Expression Pattern and Molecular Mechanism of Oxidative Stress-Related Genes in Myocardial Ischemia–Reperfusion Injury
Abstract
:1. Introduction
2. Materials and Methods
2.1. Data Resource
2.2. Identification of the DEGs in Myocardial Ischemia-Reperfusion Injury
2.3. Identification of DEGs Associated with Oxidative Stress
2.4. GO Annotation and KEGG Pathway Enrichment Analysis
2.5. Construction of the Protein–Protein Interaction Network and Identification of Hub Genes
2.6. Validation of the Relationship between Common Differentially Expressed OSRGs and Reperfusion Time
2.7. Prediction of OSRG-Related miRNAs, Transcription Factors, and Targeted Therapeutic Drugs
2.8. Further Validation of Common Differentially Expressed OSRGs in External Dataset GSE4105
2.9. Establishment of Hypoxia–Reoxygenation Model in H9C2 Myocardial Cell Line
2.10. Real-Time Quantitative Polymerase Chain Reaction (RT-qPCR)
3. Results
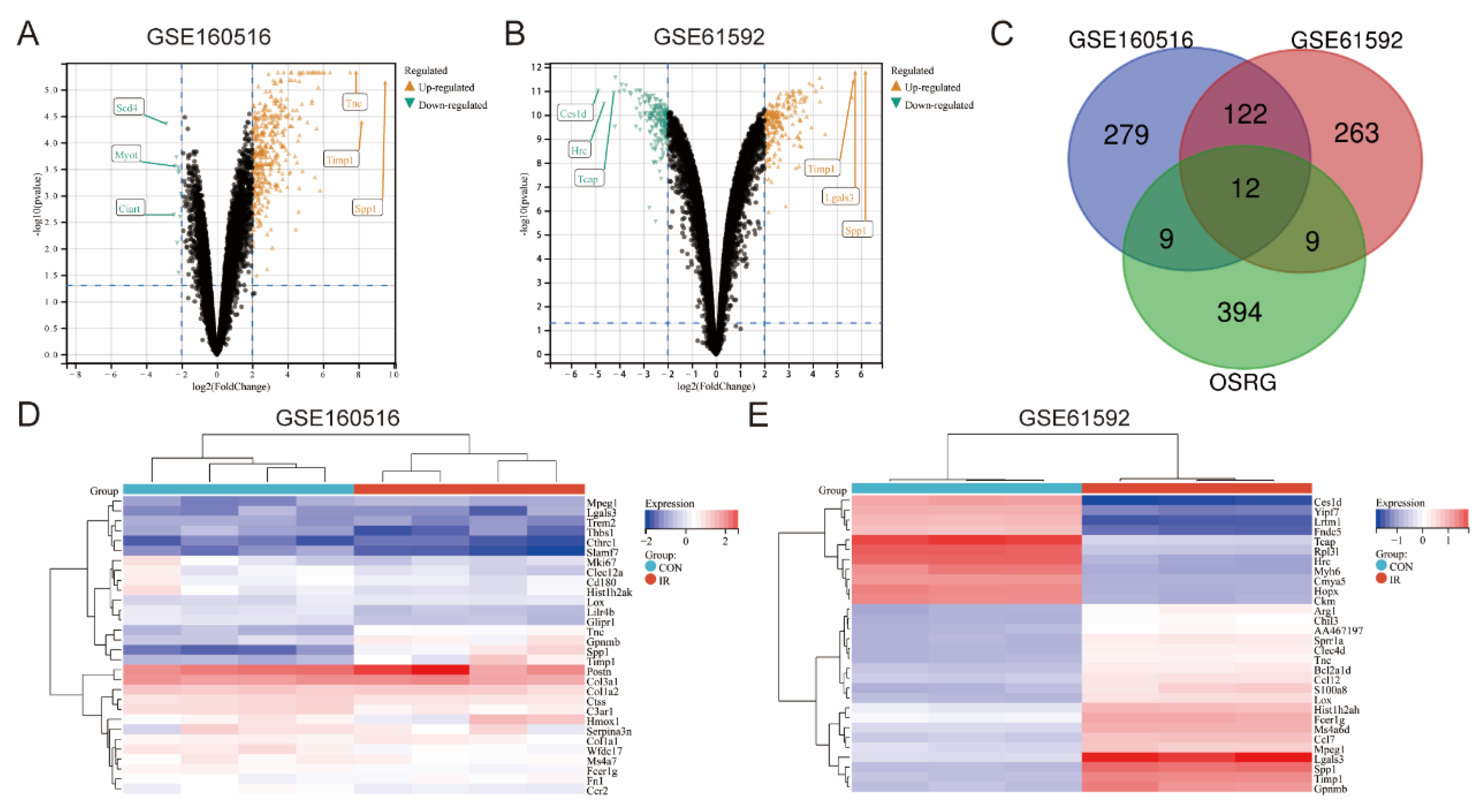
3.1. Identification of DEGs and Differentially Expressed OSRGs in Myocardial Ischemia-Reperfusion Injury
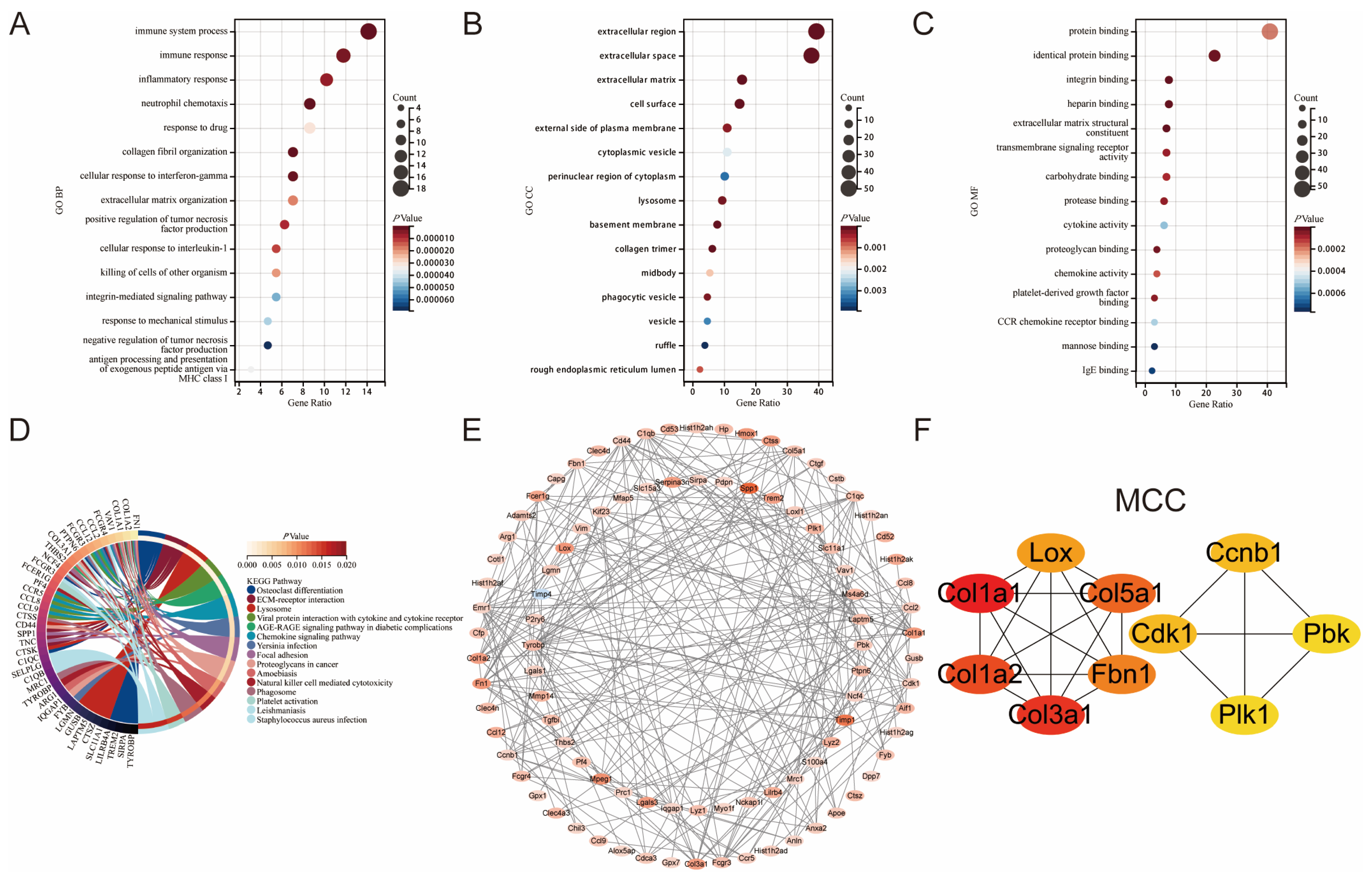
3.2. Enrichment Analysis of Common DEGs in GSE160516 and GSE61592
3.3. Construction of the Protein-Protein Interaction Network and Identification of Hub Genes in Myocardial Ischemia-Reperfusion Injury
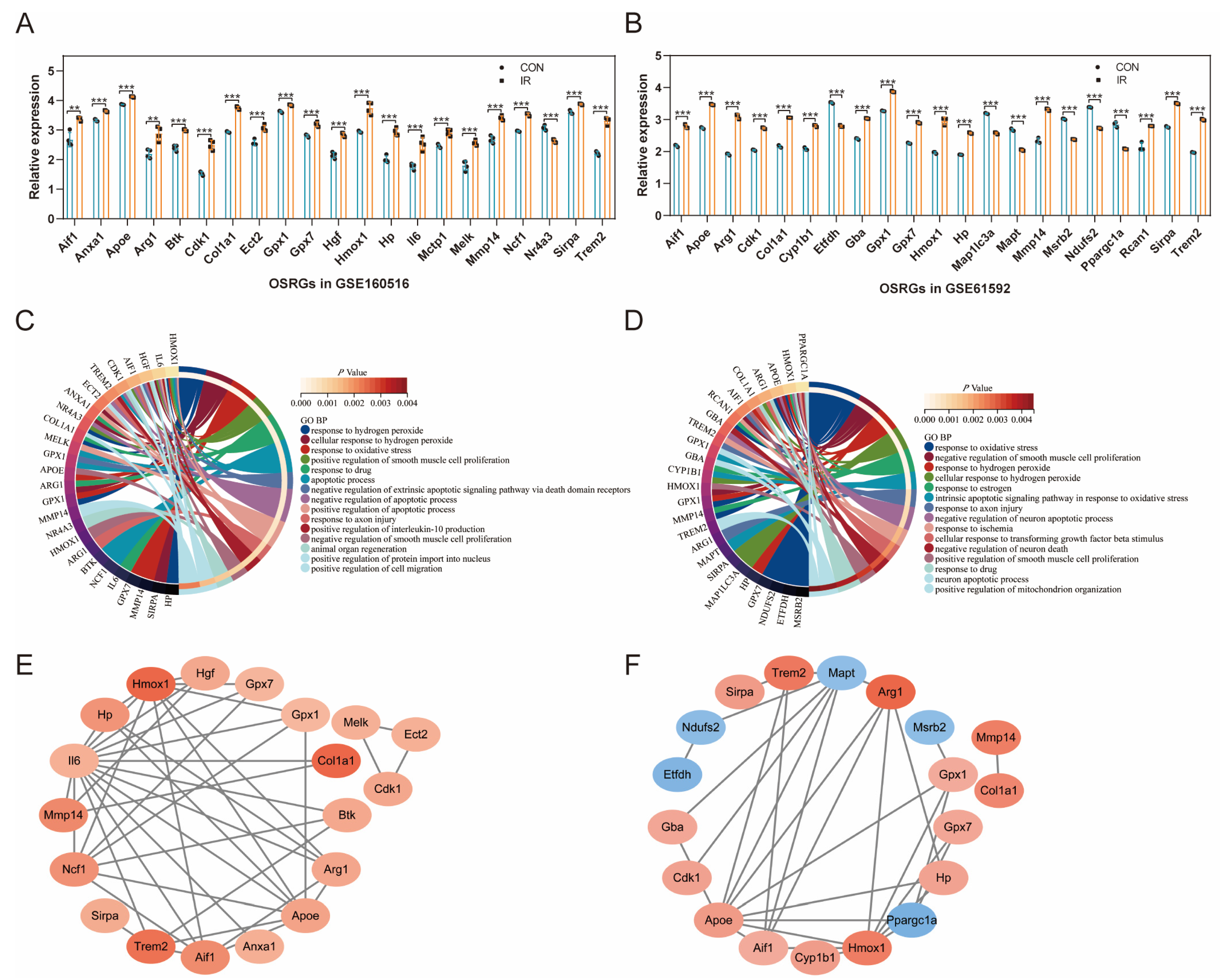
3.4. Expression Pattern of OSRGs in GSE160516 and GSE61592
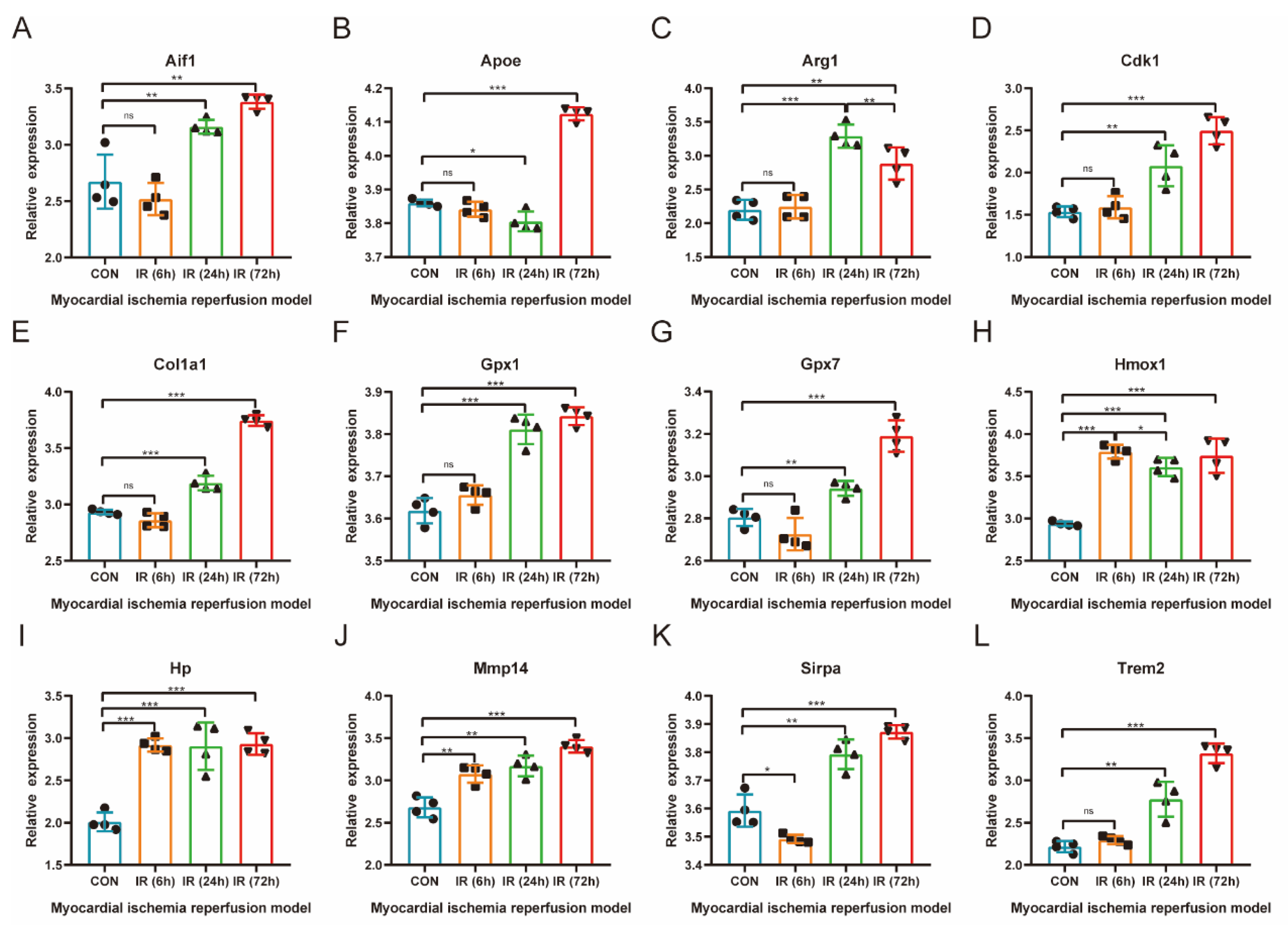
3.5. Validation of the Relationship between Common Differentially Expressed OSRGs and Reperfusion Time
3.6. Prediction of OSRGs-Related miRNAs, Transcription Factors, and Therapeutic Drugs
3.7. Further Validation of Common Differentially Expressed OSRGs in External Dataset GSE4105
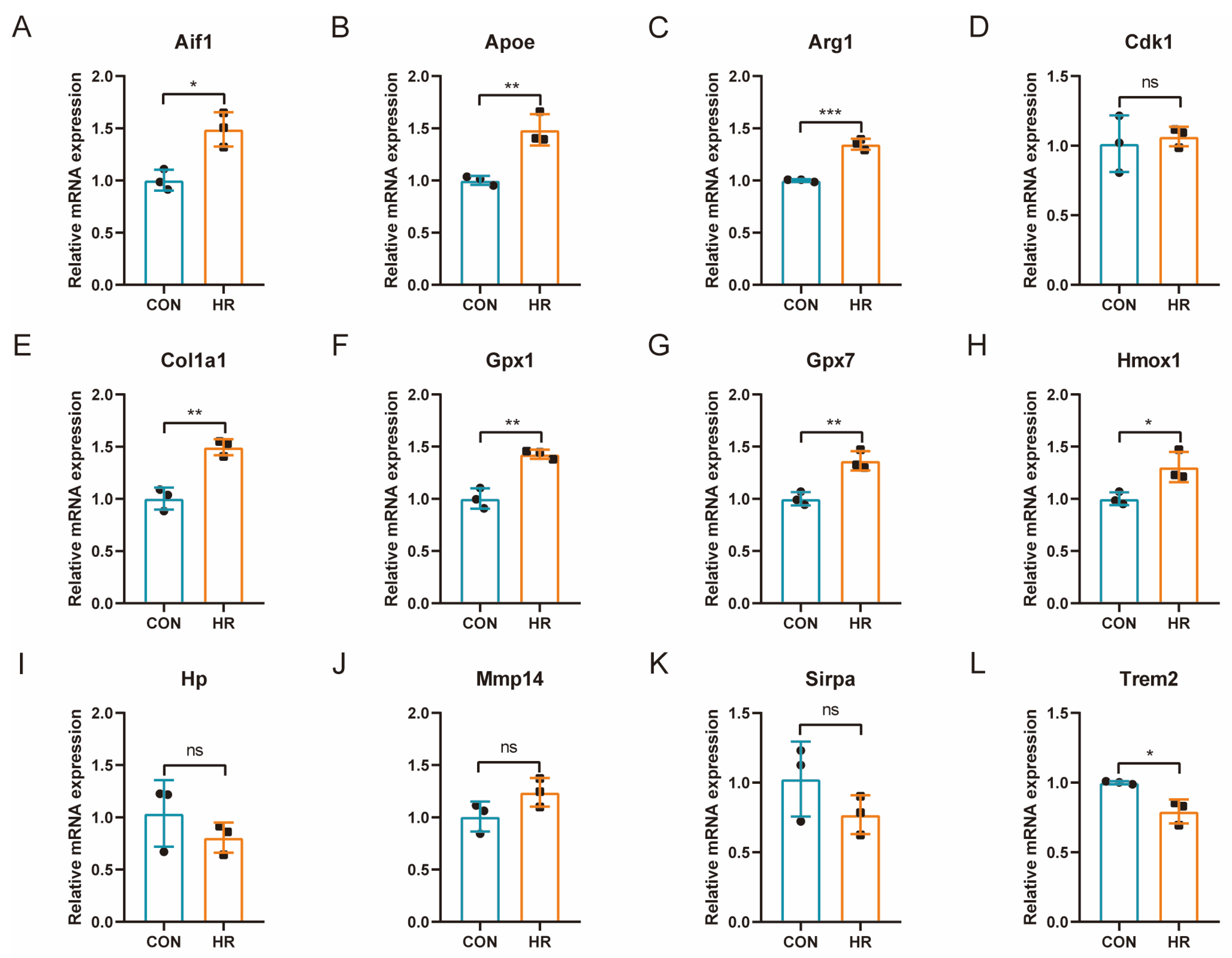
3.8. qRT-PCR Verification of OSRGs in H9C2 Cell Hypoxia–Reoxygenation Model
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AMI | Acute myocardial infarction |
| BP | Biological process |
| CC | Cellular component |
| DAMP | Damage-associated molecular pattern |
| DEG | Differentially expressed genes |
| GEO | Gene expression omnibus |
| H2O2 | Hydrogen peroxide |
| HR | Hypoxia–reoxygenation |
| IR | Ischemia–reperfusion |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| LAD | Left anterior descending |
| MF | Molecular function |
| MIRI | Myocardial ischemia–reperfusion injury |
| MSigDB | Molecular signatures database |
| OS | Oxidative stress |
| OSRG | Oxidative stress-related gene |
| PCI | Percutaneous coronary intervention |
| PPI | Protein–protein interaction |
| ROS | Reactive oxygen species |
| TF | Transcription factor |
References
- Yellon, D.M.; Hausenloy, D.J. Myocardial reperfusion injury. N. Engl. J. Med. 2007, 357, 1121–1135. [Google Scholar] [CrossRef]
- Fernandez-Jimenez, R.; Galan-Arriola, C.; Sanchez-Gonzalez, J.; Aguero, J.; Lopez-Martin, G.J.; Gomez-Talavera, S.; Garcia-Prieto, J.; Benn, A.; Molina-Iracheta, A.; Barreiro-Perez, M.; et al. Effect of Ischemia Duration and Protective Interventions on the Temporal Dynamics of Tissue Composition After Myocardial Infarction. Circ. Res. 2017, 121, 439–450. [Google Scholar] [CrossRef]
- Huang, C.; Zhou, S.; Chen, C.; Wang, X.; Ding, R.; Xu, Y.; Cheng, Z.; Ye, Z.; Sun, L.; Wang, Z.J.; et al. Biodegradable Redox-Responsive AIEgen-Based-Covalent Organic Framework Nanocarriers for Long-Term Treatment of Myocardial Ischemia/Reperfusion Injury. Small 2022, 18, e2205062. [Google Scholar] [CrossRef]
- Pan, Y.; Wang, X.; Liu, X.; Shen, L.; Chen, Q.; Shu, Q. Targeting Ferroptosis as a Promising Therapeutic Strategy for Ischemia-Reperfusion Injury. Antioxidants 2022, 11, 2196. [Google Scholar] [CrossRef]
- Tian, J.; Zheng, Y.; Mou, T.; Yun, M.; Tian, Y.; Lu, Y.; Bai, Y.; Zhou, Y.; Hacker, M.; Zhang, X.; et al. Metformin confers longitudinal cardiac protection by preserving mitochondrial homeostasis following myocardial ischemia/reperfusion injury. Eur. J. Nucl. Med. Mol. Imaging 2022, 50, 825–838. [Google Scholar] [CrossRef]
- Lillo-Moya, J.; Rojas-Sole, C.; Munoz-Salamanca, D.; Panieri, E.; Saso, L.; Rodrigo, R. Targeting Ferroptosis against Ischemia/Reperfusion Cardiac Injury. Antioxidants 2021, 10, 667. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; He, Z.; Xuan, Q.; Zhang, Y.; Xu, J.; Lin, J.; Li, H.; Chen, W.; Jiang, T. Analysis of the potential ferroptosis mechanism and multitemporal expression change of central ferroptosis-related genes in cardiac ischemia-reperfusion injury. Front. Physiol. 2022, 13, 934901. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Tang, Q.; Cao, W.; Zhou, N.; Jin, X.; Song, Z.; Zu, L.; Xu, S. Identification of a novel oxidative stress-related prognostic model in lung adenocarcinoma. Front. Pharmacol. 2022, 13, 1030062. [Google Scholar] [CrossRef]
- Kirtonia, A.; Sethi, G.; Garg, M. The multifaceted role of reactive oxygen species in tumorigenesis. Cell Mol. Life Sci. 2020, 77, 4459–4483. [Google Scholar] [CrossRef] [PubMed]
- Kazemian, S.; Ahmadi, R.; Rafiei, A.; Azadegan-Dehkordi, F.; Khaledifar, A.; Abdollahpour-Alitappeh, M.; Bagheri, N. The Serum Levels of IL-36 in Patients with Coronary Artery Disease and Their Correlation with the Serum Levels of IL-32, IL-6, TNF-alpha, and Oxidative Stress. Int. Arch Allergy Immunol. 2022, 183, 1137–1145. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.; Guo, W.; Shang, F.; Li, Y.; Li, W.; Liu, J.; Ma, C.; Teng, J. Bakuchiol Alleviates Hyperglycemia-Induced Diabetic Cardiomyopathy by Reducing Myocardial Oxidative Stress via Activating the SIRT1/Nrf2 Signaling Pathway. Oxid. Med. Cell. Longev. 2020, 2020, 3732718. [Google Scholar] [CrossRef] [PubMed]
- Szygula-Jurkiewicz, B.; Szczurek-Wasilewicz, W.; Osadnik, T.; Frycz-Kurek, A.M.; Maciol-Skurk, K.; Malyszek-Tumidajewicz, J.; Skrzypek, M.; Romuk, E.; Gasior, M.; Banach, M.; et al. Oxidative Stress Markers in Hypertrophic Cardiomyopathy. Medicina 2021, 58, 31. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Hou, D.; He, J.; Zeng, X.; Liu, R.; Liu, L.; Li, T.; Xiao, Y.; Ma, R.; Huang, H.; et al. Oxidative-Damaged Mitochondria Activate GABARAPL1-Induced NLRP3 Inflammasomes in an Autophagic-Exosome Manner after Acute Myocardial Ischemia. Oxid. Med. Cell. Longev. 2022, 2022, 7958542. [Google Scholar] [CrossRef] [PubMed]
- Yu, F.; Liu, F.; Luo, J.Y.; Zhao, Q.; Wang, H.L.; Fang, B.B.; Li, X.M.; Yang, Y.N. Targeted activation of ERK1/2 reduces ischemia and reperfusion injury in hyperglycemic myocardium by improving mitochondrial function. Ann. Transl. Med. 2022, 10, 1238. [Google Scholar] [CrossRef]
- Mahdiani, S.; Omidkhoda, N.; Rezaee, R.; Heidari, S.; Karimi, G. Induction of JAK2/STAT3 pathway contributes to protective effects of different therapeutics against myocardial ischemia/reperfusion. Biomed. Pharmacother. 2022, 155, 113751. [Google Scholar] [CrossRef]
- Guo, X.; Hong, S.; He, H.; Zeng, Y.; Chen, Y.; Mo, X.; Li, J.; Li, L.; Steinmetz, R.; Liu, Q. NFkappaB promotes oxidative stress-induced necrosis and ischemia/reperfusion injury by inhibiting Nrf2-ARE pathway. Free Radic. Biol. Med. 2020, 159, 125–135. [Google Scholar] [CrossRef]
- Meng, X.; Zhang, L.; Han, B.; Zhang, Z. PHLDA3 inhibition protects against myocardial ischemia/reperfusion injury by alleviating oxidative stress and inflammatory response via the Akt/Nrf2 axis. Environ. Toxicol. 2021, 36, 2266–2277. [Google Scholar] [CrossRef]
- Wang, Y.; Chen, Z.; Li, Y.; Ma, L.; Zou, Y.; Wang, X.; Yin, C.; Pan, L.; Shen, Y.; Jia, J.; et al. Low density lipoprotein receptor related protein 6 (LRP6) protects heart against oxidative stress by the crosstalk of HSF1 and GSK3beta. Redox Biol. 2020, 37, 101699. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhao, Q.; Li, X.; Ji, F. Dexmedetomidine reversed hypoxia/reoxygenation injury-induced oxidative stress and endoplasmic reticulum stress-dependent apoptosis of cardiomyocytes via SIRT1/CHOP signaling pathway. Mol. Cell Biochem. 2021, 476, 2803–2812. [Google Scholar] [CrossRef] [PubMed]
- Tsai, K.L.; Hsieh, P.L.; Chou, W.C.; Cheng, H.C.; Huang, Y.T.; Chan, S.H. Dapagliflozin attenuates hypoxia/reoxygenation-caused cardiac dysfunction and oxidative damage through modulation of AMPK. Cell Biosci. 2021, 11, 44. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Yang, C.; Liu, Y.J.; Zeng, P.; Gong, T.; Tao, L.; Li, X.A. Analysis of susceptibility genes and myocardial infarction risk correlation of ischemic cardiomyopathy based on bioinformatics. J. Thorac. Dis. 2022, 14, 3445–3453. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Qi, L.; Li, Y.; Zhang, S.; Lin, L.; Zhou, L.; Han, W.; Qu, X.; Cai, J.; Ye, M.; et al. Association of Pericardiac Adipose Tissue With Coronary Artery Disease. Front. Endocrinol. 2021, 12, 724859. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Wang, X. miRDB: An online database for prediction of functional microRNA targets. Nucleic. Acids Res. 2020, 48, D127–D131. [Google Scholar] [CrossRef] [PubMed]
- Miao, L.; Yin, R.X.; Zhang, Q.H.; Liao, P.J.; Wang, Y.; Nie, R.J.; Li, H. A novel circRNA-miRNA-mRNA network identifies circ-YOD1 as a biomarker for coronary artery disease. Sci. Rep. 2019, 9, 18314. [Google Scholar] [CrossRef]
- Han, H.; Cho, J.W.; Lee, S.; Yun, A.; Kim, H.; Bae, D.; Yang, S.; Kim, C.Y.; Lee, M.; Kim, E.; et al. TRRUST v2: An expanded reference database of human and mouse transcriptional regulatory interactions. Nucleic. Acids Res. 2018, 46, D380–D386. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Shao, J.; Lu, X.; Jiang, M.; Li, X.; Liu, Z.; Zhao, Y.; Zhou, J.; Lin, L.; Wang, L.; et al. Potential of immune-related genes as promising biomarkers for premature coronary heart disease through high throughput sequencing and integrated bioinformatics analysis. Front. Cardiovasc. Med. 2022, 9, 893502. [Google Scholar] [CrossRef]
- Lundberg, A.M.; Ketelhuth, D.F.; Johansson, M.E.; Gerdes, N.; Liu, S.; Yamamoto, M.; Akira, S.; Hansson, G.K. Toll-like receptor 3 and 4 signalling through the TRIF and TRAM adaptors in haematopoietic cells promotes atherosclerosis. Cardiovasc. Res. 2013, 99, 364–373. [Google Scholar] [CrossRef]
- Lee, S.M.; Hutchinson, M.; Saint, D.A. The role of Toll-like receptor 4 (TLR4) in cardiac ischaemic-reperfusion injury, cardioprotection and preconditioning. Clin. Exp. Pharmacol. Physiol. 2016, 43, 864–871. [Google Scholar] [CrossRef] [PubMed]
- Frangogiannis, N.G. Chemokines in ischemia and reperfusion. Thromb. Haemost. 2007, 97, 738–747. [Google Scholar] [CrossRef] [PubMed]
- Sui, Z.; Wang, M.M.; Xing, Y.; Qi, J.; Wang, W. Targeting MCOLN1/TRPML1 channels to protect against ischemia-reperfusion injury by restoring the inhibited autophagic flux in cardiomyocytes. Autophagy 2022, 18, 3053–3055. [Google Scholar] [CrossRef]
- Mauler, M.; Herr, N.; Schoenichen, C.; Witsch, T.; Marchini, T.; Hardtner, C.; Koentges, C.; Kienle, K.; Ollivier, V.; Schell, M.; et al. Platelet Serotonin Aggravates Myocardial Ischemia/Reperfusion Injury via Neutrophil Degranulation. Circulation 2019, 139, 918–931. [Google Scholar] [CrossRef] [PubMed]
- McDaniel, D.O.; Zhou, X.; Moore, C.K.; Aru, G. Cardiac allograft rejection correlates with increased expressions of Toll-like receptors 2 and 4 and allograft inflammatory factor 1. Transplant. Proc. 2010, 42, 4235–4237. [Google Scholar] [CrossRef] [PubMed]
- Huebener, P.; Abou-Khamis, T.; Zymek, P.; Bujak, M.; Ying, X.; Chatila, K.; Haudek, S.; Thakker, G.; Frangogiannis, N.G. CD44 is critically involved in infarct healing by regulating the inflammatory and fibrotic response. J. Immunol. 2008, 180, 2625–2633. [Google Scholar] [CrossRef] [PubMed]
- Erogbogbo, F.; May, J.; Swihart, M.; Prasad, P.N.; Smart, K.; Jack, S.E.; Korcyk, D.; Webster, M.; Stewart, R.; Zeng, I.; et al. Bioengineering silicon quantum dot theranostics using a network analysis of metabolomic and proteomic data in cardiac ischemia. Theranostics 2013, 3, 719–728. [Google Scholar] [CrossRef] [PubMed]
- Spahn, J.H.; Li, W.; Bribriesco, A.C.; Liu, J.; Shen, H.; Ibricevic, A.; Pan, J.H.; Zinselmeyer, B.H.; Brody, S.L.; Goldstein, D.R.; et al. DAP12 expression in lung macrophages mediates ischemia/reperfusion injury by promoting neutrophil extravasation. J. Immunol. 2015, 194, 4039–4048. [Google Scholar] [CrossRef]
- Zhang, X.; Cheng, L.; Xu, L.; Zhang, Y.; Yang, Y.; Fu, Q.; Mi, W.; Li, H. The lncRNA, H19 Mediates the Protective Effect of Hypoxia Postconditioning Against Hypoxia-Reoxygenation Injury to Senescent Cardiomyocytes by Targeting microRNA-29b-3p. Shock 2019, 52, 249–256. [Google Scholar] [CrossRef]
- Wang, Z.K.; Chen, R.R.; Li, J.H.; Chen, J.Y.; Li, W.; Niu, X.L.; Wang, F.F.; Wang, J.; Yang, J.X. Puerarin protects against myocardial ischemia/reperfusion injury by inhibiting inflammation and the NLRP3 inflammasome: The role of the SIRT1/NF-kappaB pathway. Int. Immunopharmacol. 2020, 89, 107086. [Google Scholar] [CrossRef]
- Xu, Y.; Wang, B.; Liu, X.; Deng, Y.; Zhu, Y.; Zhu, F.; Liang, Y.; Li, H. Sp1 Targeted PARP1 Inhibition Protects Cardiomyocytes FromMyocardial Ischemia-Reperfusion Injury via Downregulation of Autophagy. Front. Cell Dev. Biol. 2021, 9, 621906. [Google Scholar] [CrossRef]
- Heitmeier, M.R.; Payne, M.A.; Weinheimer, C.; Kovacs, A.; Hresko, R.C.; Jay, P.Y.; Hruz, P.W. Metabolic and Cardiac Adaptation to Chronic Pharmacologic Blockade of Facilitative Glucose Transport in Murine Dilated Cardiomyopathy and Myocardial Ischemia. Sci. Rep. 2018, 8, 6475. [Google Scholar] [CrossRef]
- Liu, Z.; Zhang, F.; Zhao, L.; Zhang, X.; Li, Y.; Liu, L. Protective Effect of Pravastatin on Myocardial Ischemia Reperfusion Injury by Regulation of the miR-93/Nrf2/ARE Signal Pathway. Drug Des. Devel Ther. 2020, 14, 3853–3864. [Google Scholar] [CrossRef]
- Fu, Y.; Wang, Z.; Chen, W.L.; Moore, P.K.; Zhu, Y.Z. Cardioprotective effects of nitric oxide-aspirin in myocardial ischemia-reperfused rats. Am. J. Physiol. Heart Circ. Physiol. 2007, 293, H1545–H1552. [Google Scholar] [CrossRef] [PubMed]
- Hao, J.B.; Liu, X.T.; Tang, J.; Chang, X.Y.; Honge, J.; Zhu, B.; Hao, L.R.; Zhang, L. The Effect of Allograft Inflammatory Factor-1 on Inflammation, Oxidative Stress, and Autophagy via miR-34a/ATG4B Pathway in Diabetic Kidney Disease. Oxid. Med. Cell. Longev. 2022, 2022, 1668000. [Google Scholar]
- Jiang, W.W.; Kong, L.B.; Wu, X.F.; Pu, L.Y.; Wang, X.H. Allograft inflammatory factor-1 is up-regulated in warm and cold ischemia-reperfusion injury in rat liver and may be inhibited by FK506. J. Surg. Res. 2011, 165, 158–164. [Google Scholar] [CrossRef] [PubMed]
- Krimbou, L.; Denis, M.; Haidar, B.; Carrier, M.; Marcil, M.; Genest, J.J. Molecular interactions between apoE and ABCA1: Impact on apoE lipidation. J. Lipid Res. 2004, 45, 839–848. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Zhang, S.; Ding, S.; Abudupataer, M.; Zhang, Z.; Zhu, X.; Zhang, W.; Zou, Y.; Yang, X.; Ge, J.; et al. Excessive Neutrophil Extracellular Trap Formation Aggravates Acute Myocardial Infarction Injury in Apolipoprotein E Deficiency Mice via the ROS-Dependent Pathway. Oxid. Med. Cell. Longev. 2019, 2019, 1209307. [Google Scholar] [CrossRef] [PubMed]
- Munder, M.; Mollinedo, F.; Calafat, J.; Canchado, J.; Gil-Lamaignere, C.; Fuentes, J.M.; Luckner, C.; Doschko, G.; Soler, G.; Eichmann, K.; et al. Arginase I is constitutively expressed in human granulocytes and participates in fungicidal activity. Blood 2005, 105, 2549–2556. [Google Scholar] [CrossRef]
- Fouda, A.Y.; Xu, Z.; Shosha, E.; Lemtalsi, T.; Chen, J.; Toque, H.A.; Tritz, R.; Cui, X.; Stansfield, B.K.; Huo, Y.; et al. Arginase 1 promotes retinal neurovascular protection from ischemia through suppression of macrophage inflammatory responses. Cell Death Dis. 2018, 9, 1001. [Google Scholar] [CrossRef]
- Peng, D.; Belkhiri, A.; Hu, T.; Chaturvedi, R.; Asim, M.; Wilson, K.T.; Zaika, A.; El-Rifai, W. Glutathione peroxidase 7 protects against oxidative DNA damage in oesophageal cells. Gut 2012, 61, 1250–1260. [Google Scholar] [CrossRef]
- Gozzelino, R.; Jeney, V.; Soares, M.P. Mechanisms of cell protection by heme oxygenase-1. Annu. Rev. Pharmacol. Toxicol. 2010, 50, 323–354. [Google Scholar] [CrossRef]
- Jiang, Y.; Chen, L.; Tang, Y.; Ma, G.; Shen, C.; Qi, C.; Zhu, Q.; Yao, Y.; Liu, N. HO-1 gene overexpression enhances the beneficial effects of superparamagnetic iron oxide labeled bone marrow stromal cells transplantation in swine hearts underwent ischemia/reperfusion: An MRI study. Basic Res. Cardiol. 2010, 105, 431–442. [Google Scholar] [CrossRef]
- Liu, X.; Wei, J.; Peng, D.H.; Layne, M.D.; Yet, S.F. Absence of heme oxygenase-1 exacerbates myocardial ischemia/reperfusion injury in diabetic mice. Diabetes 2005, 54, 778–784. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, J.; Luo, J.; Cai, H.; Li, C.; Lei, Z.; Lu, Y.; Ni, L.; Cao, J.; Cheng, B.; Hu, X. Expression Pattern and Molecular Mechanism of Oxidative Stress-Related Genes in Myocardial Ischemia–Reperfusion Injury. J. Cardiovasc. Dev. Dis. 2023, 10, 79. https://doi.org/10.3390/jcdd10020079
Wu J, Luo J, Cai H, Li C, Lei Z, Lu Y, Ni L, Cao J, Cheng B, Hu X. Expression Pattern and Molecular Mechanism of Oxidative Stress-Related Genes in Myocardial Ischemia–Reperfusion Injury. Journal of Cardiovascular Development and Disease. 2023; 10(2):79. https://doi.org/10.3390/jcdd10020079
Chicago/Turabian StyleWu, Jiahe, Jingyi Luo, Huanhuan Cai, Chenze Li, Zhe Lei, Yi Lu, Lihua Ni, Jianlei Cao, Bo Cheng, and Xiaorong Hu. 2023. "Expression Pattern and Molecular Mechanism of Oxidative Stress-Related Genes in Myocardial Ischemia–Reperfusion Injury" Journal of Cardiovascular Development and Disease 10, no. 2: 79. https://doi.org/10.3390/jcdd10020079
APA StyleWu, J., Luo, J., Cai, H., Li, C., Lei, Z., Lu, Y., Ni, L., Cao, J., Cheng, B., & Hu, X. (2023). Expression Pattern and Molecular Mechanism of Oxidative Stress-Related Genes in Myocardial Ischemia–Reperfusion Injury. Journal of Cardiovascular Development and Disease, 10(2), 79. https://doi.org/10.3390/jcdd10020079





