Cardiovascular and Renal Comorbidities Included into Neural Networks Predict the Outcome in COVID-19 Patients Admitted to an Intensive Care Unit: Three-Center, Cross-Validation, Age- and Sex-Matched Study
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients
2.2. Machine Learning
- Decision trees
- Random forests
- Extra trees
- Neural networks (multilayer perceptron)
- K-nearest neighbors
- Gradient boosting algorithms:
- 6.1.
- XGBoost
- 6.2.
- LightGBM
- 6.3.
- CatBoost
- Multivariable logistic regression as a reference
- Data preprocessing: missing data have been automatically inputted with median column values. Categorical data were replaced on the binary values, and multi categorical variables were converted into dummy variables.
- Power normalization: as some ML algorithms are sensitive to the data distribution, we applied Power transforms, a technique for transforming numerical input or output variables to have a Gaussian or more Gaussian-like probability distribution. This approach reduced data variability and skewness. The power transformation used in our study was based on the Yeo-Johnson transformation [40].
- Model hyperparameters were tuned by mljar-supervised “Compete” algorithm via hill climbing to fine-tune final models.
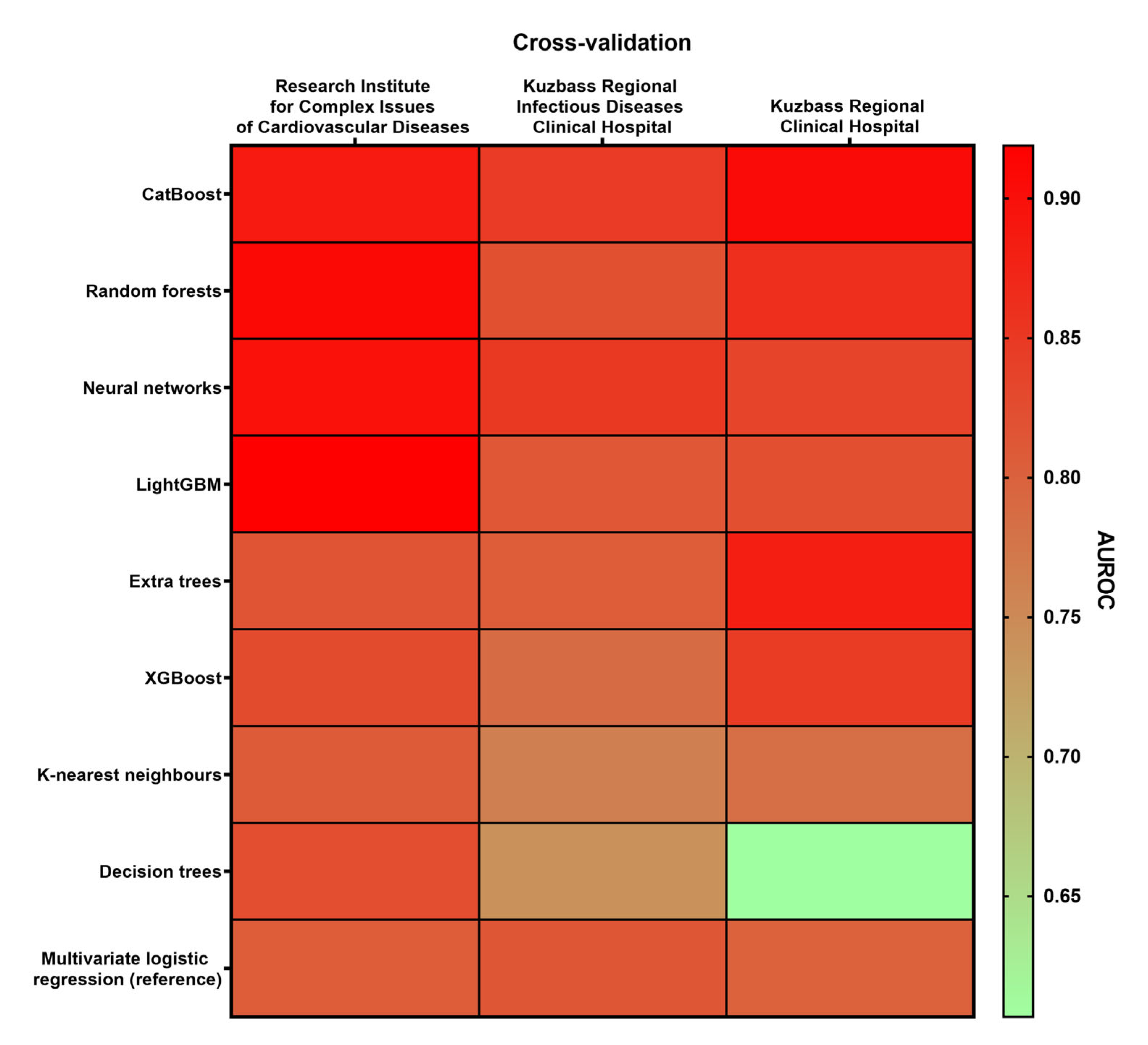
- Multicenter cross-validation. The general dataset was divided into two the learning dataset (including two sub-datasets from separate hospitals) and the test dataset (including a sub-dataset from the remaining hospital). This procedure was performed for all combinations of learning and test datasets: (1) learning dataset: Research Institute for Complex Issues of Cardiovascular Diseases and Kuzbass Regional Infectious Diseases Clinical Hospital (n = 206), cross-validation dataset: Kuzbass Regional Clinical Hospital (n = 144); (2) learning dataset: Research Institute for Complex Issues of Cardio-vascular Diseases and Kuzbass Regional Clinical Hospital (n = 244), cross-validation dataset: Kuzbass Regional Infectious Diseases Clinical Hospital (n = 106); (3) learning dataset: Kuzbass Regional Infectious Diseases Clinical Hospital and Kuzbass Regional Clinical Hospital (n = 250), cross-validation dataset: Research Institute for Complex Issues of Cardiovascular Diseases (n = 100).
- As the evaluation metrics, we used AUROC (the primary metric for optimization), %sensitivity, %specificity, and range (variability) of these parameters between the distinct study centers. For binary classifications, we used the default probability threshold of 0.5 (irrespective of the number of folds for cross-validation) and then calculated sensitivity and specificity. We deliberately excluded the optimization of the probability threshold to ensure an equal evaluation for each cross-validation fold. Such a custom cross-validation strategy included training on the data from the two clinics and cross-validation on the dataset from the remaining clinic, with the testing of all three possible combinations in this regard. As a consequence, we had three values (according to the number of cross-validation folds) for each of the selected metrics (sensitivity and specificity), which were obtained using a unified probability threshold (0.5). These metrics provided transparency and permitted the clinical interpretation of the algorithm efficiency.
- Out of all models, we selected those having the highest AUROC, %sensitivity, and %specificity (9 models in total, one per each ML algorithm: decision trees, random forests, extra trees, neural networks, k-nearest neighbors, gradient boosting (XGBoost, LightGBM, and CatBoost), and multivariate logistic regression as a reference). Optimal parameters for the best models developed by each machine learning approach are provided in Table S1.
- During the ML, we conducted a feature importance analysis using a SHAP (SHapley Additive exPlanations) technique [41], a game theoretic approach that explains the contribution of each feature to an individual predicted value (i.e., measures the impact of each factor into the output of any used ML model). For each of the selected models (n = 9 as described above), we quantified the feature importance within the [-1; 1] interval. Further, we applied a Predictor Screening tool of STATISTICA 13 software (TIBCO Software, Palo Alto, CA, USA).
- In addition to PyCharm integrated development environment, we have also used STATISTICA Automated Neural Networks (SANN) tool, which automatically generates, evaluates, and exports neural networks employing a multilayer perceptron architecture according to the input variables. The screening of the most efficient neural networks has been performed manually. When using this approach, ML and cross-validation have been carried out on a general dataset (70:30 learning:cross-validation samples proportion). In addition, the most efficient neural networks underwent cross-validation on four virtual patient samples generated by bootstrapping, a statistical procedure that resamples a single dataset by repeatedly drawing samples from the source data with replacement to create a simulated dataset.
3. Results
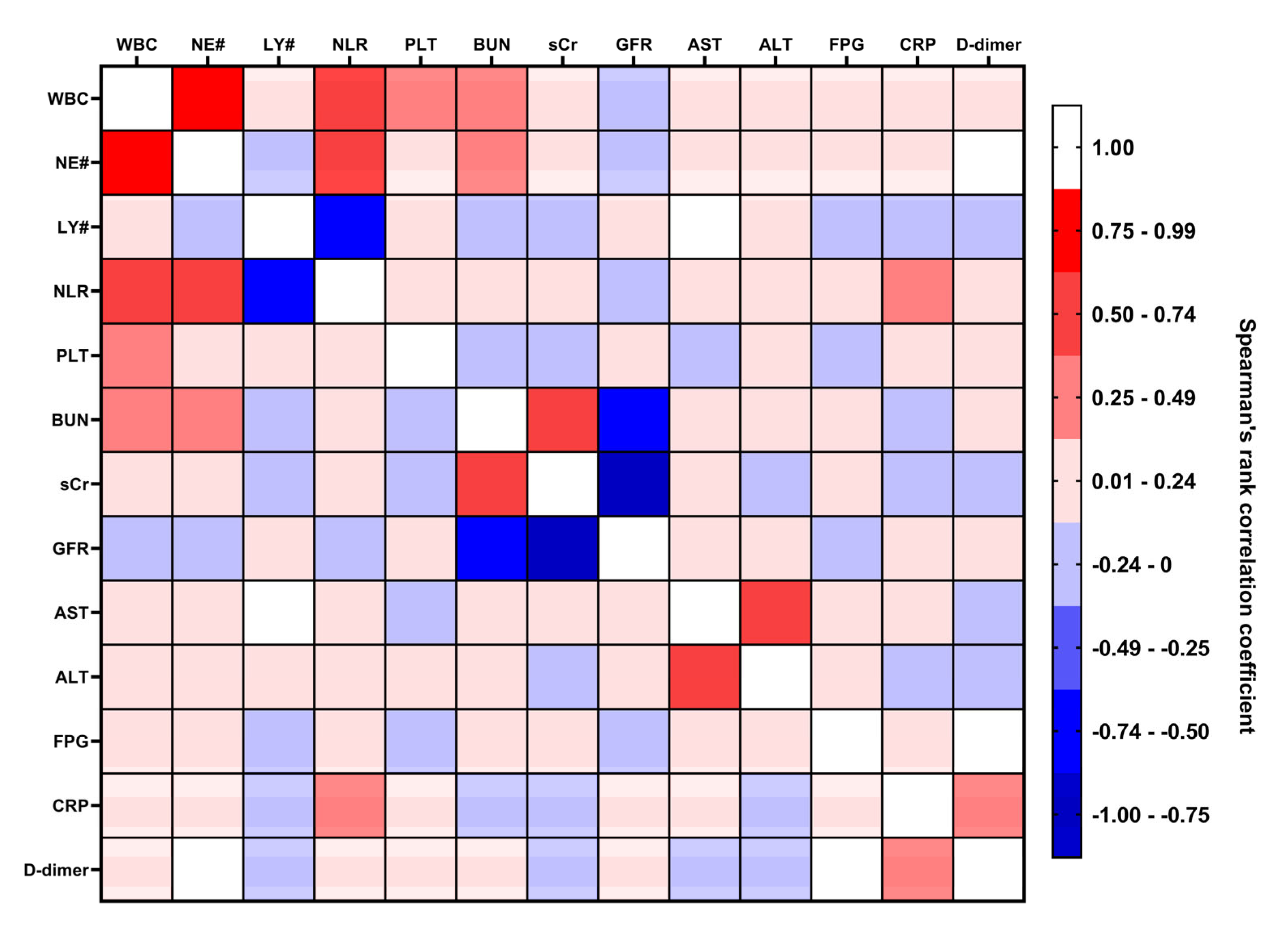
3.1. Univariate Analysis Identifies Cardiovascular Comorbidity, Immune Cell Counts, Kidney Dysfunction Markers, C-Reactive Protein, and D-Dimer Levels as the Potential Predictors of COVID-19-Related Death at the Stage of ICU Admission
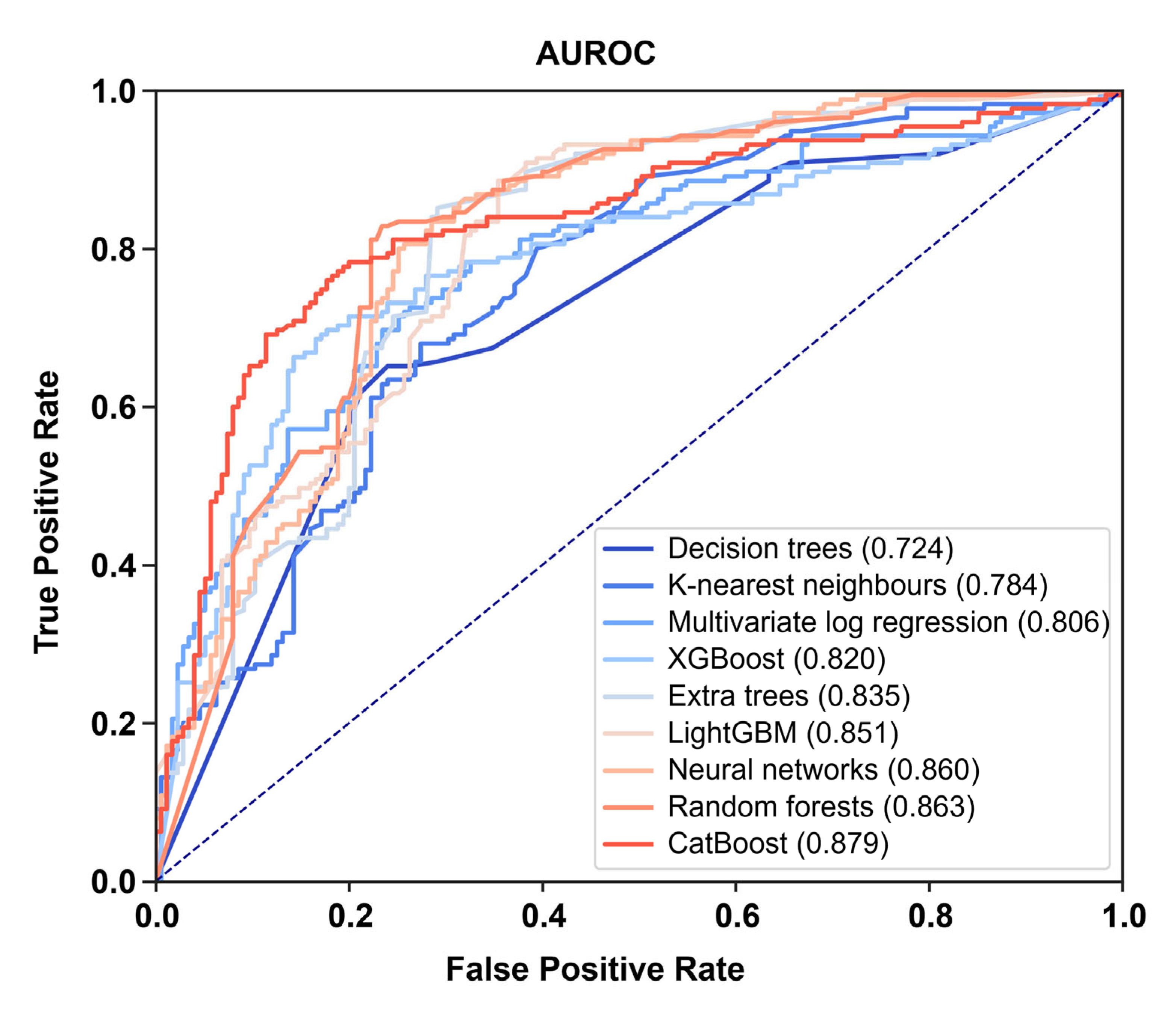
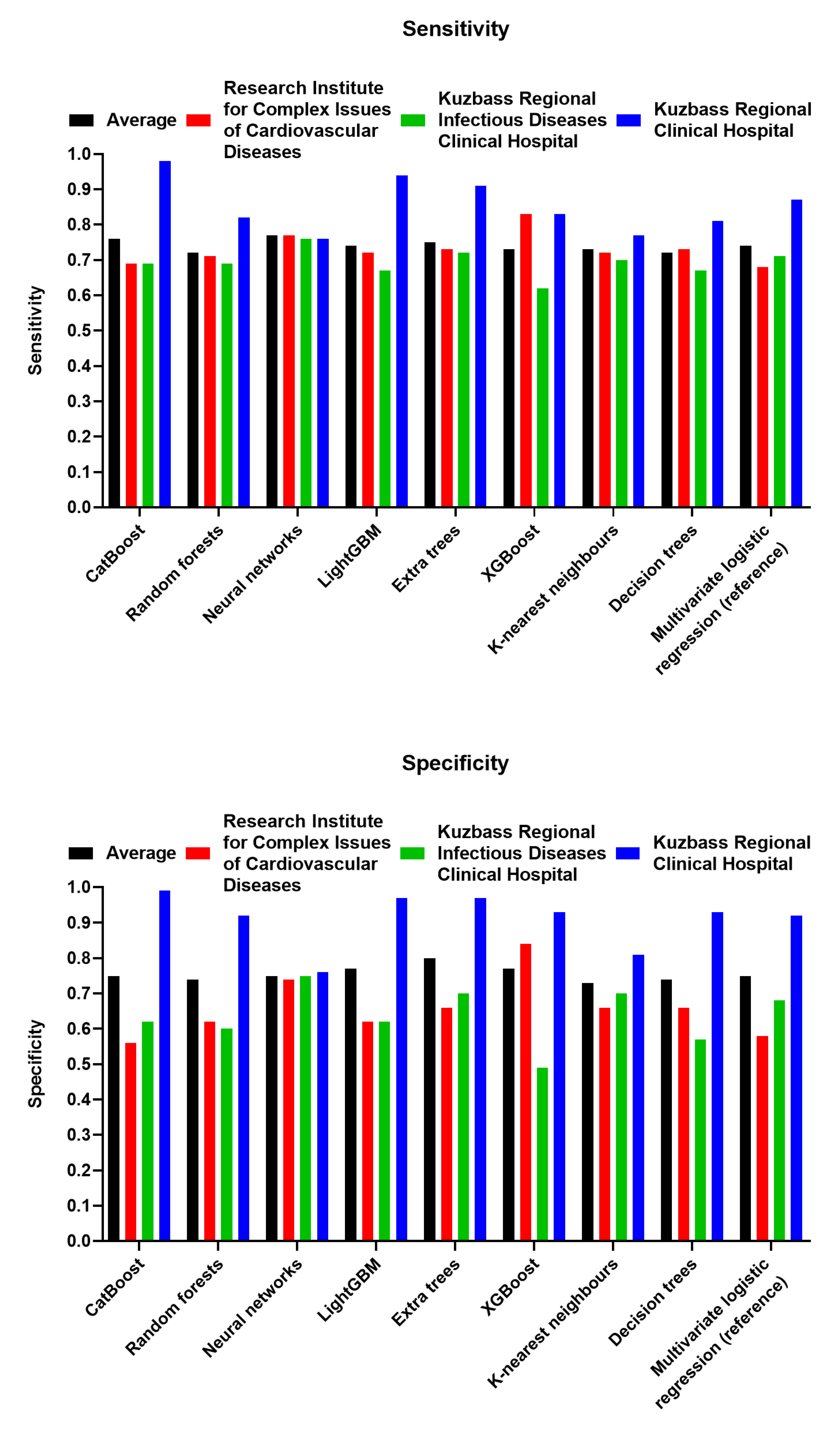
3.2. Neural Networks Represent the Most Reliable and Efficient Algorithm for the Prognostication of Patients Admitted to ICU with Severe COVID-19
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- WHO Coronavirus (COVID-19) Dashboard. Available online: https://covid19.who.int/ (accessed on 27 November 2022).
- Kamran, F.; Tang, S.; Otles, E.; McEvoy, D.S.; Saleh, S.N.; Gong, J.; Li, B.Y.; Dutta, S.; Liu, X.; Medford, R.J.; et al. Early identification of patients admitted to hospital for COVID-19 at risk of clinical deterioration: Model development and multisite external validation study. BMJ 2022, 376, e068576. [Google Scholar] [CrossRef] [PubMed]
- Yadaw, A.S.; Li, Y.C.; Bose, S.; Iyengar, R.; Bunyavanich, S.; Pandey, G. Clinical features of COVID-19 mortality: Development and validation of a clinical prediction model. Lancet Digit. Health 2020, 2, e516–e525. [Google Scholar] [CrossRef] [PubMed]
- Rasmy, L.; Nigo, M.; Kannadath, B.S.; Xie, Z.; Mao, B.; Patel, K.; Zhou, Y.; Zhang, W.; Ross, A.; Xu, H.; et al. Recurrent neural network models (CovRNN) for predicting outcomes of patients with COVID-19 on admission to hospital: Model development and validation using electronic health record data. Lancet Digit. Health 2022, 4, e415–e425. [Google Scholar] [CrossRef]
- Jiao, Z.; Choi, J.W.; Halsey, K.; Tran, T.M.L.; Hsieh, B.; Wang, D.; Eweje, F.; Wang, R.; Chang, K.; Wu, J.; et al. Prognostication of patients with COVID-19 using artificial intelligence based on chest x-rays and clinical data: A retrospective study. Lancet Digit. Health 2021, 3, e286–e294. [Google Scholar] [CrossRef] [PubMed]
- Mei, X.; Lee, H.C.; Diao, K.Y.; Huang, M.; Lin, B.; Liu, C.; Xie, Z.; Ma, Y.; Robson, P.M.; Chung, M.; et al. Artificial intelligence-enabled rapid diagnosis of patients with COVID-19. Nat. Med. 2020, 26, 1224–1228. [Google Scholar] [CrossRef]
- Meraihi, Y.; Gabis, A.B.; Mirjalili, S.; Ramdane-Cherif, A.; Alsaadi, F.E. Machine Learning-Based Research for COVID-19 Detection, Diagnosis, and Prediction: A Survey. SN Comput. Sci. 2022, 3, 286. [Google Scholar] [CrossRef]
- Ustebay, S.; Sarmis, A.; Kaya, G.K.; Sujan, M. A comparison of machine learning algorithms in predicting COVID-19 prognostics. Intern. Emerg. Med. 2022, 17, 1–11. [Google Scholar] [CrossRef]
- Fernandes, F.T.; de Oliveira, T.A.; Teixeira, C.E.; Batista, A.F.M.; Dalla Costa, G.; Chiavegatto Filho, A.D.P. A multipurpose machine learning approach to predict COVID-19 negative prognosis in São Paulo, Brazil. Sci. Rep. 2021, 11, 3343. [Google Scholar] [CrossRef]
- Jimenez-Solem, E.; Petersen, T.S.; Hansen, C.; Hansen, C.; Lioma, C.; Igel, C.; Boomsma, W.; Krause, O.; Lorenzen, S.; Selvan, R.; et al. Developing and validating COVID-19 adverse outcome risk prediction models from a bi-national European cohort of 5594 patients. Sci. Rep. 2021, 11, 3246. [Google Scholar] [CrossRef]
- Guan, X.; Zhang, B.; Fu, M.; Li, M.; Yuan, X.; Zhu, Y.; Peng, J.; Guo, H.; Lu, Y. Clinical and inflammatory features based machine learning model for fatal risk prediction of hospitalized COVID-19 patients: Results from a retrospective cohort study. Ann. Med. 2021, 53, 257–266. [Google Scholar] [CrossRef]
- An, C.; Lim, H.; Kim, D.W.; Chang, J.H.; Choi, Y.J.; Kim, S.W. Machine learning prediction for mortality of patients diagnosed with COVID-19: A nationwide Korean cohort study. Sci. Rep. 2020, 10, 18716. [Google Scholar] [CrossRef]
- Kukar, M.; Gunčar, G.; Vovko, T.; Podnar, S.; Černelč, P.; Brvar, M.; Zalaznik, M.; Notar, M.; Moškon, S.; Notar, M. COVID-19 diagnosis by routine blood tests using machine learning. Sci. Rep. 2021, 11, 10738. [Google Scholar] [CrossRef] [PubMed]
- Thell, R.; Zimmermann, J.; Szell, M.; Tomez, S.; Eisenburger, P.; Haugk, M.; Kreil, A.; Spiel, A.; Blaschke, A.; Klicpera, A.; et al. Standard blood laboratory values as a clinical support tool to distinguish between SARS-CoV-2 positive and negative patients. Sci. Rep. 2021, 11, 9365. [Google Scholar] [CrossRef] [PubMed]
- AlJame, M.; Imtiaz, A.; Ahmad, I.; Mohammed, A. Deep forest model for diagnosing COVID-19 from routine blood tests. Sci. Rep. 2021, 11, 16682. [Google Scholar] [CrossRef] [PubMed]
- Abayomi-Alli, O.O.; Damaševičius, R.; Maskeliūnas, R.; Misra, S. An Ensemble Learning Model for COVID-19 Detection from Blood Test Samples. Sensors 2022, 22, 2224. [Google Scholar] [CrossRef] [PubMed]
- Zuin, G.; Araujo, D.; Ribeiro, V.; Seiler, M.G.; Prieto, W.H.; Pintão, M.C.; Dos Santos Lazari, C.; Granato, C.F.H.; Veloso, A. Prediction of SARS-CoV-2-positivity from million-scale complete blood counts using machine learning. Commun. Med. 2022, 2, 72. [Google Scholar] [CrossRef] [PubMed]
- Bottino, F.; Tagliente, E.; Pasquini, L.; Napoli, A.D.; Lucignani, M.; Figà-Talamanca, L.; Napolitano, A. COVID Mortality Prediction with Machine Learning Methods: A Systematic Review and Critical Appraisal. J. Pers. Med. 2021, 11, 893. [Google Scholar] [CrossRef] [PubMed]
- Araújo, D.C.; Veloso, A.A.; Borges, K.B.G.; Carvalho, M.D.G. Prognosing the risk of COVID-19 death through a machine learning-based routine blood panel: A retrospective study in Brazil. Int. J. Med. Inform. 2022, 165, 104835. [Google Scholar] [CrossRef]
- Hu, C.; Liu, Z.; Jiang, Y.; Shi, O.; Zhang, X.; Xu, K.; Suo, C.; Wang, Q.; Song, Y.; Yu, K.; et al. Early prediction of mortality risk among patients with severe COVID-19, using machine learning. Int. J. Epidemiol. 2021, 49, 1918–1929. [Google Scholar] [CrossRef]
- Vaid, A.; Somani, S.; Russak, A.J.; De Freitas, J.K.; Chaudhry, F.F.; Paranjpe, I.; Johnson, K.W.; Lee, S.J.; Miotto, R.; Richter, F.; et al. Machine Learning to Predict Mortality and Critical Events in a Cohort of Patients With COVID-19 in New York City: Model Development and Validation. J. Med. Internet Res. 2020, 22, e24018. [Google Scholar] [CrossRef]
- Bertsimas, D.; Lukin, G.; Mingardi, L.; Nohadani, O.; Orfanoudaki, A.; Stellato, B.; Wiberg, H.; Gonzalez-Garcia, S.; Parra-Calderón, C.L.; Robinson, K.; et al. COVID-19 mortality risk assessment: An international multi-center study. PLoS ONE 2020, 15, e0243262. [Google Scholar] [CrossRef]
- Booth, A.L.; Abels, E.; McCaffrey, P. Development of a prognostic model for mortality in COVID-19 infection using machine learning. Mod. Pathol. 2021, 34, 522–531. [Google Scholar] [CrossRef] [PubMed]
- Ko, H.; Chung, H.; Kang, W.S.; Park, C.; Kim, D.W.; Kim, S.E.; Chung, C.R.; Ko, R.E.; Lee, H.; Seo, J.H.; et al. An Artificial Intelligence Model to Predict the Mortality of COVID-19 Patients at Hospital Admission Time Using Routine Blood Samples: Development and Validation of an Ensemble Model. J. Med. Internet Res. 2020, 22, e25442. [Google Scholar] [CrossRef] [PubMed]
- Jamshidi, E.; Asgary, A.; Tavakoli, N.; Zali, A.; Setareh, S.; Esmaily, H.; Jamaldini, S.H.; Daaee, A.; Babajani, A.; Sendani Kashi, M.A.; et al. Using Machine Learning to Predict Mortality for COVID-19 Patients on Day 0 in the ICU. Front. Digit. Health 2022, 3, 681608. [Google Scholar] [CrossRef]
- Afrash, M.R.; Shanbehzadeh, M.; Kazemi-Arpanahi, H. Predicting Risk of Mortality in COVID-19 Hospitalized Patients using Hybrid Machine Learning Algorithms. J. Biomed. Phys. Eng. 2022, 12, 611–626. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Lin, Y.; Zhu, T.; Fan, M.; Xu, S.; Qiu, W.; Chen, C.; Li, L.; Wang, Y.; Yan, J.; et al. Development and external evaluation of predictions models for mortality of COVID-19 patients using machine learning method. Neural Comput. Appl. 2021, 33, 1–10. [Google Scholar] [CrossRef]
- Farr, R.J.; Rootes, C.L.; Rowntree, L.C.; Nguyen, T.H.O.; Hensen, L.; Kedzierski, L.; Cheng, A.C.; Kedzierska, K.; Au, G.G.; Marsh, G.A.; et al. Altered microRNA expression in COVID-19 patients enables identification of SARS-CoV-2 infection. PLoS Pathog. 2021, 17, e1009759. [Google Scholar] [CrossRef]
- Fraser, D.D.; Patterson, E.K.; Slessarev, M.; Gill, S.E.; Martin, C.; Daley, M.; Miller, M.R.; Patel, M.A.; Dos Santos, C.C.; Bosma, K.J.; et al. Endothelial Injury and Glycocalyx Degradation in Critically Ill Coronavirus Disease 2019 Patients: Implications for Microvascular Platelet Aggregation. Crit. Care Explor. 2020, 2, e0194. [Google Scholar] [CrossRef]
- Fraser, D.D.; Cepinskas, G.; Slessarev, M.; Martin, C.; Daley, M.; Miller, M.R.; O’Gorman, D.B.; Gill, S.E.; Patterson, E.K.; Dos Santos, C.C. Inflammation Profiling of Critically Ill Coronavirus Disease 2019 Patients. Crit. Care Explor. 2020, 2, e0144. [Google Scholar] [CrossRef]
- Fraser, D.D.; Slessarev, M.; Martin, C.M.; Daley, M.; Patel, M.A.; Miller, M.R.; Patterson, E.K.; O’Gorman, D.B.; Gill, S.E.; Wishart, D.S.; et al. Metabolomics Profiling of Critically Ill Coronavirus Disease 2019 Patients: Identification of Diagnostic and Prognostic Biomarkers. Crit. Care Explor. 2020, 2, e0272. [Google Scholar] [CrossRef]
- Fraser, D.D.; Cepinskas, G.; Patterson, E.K.; Slessarev, M.; Martin, C.; Daley, M.; Patel, M.A.; Miller, M.R.; O’Gorman, D.B.; Gill, S.E.; et al. Novel Outcome Biomarkers Identified with Targeted Proteomic Analyses of Plasma from Critically Ill Coronavirus Disease 2019 Patients. Crit. Care Explor. 2020, 2, e0189. [Google Scholar] [CrossRef]
- Byeon, S.K.; Madugundu, A.K.; Garapati, K.; Ramarajan, M.G.; Saraswat, M.; Kumar-M, P.; Hughes, T.; Shah, R.; Patnaik, M.M.; Chia, N.; et al. Development of a multiomics model for identification of predictive biomarkers for COVID-19 severity: A retrospective cohort study. Lancet Digit. Health 2022, 4, e632–e645. [Google Scholar] [CrossRef]
- Fraser, D.D.; Cepinskas, G.; Slessarev, M.; Martin, C.M.; Daley, M.; Patel, M.A.; Miller, M.R.; Patterson, E.K.; O’Gorman, D.B.; Gill, S.E.; et al. Detection and Profiling of Human Coronavirus Immunoglobulins in Critically Ill Coronavirus Disease 2019 Patients. Crit. Care Explor. 2021, 3, e0369. [Google Scholar] [CrossRef] [PubMed]
- Hahm, C.R.; Lee, Y.K.; Oh, D.H.; Ahn, M.Y.; Choi, J.P.; Kang, N.R.; Oh, J.; Choi, H.; Kim, S. Factors Associated with Worsening Oxygenation in Patients with Non-severe COVID-19 Pneumonia. Tuberc. Respir. Dis. 2021, 84, 115–124. [Google Scholar] [CrossRef]
- Juneja, G.K.; Castelo, M.; Yeh, C.H.; Cerroni, S.E.; Hansen, B.E.; Chessum, J.E.; Abraham, J.; Cani, E.; Dwivedi, D.J.; Fraser, D.D.; et al. Biomarkers of coagulation, endothelial function, and fibrinolysis in critically ill patients with COVID-19: A single-center prospective longitudinal study. J. Thromb. Haemost. 2021, 19, 1546–1557. [Google Scholar] [CrossRef] [PubMed]
- Soya, E.; Ekenel, N.; Savas, R.; Toprak, T.; Bewes, J.; Doganay, O. Pixel-based analysis of pulmonary changes on CT lung images due to COVID-19 pneumonia. J. Clin. Imaging Sci. 2022, 12, 6. [Google Scholar] [CrossRef] [PubMed]
- Attia, Z.I.; Kapa, S.; Dugan, J.; Pereira, N.; Noseworthy, P.A.; Jimenez, F.L.; Cruz, J.; Carter, R.E.; DeSimone, D.C.; Signorino, J.; et al. Rapid Exclusion of COVID Infection with the Artificial Intelligence Electrocardiogram. Mayo Clin. Proc. 2021, 96, 2081–2094. [Google Scholar] [CrossRef]
- Uddin, S.; Khan, A.; Hossain, M.E.; Moni, M.A. Comparing different supervised machine learning algorithms for disease prediction. BMC Med. Inform. Decis. Mak. 2019, 19, 281. [Google Scholar] [CrossRef]
- Yeo, I.-K. A new family of power transformations to improve normality or symmetry. Biometrika 2000, 87, 954–959. [Google Scholar] [CrossRef]
- Ning, Y.; Ong, M.E.H.; Chakraborty, B.; Goldstein, B.A.; Ting, D.S.W.; Vaughan, R.; Liu, N. Shapley variable importance cloud for interpretable machine learning. Patterns 2022, 3, 100452. [Google Scholar] [CrossRef]
- Gomes, R.; Kamrowski, C.; Langlois, J.; Rozario, P.; Dircks, I.; Grottodden, K.; Martinez, M.; Tee, W.Z.; Sargeant, K.; LaFleur, C.; et al. A Comprehensive Review of Machine Learning Used to Combat COVID-19. Diagnostics 2022, 12, 1853. [Google Scholar] [CrossRef]
- Dogan, O.; Tiwari, S.; Jabbar, M.A.; Guggari, S. A systematic review on AI/ML approaches against COVID-19 outbreak. Complex Intell. Syst. 2021, 7, 2655–2678. [Google Scholar] [CrossRef]
- Gupta, R.K.; Marks, M.; Samuels, T.H.A.; Luintel, A.; Rampling, T.; Chowdhury, H.; Quartagno, M.; Nair, A.; Lipman, M.; Abubakar, I.; et al. Systematic evaluation and external validation of 22 prognostic models among hospitalised adults with COVID-19: An observational cohort study. Eur. Respir. J. 2020, 56, 2003498. [Google Scholar] [CrossRef]
- Khan, M.; Mehran, M.T.; Haq, Z.U.; Ullah, Z.; Naqvi, S.R.; Ihsan, M.; Abbass, H. Applications of artificial intelligence in COVID-19 pandemic: A comprehensive review. Expert Syst. Appl. 2021, 185, 115695. [Google Scholar] [CrossRef]
- Syeda, H.B.; Syed, M.; Sexton, K.W.; Syed, S.; Begum, S.; Syed, F.; Prior, F.; Yu, F., Jr. Role of Machine Learning Techniques to Tackle the COVID-19 Crisis: Systematic Review. JMIR Med. Inform. 2021, 9, e23811. [Google Scholar] [CrossRef] [PubMed]
- Kutikhin, A.G.; Shishkova, D.K.; Velikanova, E.A.; Sinitsky, M.Y.; Sinitskaya, A.V.; Markova, V.E. Endothelial Dysfunction in the Context of Blood-Brain Barrier Modeling. J. Evol. Biochem. Physiol. 2022, 58, 781–806. [Google Scholar] [CrossRef]
- Bogdanov, L.A.; Velikanova, E.A.; Kanonykina, A.Y.; Frolov, A.V.; Shishkova, D.K.; Lazebnaya, A.I.; Kutikhin, A.G. Vascular smooth muscle cell contractile proteins as universal markers of vessels of microcirculatory bed. Compl. Iss. Cardiovasc. Dis. 2022, 11, 162–176. [Google Scholar] [CrossRef]
- Shishkova, D.K.; Sinitskaya, A.V.; Sinitsky, M.Y.; Matveeva, V.G.; Velikanova, E.A.; Markova, V.E.; Kutikhin, A.G. Spontaneous endothelial-to-mesenchymal transition in human primary umbilical vein endothelial cells. Compl. Iss. Cardiovasc. Dis. 2022, 11, 97–114. [Google Scholar] [CrossRef]
- Vincent, J.L.; Levi, M.; Hunt, B.J. Prevention and management of thrombosis in hospitalised patients with COVID-19 pneumonia. Lancet Respir. Med. 2022, 10, 214–220. [Google Scholar] [CrossRef] [PubMed]
- Pelle, M.C.; Zaffina, I.; Lucà, S.; Forte, V.; Trapanese, V.; Melina, M.; Giofrè, F.; Arturi, F. Endothelial Dysfunction in COVID-19: Potential Mechanisms and Possible Therapeutic Options. Life 2022, 12, 1605. [Google Scholar] [CrossRef] [PubMed]
- Six, I.; Guillaume, N.; Jacob, V.; Mentaverri, R.; Kamel, S.; Boullier, A.; Slama, M. The Endothelium and COVID-19: An Increasingly Clear Link Brief Title: Endotheliopathy in COVID-19. Int. J. Mol. Sci. 2022, 23, 6196. [Google Scholar] [CrossRef] [PubMed]
- Ambrosino, P.; Calcaterra, I.L.; Mosella, M.; Formisano, R.; D’Anna, S.E.; Bachetti, T.; Marcuccio, G.; Galloway, B.; Mancini, F.P.; Papa, A.; et al. Endothelial Dysfunction in COVID-19: A Unifying Mechanism and a Potential Therapeutic Target. Biomedicines 2022, 10, 812. [Google Scholar] [CrossRef] [PubMed]
- Chee, M.L.; Ong, M.E.H.; Siddiqui, F.J.; Zhang, Z.; Lim, S.L.; Ho, A.F.W.; Liu, N. Artificial Intelligence Applications for COVID-19 in Intensive Care and Emergency Settings: A Systematic Review. Int. J. Environ. Res. Public Health 2021, 18, 4749. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Cai, G.Y.; Fang, W.; Li, H.Y.; Wang, S.Y.; Chen, L.; Yu, Y.; Liu, D.; Xu, S.; Cui, P.F.; et al. Machine learning based early warning system enables accurate mortality risk prediction for COVID-19. Nat. Commun. 2020, 11, 5033. [Google Scholar] [CrossRef]
- Abdulaal, A.; Patel, A.; Charani, E.; Denny, S.; Alqahtani, S.A.; Davies, G.W.; Mughal, N.; Moore, L.S.P. Comparison of deep learning with regression analysis in creating predictive models for SARS-CoV-2 outcomes. BMC Med. Inform. Decis. Mak. 2020, 20, 299. [Google Scholar] [CrossRef] [PubMed]
- Abdulaal, A.; Patel, A.; Charani, E.; Denny, S.; Mughal, N.; Moore, L. Prognostic Modeling of COVID-19 Using Artificial Intelligence in the United Kingdom: Model Development and Validation. J. Med. Internet Res. 2020, 22, e20259. [Google Scholar] [CrossRef]
- Italia, L.; Tomasoni, D.; Bisegna, S.; Pancaldi, E.; Stretti, L.; Adamo, M.; Metra, M. COVID-19 and Heart Failure: From Epidemiology During the Pandemic to Myocardial Injury, Myocarditis, and Heart Failure Sequelae. Front. Cardiovasc. Med. 2021, 8, 713560. [Google Scholar] [CrossRef]
- Sokolski, M.; Reszka, K.; Suchocki, T.; Adamik, B.; Doroszko, A.; Drobnik, J.; Gorka-Dynysiewicz, J.; Jedrzejczyk, M.; Kaliszewski, K.; Kilis-Pstrusinska, K.; et al. History of Heart Failure in Patients Hospitalized Due to COVID-19: Relevant Factor of In-Hospital Complications and All-Cause Mortality up to Six Months. J. Clin. Med. 2022, 11, 241. [Google Scholar] [CrossRef]
- Cheng, J.; Sollee, J.; Hsieh, C.; Yue, H.; Vandal, N.; Shanahan, J.; Choi, J.W.; Tran, T.M.L.; Halsey, K.; Iheanacho, F.; et al. COVID-19 mortality prediction in the intensive care unit with deep learning based on longitudinal chest X-rays and clinical data. Eur. Radiol. 2022, 32, 4446–4456. [Google Scholar] [CrossRef]
- Kar, S.; Chawla, R.; Haranath, S.P.; Ramasubban, S.; Ramakrishnan, N.; Vaishya, R.; Sibal, A.; Reddy, S. Multivariable mortality risk prediction using machine learning for COVID-19 patients at admission (AICOVID). Sci. Rep. 2021, 11, 12801. [Google Scholar] [CrossRef]
- Herzog, A.L.; von Jouanne-Diedrich, H.K.; Wanner, C.; Weismann, D.; Schlesinger, T.; Meybohm, P.; Stumpner, J. COVID-19 and the kidney: A retrospective analysis of 37 critically ill patients using machine learning. PLoS ONE 2021, 16, e0251932. [Google Scholar] [CrossRef] [PubMed]
- Rechtman, E.; Curtin, P.; Navarro, E.; Nirenberg, S.; Horton, M.K. Vital signs assessed in initial clinical encounters predict COVID-19 mortality in an NYC hospital system. Sci. Rep. 2020, 10, 21545. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.S.; Ge, P.; Jiang, C.; Zhang, Y.; Li, X.; Zhao, Z.; Zhang, L.; Duong, T.Q. Deep-learning artificial intelligence analysis of clinical variables predicts mortality in COVID-19 patients. J. Am. Coll. Emerg. Physicians Open 2020, 1, 1364–1373. [Google Scholar] [CrossRef] [PubMed]
- Fu, L.; Wang, B.; Yuan, T.; Chen, X.; Ao, Y.; Fitzpatrick, T.; Li, P.; Zhou, Y.; Lin, Y.F.; Duan, Q.; et al. Clinical characteristics of coronavirus disease 2019 (COVID-19) in China: A systematic review and meta-analysis. J. Infect. 2020, 80, 656–665. [Google Scholar] [CrossRef]
- Lippi, G.; Plebani, M. Laboratory abnormalities in patients with COVID-2019 infection. Clin. Chem. Lab. Med. 2020, 58, 1131–1134. [Google Scholar] [CrossRef]
- Kim, K.M.; Evans, D.S.; Jacobson, J.; Jiang, X.; Browner, W.; Cummings, S.R. Rapid prediction of in-hospital mortality among adults with COVID-19 disease. PLoS ONE 2022, 17, e0269813. [Google Scholar] [CrossRef]
- Lasso, G.; Khan, S.; Allen, S.A.; Mariano, M.; Florez, C.; Orner, E.P.; Quiroz, J.A.; Quevedo, G.; Massimi, A.; Hegde, A.; et al. Longitudinally monitored immune biomarkers predict the timing of COVID-19 outcomes. PLoS Comput. Biol. 2022, 18, e1009778. [Google Scholar] [CrossRef]
- Murri, R.; Lenkowicz, J.; Masciocchi, C.; Iacomini, C.; Fantoni, M.; Damiani, A.; Marchetti, A.; Sergi, P.D.A.; Arcuri, G.; Cesario, A.; et al. A machine-learning parsimonious multivariable predictive model of mortality risk in patients with COVID-19. Sci. Rep. 2021, 11, 21136. [Google Scholar] [CrossRef]
- Snider, J.M.; You, J.K.; Wang, X.; Snider, A.J.; Hallmark, B.; Zec, M.M.; Seeds, M.C.; Sergeant, S.; Johnstone, L.; Wang, Q.; et al. Group IIA secreted phospholipase A2 is associated with the pathobiology leading to COVID-19 mortality. J. Clin. Investig. 2021, 131, e149236. [Google Scholar] [CrossRef]
- Shiri, I.; Sorouri, M.; Geramifar, P.; Nazari, M.; Abdollahi, M.; Salimi, Y.; Khosravi, B.; Askari, D.; Aghaghazvini, L.; Hajianfar, G.; et al. Machine learning-based prognostic modeling using clinical data and quantitative radiomic features from chest CT images in COVID-19 patients. Comput. Biol. Med. 2021, 132, 104304. [Google Scholar] [CrossRef]

| Feature | Research Institute for Complex Issues of Cardiovascular Diseases (n = 100) | Kuzbass Regional Infectious Diseases Clinical Hospital (n = 106) | Kuzbass Regional Clinical Hospital (n = 144) | FDR-Corrected p Value | Average (n = 350) |
|---|---|---|---|---|---|
| Clinical data | |||||
| Sex, M/F, n (%) | 50/50 (50.00%/50.00%) | 53/53 (50.00%/50.00%) | 72/72 (50.00%/50.00%) | 1.00 | 175/175 (50.00%/50.00%) |
| Age, years, Me [IQR] | 73.00 [67.00–80.75] | 68.50 [62.75–79.00] | 64.00 [56.25–69.00] | 0.0001 | 68.00 [61.00–75.00] |
| AH, n (%) | 98/100 (98.00%) | 90/106 (84.90%) | 115/144 (79.86%) | 0.0001 | 297/350 (84.86%) |
| DM, n (%) | 35/100 (35.00%) | 36/106 (33.96%) | 63/144 (43.75%) | 0.21 | 131/350 (37.43%) |
| CAD/CHF, n (%) | 86/100 (86.00%) | 55/106 (51.89%) | 91/144 (63.19%) | 0.0001 | 232/350 (66.29%) |
| COPD/asthma, n (%) | 12/100 (12.00%) | 7/106 (6.60%) | 17/144 (11.80%) | 0.33 | 36/350 (10.29%) |
| Stage 3–5 CKD, n (%) | 34/100 (34.00%) | 10/106 (9.43%) | 38/144 (26.39%) | 0.0001 | 82/350 (23.43%) |
| Complete blood count measurements | |||||
| WBC, × 109/L, Me [IQR] | 7.30 [5.25–11.68] | 8.55 [5.50–13.45] | 10.75 [7.82–14.20] | 0.0001 | 9.10 [6.00–12.90] |
| NE#, × 109/L, Me [IQR] | 5.40 [3.25–9.10] | 7.55 [4.30–11.50] | 9.20 [6.60–12.40] | 0.0001 | 7.80 [4.50–11.30] |
| LY#, × 109/L, Me [IQR] | 1.20 [0.60–1.90] | 0.90 [0.40–1.20] | 0.80 [0.50–1.20] | 0.0003 | 0.90 [0.50–1.30] |
| NLR, Me [IQR] | 4.75 [2.20–12.40] | 9.40 [5.30–17.55] | 11.60 [6.87–17.60] | 0.0001 | 9.10 [4.70–16.33] |
| PLT, × 109/L, Me [IQR] | 193.5 [154.8–266.8] | 188.5 [156.0–253.5] | 239.5 [178.3–297.5] | 0.0003 | 216.0 [160.0–277.0] |
| Biochemical profiling | |||||
| BUN, mmol/L, Me [IQR] | 6.95 [5.97–9.32] | 7.80 [5.60–13.83] | 8.45 [6.00–12.05] | 0.09 | 7.80 [5.90–11.75] |
| sCr, µmol/L, Me [IQR] | 87.00 [74.00–107.00] | 97.00 [76.75–126.80] | 78.50 [65.25–106.50] | 0.0001 | 85.00 [69.75–112.30] |
| GFR (CKD-EPI), mL/min/1.73 m2, Me [IQR] | 69.00 [51.00–85.00] | 60.00 [44.75–79.50] | 86.50 [57.25–98.75] | 0.0001 | 73.00 [50.75–93.00] |
| AST, U/L, Me [IQR] | 22.50 [18.25–35.00] | 39.00 [30.75–58.00] | 42.00 [27.00–66.75] | 0.0001 | 36.00 [25.00–55.25] |
| ALT, U/L, Me [IQR] | 21.50 [16.00–30.00] | 31.00 [25.75–47.00] | 38.50 [22.25–60.75] | 0.0001 | 30.00 [20.00–48.25] |
| FPG, mmol/L, Me [IQR] | 6.20 [5.30–7.37] | 7.10 [5.57–11.63] | 7.20 [5.60–9.45] | 0.0017 | 6.70 [5.50–8.92] |
| CRP, mg/L, Me [IQR] | 52.00 [17.25–165.00] | 41.50 [13.00–109.30] | 101.00 [47.75–164.80] | 0.0001 | 68.50 [22.75–140.00] |
| D-dimer, ng/mL, Me [IQR] | 2685 [856–6701] | 1011 [325–1379] | 2974 [1406–5528] | 0.0001 | 1802 [840–4320] |
| Outcome | |||||
| In-hospital death/hospital discharge, n (%) | 50/50 (50.00%/50.00%) | 53/53 (50.00%/50.00%) | 72/72 (50.00%/50.00%) | 1.00 | 175/175 (50.00%/50.00%) |
| Feature | In-Hospital Death (n = 175) | Hospital Discharge (n = 175) | p Value |
|---|---|---|---|
| Clinical data | |||
| Sex, M/F, n (%) | 79/96 (45.14%/54.86%) | 79/96 (45.14%/54.86%) | N/A |
| Age, years, Me [IQR] | 68.00 [61.00–75.00] | 68.00 [61.00–75.00] | N/A |
| AH, n (%) | 161/175 (92.00%) | 142/175 (81.14%) | 0.003 |
| DM, n (%) | 70/175 (40.00%) | 64/175 (36.57%) | 0.51 |
| CAD/CHF, n (%) | 153/175 (87.43%) | 79/175 (45.14%) | 0.0001 |
| COPD/asthma, n (%) | 16/175 (9.14%) | 20/175 (11.43%) | 0.48 |
| Stage 3–5 CKD, n (%) | 45/175 (25.71%) | 37/175 (21.14%) | 0.31 |
| Complete blood count measurements | |||
| WBC, × 109/L, Me [IQR] | 10.00 [6.70–14.30] | 8.70 [5.60–11.70] | 0.004 |
| NE#, × 109/L, Me [IQR] | 8.70 [5.50–12.80] | 6.80 [3.80–9.90] | 0.0001 |
| LY#, × 109/L, Me [IQR] | 0.70 [0.50–1.20] | 1.00 [0.70–1.50] | 0.0004 |
| NLR, Me [IQR] | 11.40 [6.80–20.60] | 6.90 [3.10–13.60] | 0.0001 |
| PLT, × 109/L, Me [IQR] | 208.0 [156.0–269.0] | 219.0 [167.0–284.0] | 0.09 |
| Biochemical profiling | |||
| BUN, mmol/L, Me [IQR] | 8.30 [6.50–13.10] | 7.40 [5.60–10.60] | 0.007 |
| sCr, µmol/L, Me [IQR] | 89.0 [72.0–120.0] | 83.0 [68.0–107.0] | 0.014 |
| GFR (CKD-EPI), mL/min/1.73 m2, Me [IQR] | 69.00 [48.00–90.00] | 75.00 [54.00–94.00] | 0.05 |
| AST, U/L, Me [IQR] | 37.00 [25.00–61.00] | 35.00 [23.00–50.00] | 0.07 |
| ALT, U/L, Me [IQR] | 28.00 [20.00–46.00] | 30.00 [20.00–50.00] | 0.54 |
| FPG, mmol/L, Me [IQR] | 7.10 [5.50–9.90] | 6.40 [5.50–8.30] | 0.13 |
| CRP, mg/L, Me [IQR] | 101.0 [50.0–164.0] | 37.0 [10.0–109.0] | 0.0001 |
| D-dimer, ng/mL, Me [IQR] | 2770 [1194–5001] | 1263 [565–3463] | 0.0001 |
| Machine Learning Algorithm | AUROC | ||||
|---|---|---|---|---|---|
| Research Institute for Complex Issues of Cardiovascular Diseases (n = 100) | Kuzbass Regional Infectious Diseases Clinical Hospital (n = 106) | Kuzbass Regional Clinical Hospital (n = 144) | Average | Range | |
| Decision trees | 0.824 | 0.740 | 0.607 | 0.724 | 0.217 |
| Random forests | 0.908 | 0.821 | 0.861 | 0.863 | 0.087 |
| Extra trees | 0.817 | 0.806 | 0.882 | 0.835 | 0.076 |
| Neural networks | 0.898 | 0.849 | 0.834 | 0.860 | 0.064 |
| K-nearest neighbors | 0.807 | 0.763 | 0.782 | 0.784 | 0.044 |
| XGBoost | 0.827 | 0.787 | 0.845 | 0.820 | 0.058 |
| LightGBM | 0.919 | 0.812 | 0.822 | 0.851 | 0.107 |
| CatBoost | 0.887 | 0.846 | 0.905 | 0.879 | 0.059 |
| Multivariate logistic regression (reference) | 0.805 | 0.813 | 0.799 | 0.806 | 0.014 |
| Machine Learning Algorithm | Average Sens. | Average Spec. | Range (Sens.) | Range (Spec.) | Rank (Average Sens.) | Rank (Average Spec.) | Rank (Total) |
|---|---|---|---|---|---|---|---|
| Decision trees | 0.72 | 0.74 | 0.14 | 0.36 | 6 | 4 | 10 |
| Random forests | 0.72 | 0.74 | 0.13 | 0.31 | 6 | 4 | 10 |
| Extra trees | 0.75 | 0.80 | 0.19 | 0.31 | 3 | 1 | 4 |
| Neural networks | 0.77 | 0.75 | 0.01 | 0.02 | 1 | 3 | 4 |
| K-nearest neighbors | 0.73 | 0.73 | 0.07 | 0.15 | 5 | 5 | 10 |
| XGBoost | 0.73 | 0.77 | 0.21 | 0.44 | 5 | 2 | 7 |
| LightGBM | 0.74 | 0.77 | 0.26 | 0.35 | 4 | 2 | 6 |
| CatBoost | 0.76 | 0.75 | 0.28 | 0.43 | 2 | 3 | 5 |
| Multivariate logistic regression (reference) | 0.74 | 0.75 | 0.18 | 0.34 | 4 | 3 | 7 |
| Predictor | Gini | Information Value | Cramer’s V |
|---|---|---|---|
| CAD/CHF | 0.40 | 0.90 | 0.45 |
| CRP | 0.41 | 0.84 | 0.41 |
| LY# | 0.45 | 0.39 | 0.30 |
| NLR | 0.46 | 0.36 | 0.29 |
| D-dimer | 0.47 | 0.26 | 0.25 |
| FPG | 0.47 | 0.20 | 0.22 |
| NE# | 0.48 | 0.18 | 0.21 |
| PLT | 0.48 | 0.16 | 0.18 |
| WBC | 0.48 | 0.13 | 0.18 |
| BUN | 0.49 | 0.11 | 0.16 |
| AH | 0.49 | 0.11 | 0.16 |
| GFR | 0.49 | 0.11 | 0.16 |
| sCr | 0.49 | 0.10 | 0.15 |
| AST | 0.49 | 0.08 | 0.14 |
| ALT | 0.50 | 0.04 | 0.10 |
| Stage 3–5 CKD | 0.50 | 0.01 | 0.05 |
| COPD/asthma | 0.50 | 0.01 | 0.04 |
| DM | 0.50 | 0.00 | 0.04 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ovcharenko, E.; Kutikhin, A.; Gruzdeva, O.; Kuzmina, A.; Slesareva, T.; Brusina, E.; Kudasheva, S.; Bondarenko, T.; Kuzmenko, S.; Osyaev, N.; et al. Cardiovascular and Renal Comorbidities Included into Neural Networks Predict the Outcome in COVID-19 Patients Admitted to an Intensive Care Unit: Three-Center, Cross-Validation, Age- and Sex-Matched Study. J. Cardiovasc. Dev. Dis. 2023, 10, 39. https://doi.org/10.3390/jcdd10020039
Ovcharenko E, Kutikhin A, Gruzdeva O, Kuzmina A, Slesareva T, Brusina E, Kudasheva S, Bondarenko T, Kuzmenko S, Osyaev N, et al. Cardiovascular and Renal Comorbidities Included into Neural Networks Predict the Outcome in COVID-19 Patients Admitted to an Intensive Care Unit: Three-Center, Cross-Validation, Age- and Sex-Matched Study. Journal of Cardiovascular Development and Disease. 2023; 10(2):39. https://doi.org/10.3390/jcdd10020039
Chicago/Turabian StyleOvcharenko, Evgeny, Anton Kutikhin, Olga Gruzdeva, Anastasia Kuzmina, Tamara Slesareva, Elena Brusina, Svetlana Kudasheva, Tatiana Bondarenko, Svetlana Kuzmenko, Nikolay Osyaev, and et al. 2023. "Cardiovascular and Renal Comorbidities Included into Neural Networks Predict the Outcome in COVID-19 Patients Admitted to an Intensive Care Unit: Three-Center, Cross-Validation, Age- and Sex-Matched Study" Journal of Cardiovascular Development and Disease 10, no. 2: 39. https://doi.org/10.3390/jcdd10020039
APA StyleOvcharenko, E., Kutikhin, A., Gruzdeva, O., Kuzmina, A., Slesareva, T., Brusina, E., Kudasheva, S., Bondarenko, T., Kuzmenko, S., Osyaev, N., Ivannikova, N., Vavin, G., Moses, V., Danilov, V., Komossky, E., & Klyshnikov, K. (2023). Cardiovascular and Renal Comorbidities Included into Neural Networks Predict the Outcome in COVID-19 Patients Admitted to an Intensive Care Unit: Three-Center, Cross-Validation, Age- and Sex-Matched Study. Journal of Cardiovascular Development and Disease, 10(2), 39. https://doi.org/10.3390/jcdd10020039







