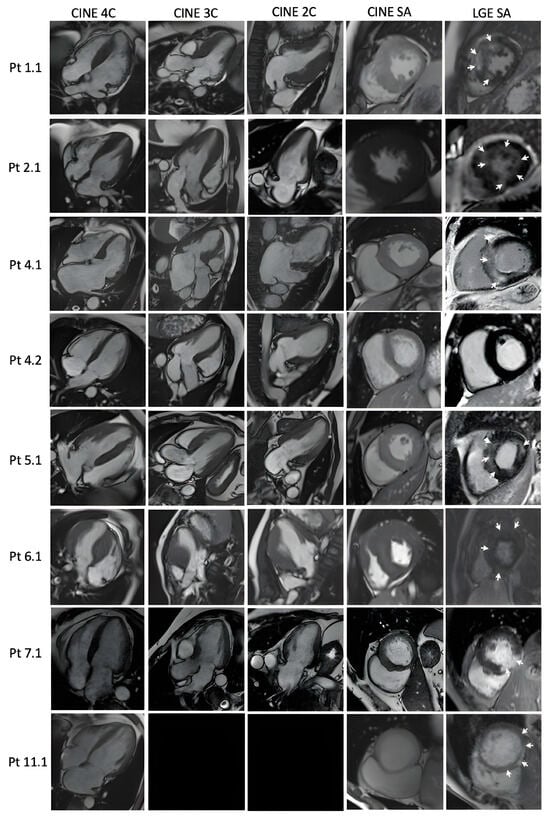
Figure 2.
Cardiac magnetic resonance findings in PLN patients: Diastolic frames of cine images in 4, 3, 2 chambers and short axis, and late gadolinium enhancement images in short axis, respectively. Black rectangles: the image is not available. White arrows demonstrate areas of LGE.
3.2. Genotype-Phenotype Correlation and PNL-Other Sarcomeric Gene Comparison
Phenotype-positive PLN carriers were included in the PLN group and phenotype-positive MYBPC3/MYH7 carriers matched for age were included in the MYBPC3/MYH7 group. There was no difference between groups with respect to gender, age at diagnosis, incidence of AF and the frequency of symptoms. Echocardiographic and CMR features did not show any statistical difference between groups, except for left ventricle volumes (LVEDVi and LVESVi), PLN patients had larger LVs compared to the MYBPC3/MYH7 group (p = 0.045 and 0.050, respectively). Regarding ECG findings, the PLN group showed more frequent pathological Q waves (p = 0.030), but conduction anomalies (bundle branch blocks) were rarely seen in these patients (p = 0.047). Patients from the PLN group experienced more frequent non-sustained VT than the MYBPC3/MYH7 group (p = 0.027). We did not detect significant differences in treatment between the two groups (Table 3).

Table 3.
Overall characteristics of the PLN and MYH7/MYBPC3 patient groups. Values are means ± standard deviation, medians with interquartile ranges and numbers (percentages). P values were calculated by the 2-sided unpaired Mann–Whitney U-test for continuous variables, and by chi-square (χ2) test for categorical variables. Bold: statistically significant results.
Patient phenotypes, clinical course and family pedigree charts for each PLN p.Leu39* individual are described in the Case Series from Section S1 in the Supplementary Material.
4. Discussion
Phospholamban has emerged as a critical regulator of Ca2+ homeostasis. Both the inhibition and overexpression of phospholamban have been associated with the development of primary cardiomyopathies in humans [9,17,18]. The human PLN gene is located on chromosome 6, and the amino acid sequence of PLN is highly conserved in all species [19].
Worldwide, several PLN variants have strong association with variable cardiac phenotypes ranging from dilated cardiomyopathy [8,17] to hypertrophic [9,10,11,12,13,18] and even arrhythmogenic cardiomyopathy (ACM) [20,21], but the causative defects leading to cardiomyopathies remain incompletely elucidated.
Different studies suggest that the p.Leu39* PLN mutant is expressed but mis-located within the cardiomyocytes [22,23]. The mis-location of PLN was associated to decreased SERCA2a expression and impaired Ca2+ handling in human pathophysiology. Calcium plays a crucial role in cardiomyocytes by acting as a signal that controls the contraction-relaxation cycle and cardiac hypertrophy. Increased Ca2+ levels in myoplasm contribute to the development and progression of hypertrophy [24].
In this study, we summarized the clinical findings of 16 patients carrying the PLN p.Leu39* variant in Romania, including 13 phenotype-positive individuals and 3 asymptomatic carriers. The phenotype of p.Leu39* variant carriers in the PLN gene has been shown to vary considerably, ranging from the typical HCM phenotype, to a mixed phenotype (dilated and hypertrophied LV) and DCM phenotype. Cardiomyopathy patients often present with highly variable phenotypic expressions in terms of structural and functional parameters of the heart and clinical course. It is common to observe patients from the same family, who share a gene variant, that exhibit variable phenotypes ranging from almost asymptomatic forms to the development of heart failure or to severe arrhythmias/sudden death. As was presented above, the PLN group presented incomplete and age-dependent penetrance, the onset of the symptoms having a considerably large range (from 4 to 66 years old) or the individual never developing the disease (as was the case with patient 6.2). While most of the patients had a “benign” evolution of the disease, with no sustained malignant arrhythmias and no family history of SCD, there is also the case of patient 9.1 and her three siblings who suffered youthful disease-related deaths. It is well known that patient-specific factors beyond the single pathogenic variant or environmental modifiers, can dramatically modulate the phenotype in different individuals [25,26].
In agreement with previous findings [12], the majority of our PLN p.Leu39* patients demonstrated cardiac hypertrophy, supporting the hypothesis that p.Leu39* is an HCM-predisposing variant. Moreover, two patients (Pt 4.1 and Pt 10.1) were firstly diagnosed with LV hypertrophy in the presence of a normal LV systolic function, but several years later after the diagnosis the LVEF started declining, the echocardiographic aspect could be linked to a progression to end-stage dilated HCM.
Haghighi et al. identified two probands with DCM and PLN p.Leu39* variant, diagnosed at 27, respectively 28 years old [9]. Nine individuals from both families of probands underwent echocardiographic examination, revealing that they all had normal LVEF, but four of them exhibited left ventricular hypertrophy. Because of the presence of hypertrophy in some members of both families, we could assume that the dilated phenotype in the probands might have been a consequence of evolution from a hypertrophic phenotype to a burnout stage. However, genome-wide association studies demonstrated that many loci are associated with both HCM and DCM [27]. Despite the genetic overlap, distinct disorders might develop through opposing genetic effects [28] between PLN function and HCM phenotype, and a possible evolution to a burn-out stage.
Abrams et al. reported the existence of a PLN p.Leu39* variant in a patient without any imaging evidence of structural heart disease, but with an arrhythmogenic phenotype, developing ventricular tachycardia and ventricular fibrillation storm [29]. It is well known that the PLN p.Arg14del founder variant (NM_002667.5) is characterized by life-threatening ventricular arrhythmias and sudden cardiac death [30,31,32], highlighting a potentially primary arrhythmogenic nature of this variant. During the follow-up, we demonstrated that in our PLN cohort, non-sustained VT is more frequent than in those with sarcomeric gene variants (p = 0.027), even the HCM Risk SCD scores were similar (Table S1) which emphasize the importance of detecting this variant. Three patients (Pt 3.1, Pt 7.1, Pt 9.1) had a positive family history for SCD in first- and second-degree relatives, some of whom died at very young ages (18, 21, 25, 30, 58 years old). Another patient (Pt 5.1) had two sons who died very early in life, but this could be attributed to a potassium voltage-gated channel [KCNQ1] c.604G>A variant (NM_000218.3), which was detected in the father in addition to the PLN variant.
Low QRS voltage is a common finding in PLN-related DCM; however, the study cohorts consisted only in patients with the p.Arg14del variant [30,33,34]. Similarly, in this p.Leu39* cohort, both patients diagnosed with a dilated phenotype (Pt 9.1, Pt 11.1) experienced low voltage electrocardiogram. Additionally, Pt 10.1 diagnosed with HCM with evolution to end-stage disease due to the extensive fibrosis, had a similar ECG pattern. One more patient (Pt 7.1) with an eccentric LV hypertrophy and a maximum wall thickness (MWT) of 17 mm showed disproportion to electrocardiographic QRS voltage. Another ECG abnormality found in the study group is the presence of pathological Q waves, which are substantially more common than in the MYBPC3/MYH7 group (p = 0.030). A distinct fibrosis pattern was reported for the PLN p.Arg14del, with extensive subepicardial fibrosis in the posterolateral wall [35], but this pattern could not be confirmed for our PLN p.Leu39* cohort with a hypertrophic phenotype, where the LGE involved mostly hypertrophied segments, similar to sarcomeric variants, but was present for the dilated phenotype (Pt. 11.1).
Previously published studies reported PLN variants as a rare cause of hypertrophic or dilated cardiomyopathies [18,36], with the exception of p.Arg14del variant which was found with higher frequency among patients with DCM and ACM [31,37]. While PLN- disease-causing variants count for a small percentage of patients with HCM among different publications [10,12], in this cohort the percentage of PLN carriers was relatively high (4.8%), highlighting the importance for routinely screening for PLN, alongside historically validated genes for HCM.
Furthermore, this is the first study to analyze the differences in clinical and paraclinical characteristics between patients with PLN p.Leu39* and those with the most commonly HCM-associated variants (MYBPC3/MYH7). After matching the two groups by age, we observed that the evolution of the PLN-related disease was similar to that related to sarcomeric genes variants, but the PLN patients are at greater risk for ventricular arrhythmia.
The main limitation of this study is the sample size, which was small. However, this is common in many studies dealing with rare diseases. Despite the fact that genetic results came from different laboratories, and employed gene panels of different size, the methodology is similar and all panels include the core sarcomeric genes associated with cardiomyopathies. To our knowledge, this is the largest cohort of patients carrying this pathogenic variant. Thus, future studies identifying the PLN p.Leu39* variant are required to elucidate the potential association.
5. Conclusions
Our results demonstrate that PLN mutant p.Leu39*-related cardiomyopathy is mainly characterized by a hypertrophic phenotype with a potential to evolve towards a dilated phenotype. We hypothesized that the dilated phenotype could be related to HCM evolution into the end-stage phase, but further studies are required for a better understanding of the observed phenotype in human carriers. In addition, the genotype–phenotype relationship in PLN patients is not distinctly different from those caused by other causal genes (MYBPC3/MYH7), but these patients could carry a high risk for ventricular arrhythmia. Our findings contribute to a better understanding of the cardiac effects and consequences of the chronic expression of the p.Leu39* mutant in humans.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/jcdd11020041/s1, Figure S1: Echocardiographic findings in PLN patients; Section S1: case series; Table S1: Assessment of risk of sudden cardiac death in PLN patients with a hypertrophied or mixed phenotype compared to the age-matched MYBPC3/MYH7 patients.
Author Contributions
A.S.A., R.J. drafted the manuscript. A.S.A., L.V., R.S., C.R., E.C. and R.J. were involved in patient selection for genetic testing, clinical management and family screening. A.C.-E. was involved in genetic testing. S.O. was involved in cardiac magnetic resonance imaging, A.S.A. and L.V. were involved in data extraction from document files. A.S.A. and R.D.A. analyzed the data and performed statistical analyses. All authors discussed the results and contributed to the final manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
This study did not require specific ethical approval. Being an observational study, publication of anonymized data was based on the individual informed consent for utilization of data for research.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are contained within the article and supplementary materials.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Maron, B.J.; Shirani, J.; Poliac, L.C.; Mathenge, R.; Roberts, W.C.; Mueller, F.O. Sudden death in young competitive athletes. Clinical, demographic, and pathological profiles. JAMA 1996, 276, 199–204. [Google Scholar] [CrossRef]
- Weintraub, R.G.; Semsarian, C.; Macdonald, P. Dilated cardiomyopathy. Lancet 2017, 390, 400–414. [Google Scholar] [CrossRef]
- Gerull, B.; Klaassen, S.; Brodehl, A. The Genetic Landscape of Cardiomyopathies. In Genetic Causes of Cardiac Disease; Springer: Cham, Switzerland, 2019; pp. 45–91. [Google Scholar]
- Meldolesi, J.; Pozzan, T. The endoplasmic reticulum Ca2+ store: A view from the lumen. Trends Biochem. Sci. 1998, 23, 10–14. [Google Scholar] [CrossRef]
- Frank, K.; Kranias, E.G. Phospholamban and cardiac contractility. Ann. Med. 2000, 32, 572–578. [Google Scholar] [CrossRef] [PubMed]
- Koss, K.L.; Kranias, E.G. Phospholamban: A prominent regulator of myocardial contractility. Circ. Res. 1996, 79, 1059–1063. [Google Scholar] [CrossRef] [PubMed]
- Simmerman, H.K.; Jones, L.R. Phospholamban: Protein structure, mechanism of action, and role in cardiac function. Physiol. Rev. 1998, 78, 921–947. [Google Scholar] [CrossRef] [PubMed]
- Sanoudou, D.; Kolokathis, F.; Arvanitis, D.; Al-Shafai, K.; Krishnamoorthy, N.; Buchan, R.J.; Walsh, R.; Tsiapras, D.; Barton, P.J.R.; Cook, S.A.; et al. Genetic modifiers to the PLN L39X mutation in a patient with DCM and sustained ventricular tachycardia? Glob. Cardiol. Sci. Pract. 2015, 2015, 29. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Haghighi, K.; Kolokathis, F.; Pater, L.; Lynch, R.A.; Asahi, M.; Gramolini, A.O.; Fan, G.C.; Tsiapras, D.; Hahn, H.S.; Adamopoulos, S.; et al. Human phospholamban null results in lethal dilated cardiomyopathy revealing a critical difference between mouse and human. J. Clin. Investig. 2003, 111, 869–876. [Google Scholar] [CrossRef] [PubMed]
- Chiu, C.; Tebo, M.; Ingles, J.; Yeates, L.; Arthur, J.W.; Lind, J.M.; Semsarian, C. Genetic screening of calcium regulation genes in familial hypertrophic cardiomyopathy. J. Mol. Cell. Cardiol. 2007, 43, 337–343. [Google Scholar] [CrossRef] [PubMed]
- Walsh, R.; Thomson, K.L.; Ware, J.S.; Funke, B.H.; Woodley, J.; McGuire, K.J.; Mazzarotto, F.; Blair, E.; Seller, A.; Taylor, J.C.; et al. Reassessment of Mendelian gene pathogenicity using 7855 cardiomyopathy cases and 60,706 reference samples. Genet. Med. 2017, 19, 192–203. [Google Scholar] [CrossRef] [PubMed]
- Landstrom, A.P.; Adekola, B.A.; Bos, J.M.; Ommen, S.R.; Ackerman, M.J. PLN-encoded phospholamban mutation in a large cohort of hypertrophic cardiomyopathy cases: Summary of the literature and implications for genetic testing. Am. Heart J. 2011, 161, 165–171. [Google Scholar] [CrossRef] [PubMed]
- Alfares, A.A.; Kelly, M.A.; McDermott, G.; Funke, B.H.; Lebo, M.S.; Baxter, S.B.; Shen, J.; McLaughlin, H.M.; Clark, E.H.; Babb, L.J.; et al. Results of clinical genetic testing of 2912 probands with hypertrophic cardiomyopathy: Expanded panels offer limited additional sensitivity. Genet. Med. 2015, 17, 880–888. [Google Scholar] [CrossRef] [PubMed]
- Mellor, G.; Laksman, Z.W.M.; Tadros, R.; Roberts, J.D.; Gerull, B.; Simpson, C.S.; Klein, G.J.; Champagne, J.; Talajic, M.; Gardner, M.; et al. Genetic Testing in the Evaluation of Unexplained Cardiac Arrest: From the CASPER (Cardiac Arrest Survivors with Preserved Ejection Fraction Registry). Circ. Cardiovasc. Genet. 2017, 10, e001686. [Google Scholar] [CrossRef] [PubMed]
- Arbelo, E.; Protonotarios, A.; Gimeno, J.R.; Arbustini, E.; Barriales-Villa, R.; Basso, C.; Bezzina, C.R.; Biagini, E.; Blom, N.A.; de Boer, R.A.; et al. 2023 ESC Guidelines for the management of cardiomyopathies. Eur. Heart J. 2023, 44, 3503–3626. [Google Scholar] [CrossRef] [PubMed]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015, 17, 405–424. [Google Scholar] [CrossRef] [PubMed]
- Schmitt, J.P.; Kamisago, M.; Asahi, M.; Li, G.H.; Ahmad, F.; Mende, U.; Kranias, E.G.; MacLennan, D.H.; Seidman, J.G.; Seidman, C.E. Dilated Cardiomyopathy and Heart Failure Caused by a Mutation in Phospholamban. Science 2003, 299, 1410–1413. [Google Scholar] [CrossRef] [PubMed]
- Medin, M.; Hermida-Prieto, M.; Monserrat, L.; Laredo, R.; Rodriguez-Rey, J.C.; Fernandez, X.; Castro-Beiras, A. Mutational screening of phospholamban gene in hypertrophic and idiopathic dilated cardiomyopathy and functional study of the PLN–42 C>G mutation. Eur. J. Heart Fail. 2007, 9, 37–43. [Google Scholar] [CrossRef] [PubMed]
- Fujii, J.; Zarain-Herzberg, A.; Willard, H.F.; Tada, M.; MacLennan, D.H. Structure of the rabbit phospholamban gene, cloning of the human cDNA, and assignment of the gene to human chromosome 6. J. Biol. Chem. 1991, 266, 11669–11675. [Google Scholar] [CrossRef]
- Te Rijdt, W.P.; Asimaki, A.; Jongbloed, J.D.H.; Hoorntje, E.T.; Lazzarini, E.; van der Zwaag, P.A.; de Boer, R.A.; van Tintelen, J.P.; Saffitz, J.E.; Berg, M.P.v.D.; et al. Distinct molecular signature of phospholamban p.Arg14del arrhythmogenic cardiomyopathy. Cardiovasc. Pathol. 2019, 40, 2–6. [Google Scholar] [CrossRef]
- van der Zwaag, P.A.; van Rijsingen, I.A.W.; de Ruiter, R.; Nannenberg, E.A.; Groeneweg, J.A.; Post, J.G.; Hauer, R.N.; van Gelder, I.C.; van den Berg, M.P.; van der Harst, P.; et al. Recurrent and founder mutations in the Netherlands—Phospholamban p.Arg14del mutation causes arrhythmogenic cardiomyopathy. Neth. Heart J. 2013, 21, 286–293. [Google Scholar] [CrossRef]
- Bailey, R.A.; Stillitano, F.; Turnbull, I.; Haghigi, K.; Fish, K.; Akar, F.; Dubois, N.; Wickramasinghe, N.; Hajjar, R.J.; Gelb, B.D.; et al. Abstract 530: Mechanisms Underlying Phospholamban L39 Stop (PLN L39X) Cardiomyopathy. Circ. Res. 2020, 127 (Suppl. S1), A530. [Google Scholar] [CrossRef]
- Devendran, A.; Bailey, R.; Kar, S.; Stillitano, F.; Turnbull, I.; Fish, K.; Dubois, N.; Wickramasinghe, N.; Hajjar, R.; Costa, K.; et al. Abstract P482: Elucidating and Characterizing the Molecular Mechanistic Role of Phospholamban L39 Stop in the Pathophysiology of Cardiomyopathy Using Patient-derived Human Induced Pluripotent Stem Cells and Humanized Knock-in Mouse Model Systems. Circ. Res. 2021, 129 (Suppl. S1), AP482. [Google Scholar] [CrossRef]
- Nakayama, H.; Otsu, K.; Yamaguchi, O.; Nishida, K.; Date, M.; Hongo, K.; Kusakari, Y.; Toyofuku, T.; Hikoso, S.; Kashiwase, K.; et al. Cardiac-specific overexpression of a high Ca2+ affinity mutant of SERCA2a attenuates in vivo pressure overload cardiac hypertrophy. FASEB J. 2003, 17, 61–63. [Google Scholar] [CrossRef] [PubMed]
- Marian, A.J. Hypertrophic cardiomyopathy: From genetics to treatment. Eur. J. Clin. Investig. 2010, 40, 360–369. [Google Scholar] [CrossRef] [PubMed]
- Arad, M. Phenotypic diversity in hypertrophic cardiomyopathy. Hum. Mol. Genet. 2002, 11, 2499–2506. [Google Scholar] [CrossRef] [PubMed]
- Walsh, R.; Offerhaus, J.A.; Tadros, R.; Bezzina, C.R. Minor hypertrophic cardiomyopathy genes, major insights into the genetics of cardiomyopathies. Nat. Rev. Cardiol. 2022, 19, 151–167. [Google Scholar] [CrossRef] [PubMed]
- Tadros, R.; Francis, C.; Xu, X.; Vermeer, A.M.C.; Harper, A.R.; Huurman, R.; Bisabu, K.K.; Walsh, R.; Hoorntje, E.T.; Rijdt, W.P.T.; et al. Shared genetic pathways contribute to risk of hypertrophic and dilated cardiomyopathies with opposite directions of effect. Nat. Genet. 2021, 53, 128–134. [Google Scholar] [CrossRef] [PubMed]
- Abrams, J.; Pitt, G.; Elemento, O.; Cheung, J.W. Phospholamban mutation leading to recurrent short-coupled polymorphic ventricular tachycardia and fibrillation in patient without apparent structural heart disease. J. Am. Coll. Cardiol. 2023, 81, 2452. [Google Scholar] [CrossRef]
- van der Zwaag, P.A.; van Rijsingen, I.A.W.; Asimaki, A.; Jongbloed, J.D.H.; van Veldhuisen, D.J.; Wiesfeld, A.C.P.; Cox, M.G.; van Lochem, L.T.; de Boer, R.A.; Hofstra, R.M.; et al. Phospholamban R14del mutation in patients diagnosed with dilated cardiomyopathy or arrhythmogenic right ventricular cardiomyopathy: Evidence supporting the concept of arrhythmogenic cardiomyopathy. Eur. J. Heart Fail. 2012, 14, 1199–1207. [Google Scholar] [CrossRef]
- van Rijsingen, I.A.W.; van der Zwaag, P.A.; Groeneweg, J.A.; Nannenberg, E.A.; Jongbloed, J.D.H.; Zwinderman, A.H.; Pinto, Y.M.; Deprez, R.H.L.D.; Post, J.G.; Tan, H.L.; et al. Outcome in Phospholamban R14del Carriers. Circ. Cardiovasc. Genet. 2014, 7, 455–465. [Google Scholar] [CrossRef]
- Vafiadaki, E.; Haghighi, K.; Arvanitis, D.A.; Kranias, E.G.; Sanoudou, D. Aberrant PLN-R14del Protein Interactions Intensify SERCA2a Inhibition, Driving Impaired Ca2+ Handling and Arrhythmogenesis. Int. J. Mol. Sci. 2022, 23, 6947. [Google Scholar] [CrossRef] [PubMed]
- Verstraelen, T.E.; van Lint, F.H.M.; Bosman, L.P.; de Brouwer, R.; Proost, V.M.; Abeln, B.G.S.; Taha, K.; Zwinderman, A.H.; Dickhoff, C.; Oomen, T.; et al. Prediction of ventricular arrhythmia in phospholamban p.Arg14del mutation carriers–reaching the frontiers of individual risk prediction. Eur. Heart J. 2021, 42, 2842–2850. [Google Scholar] [CrossRef] [PubMed]
- Groeneweg, J.A.; van der Zwaag, P.A.; Olde Nordkamp, L.R.A.; Bikker, H.; Jongbloed, J.D.H.; Jongbloed, R.; Wiesfeld, A.C.; Cox, M.G.; van der Heijden, J.F.; Atsma, D.E.; et al. Arrhythmogenic Right Ventricular Dysplasia/Cardiomyopathy According to Revised 2010 Task Force Criteria With Inclusion of Non-Desmosomal Phospholamban Mutation Carriers. Am. J. Cardiol. 2013, 112, 1197–1206. [Google Scholar] [CrossRef] [PubMed]
- Sepehrkhouy, S.; Gho, J.M.I.H.; van Es, R.; Harakalova, M.; de Jonge, N.; Dooijes, D.; van der Smagt, J.J.; Buijsrogge, M.P.; Hauer, R.N.; Goldschmeding, R.; et al. Distinct fibrosis pattern in desmosomal and phospholamban mutation carriers in hereditary cardiomyopathies. Heart Rhythm. 2017, 14, 1024–1032. [Google Scholar] [CrossRef]
- Fish, M.; Shaboodien, G.; Kraus, S.; Sliwa, K.; Seidman, C.E.; Burke, M.A.; Crotti, L.; Schwartz, P.J.; Mayosi, B.M. Mutation analysis of the phospholamban gene in 315 South Africans with dilated, hypertrophic, peripartum and arrhythmogenic right ventricular cardiomyopathies. Sci. Rep. 2016, 6, 22235. [Google Scholar] [CrossRef]
- van der Heijden, J.F.; Hassink, R.J. The phospholamban p.Arg14del founder mutation in Dutch patients with arrhythmogenic cardiomyopathy. Neth. Heart J. 2013, 21, 284–285. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).