Identification of Riboflavin Metabolism Pathway in HepG2 Cells Expressing Genotype IV Swine Hepatitis E Virus ORF3 Protein
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Overexpression of SHEV ORF3 Genotype IV in HepG2 Cells by Recombinant Adenovirus AD-ORF3 and High-Throughput Sequencing of circRNAs and Transcriptome
2.2. Bioinformatics Analysis
2.3. Prediction of the circRNA-miRNA Regulatory Network of the Influence of SHEV ORF3 Genotype IV on the Riboflavin Metabolism Pathway in HepG2 Cells
3. Results
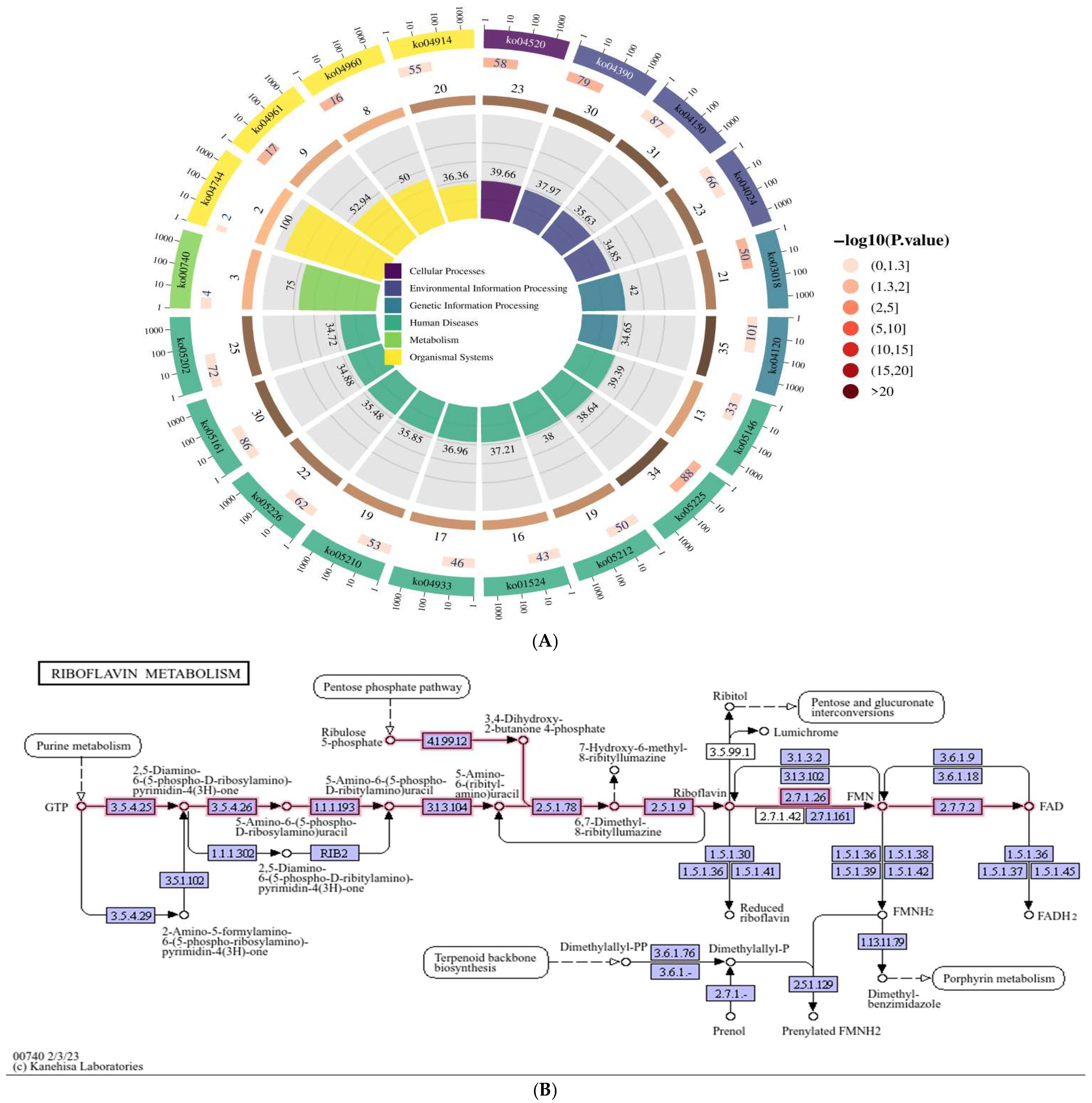
3.1. KEGG Functional Enrichment Analysis: Based on the circRNA Transcriptome Sequencing Data of HepG2 Cells Expressing Genotype IV SHEV ORF3, We Searched for the Riboflavin Metabolic Pathway
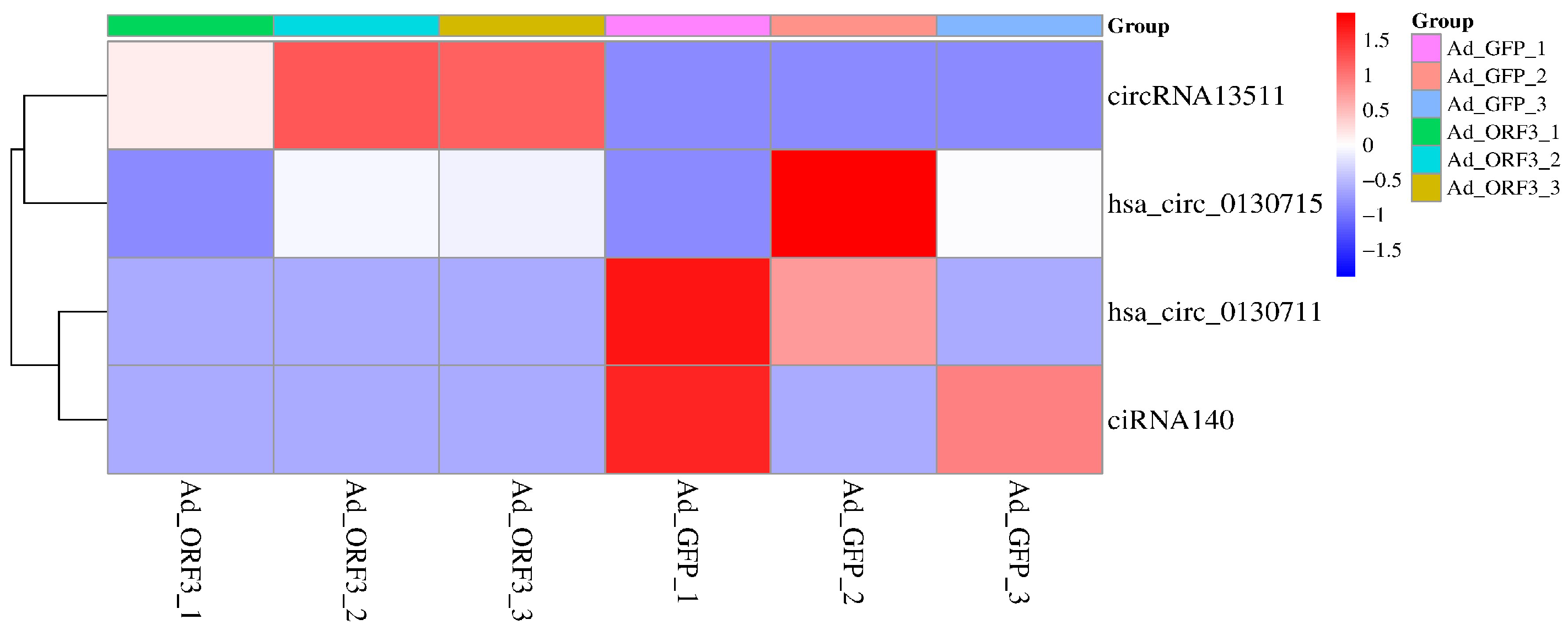
3.2. Screening of Significantly Differentially Expressed circRNAs in the Riboflavin Metabolism Pathway (ko00740)
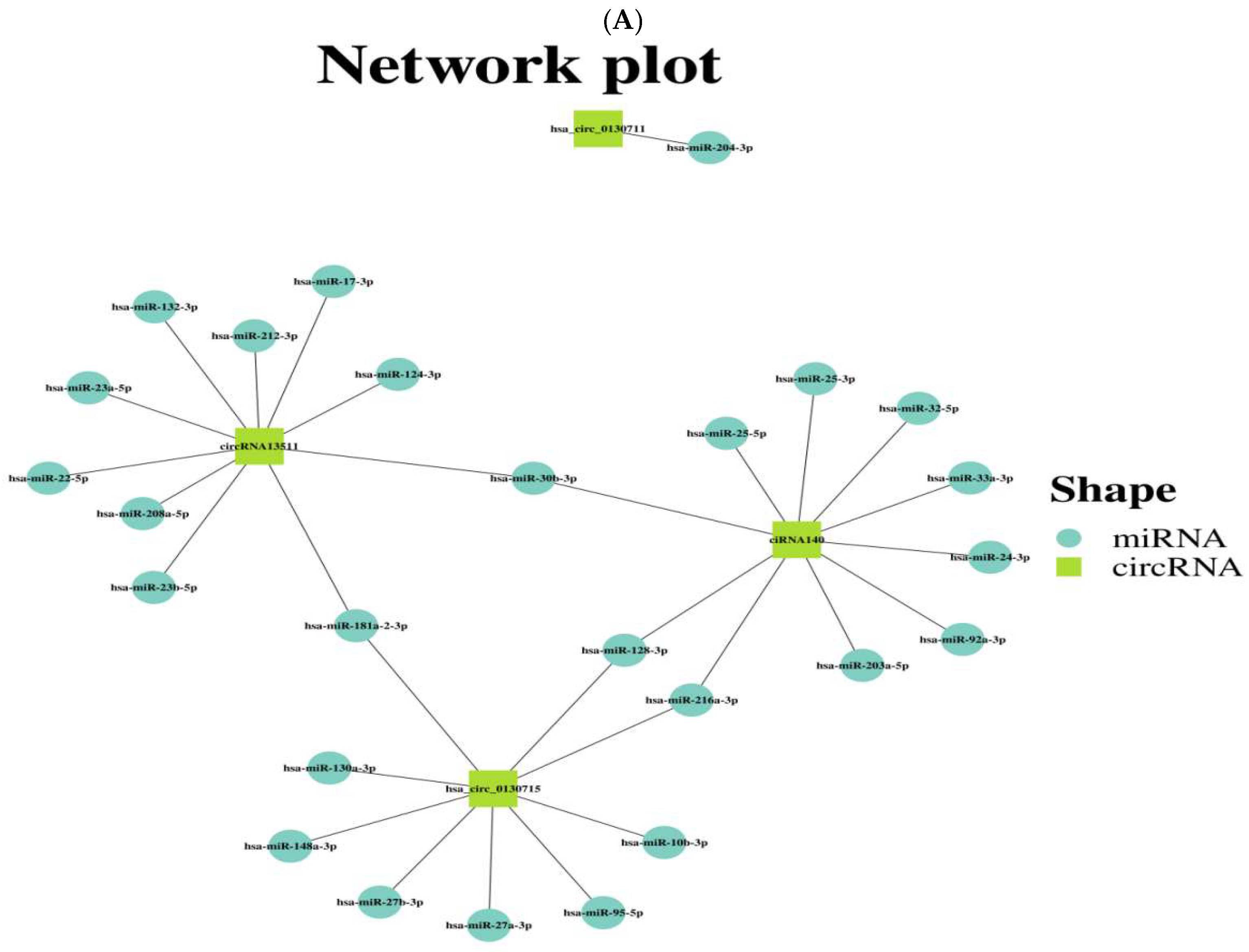
3.3. Predicting the Potential Target miRNAs of the Four Validated circRNAs and Preliminarily Exploring Their circRNA-miRNA Network
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Portais, A.; Le Marechal, M.; Nemoz, B.; Dartevel, A.; Rigault, G.; Pavese, P.; Pierre, I.; Terzi, N. Measles-associated pneumonia in an immunocompromised patient: Persistent shortcomings in vaccination guidelines. Infect. Dis. Now 2021, 51, 316–318. [Google Scholar] [CrossRef]
- Zhou, X.; Lu, H. Advances in Acute Severe Hepatitis of Unknown Etiology in Children. Zoonoses 2022, 2, 2737–7466. [Google Scholar] [CrossRef]
- Songtanin, B.; Molehin, A.J.; Brittan, K.; Manatsathit, W.; Nugent, K. Hepatitis E Virus Infections: Epidemiology, Genetic Diversity, and Clinical Considerations. Viruses 2023, 15, 1389. [Google Scholar] [CrossRef] [PubMed]
- Aslan, A.T.; Balaban, H.Y. Hepatitis E virus: Epidemiology, diagnosis, clinical manifestations, and treatment. World J. Gastroenterol. 2020, 26, 5543–5560. [Google Scholar] [CrossRef]
- Yang, Y.; Liu, B.; Tian, J.; Teng, X.; Liu, T. Vital role of autophagy flux inhibition of placental trophoblast cells in pregnancy disorders induced by HEV infection. Emerg. Microbes Infect. 2023, 12, 2276336. [Google Scholar] [CrossRef] [PubMed]
- Sooryanarain, H.; Meng, X.J. Hepatitis E virus: Reasons for emergence in humans. Curr. Opin. Virol. 2019, 34, 10–17. [Google Scholar] [CrossRef]
- Wolff, A.; Günther, T.; Johne, R. Stability of Hepatitis E Virus After Drying on Different Surfaces. Food Environ. Virol. 2022, 14, 138–148. [Google Scholar] [CrossRef]
- Li, X.; Sun, X.; Pinpin, J.; Zhao, Q.; Sun, Y. Multifunctional ORF3 protein of hepatitis E virus. J. Med. Virol. 2024, 96, e29691. [Google Scholar] [CrossRef]
- Koonin, E.V.; Gorbalenya, A.E.; Purdy, M.A.; Rozanov, M.N.; Reyes, G.R.; Bradley, D.W. Computer-assisted assignment of functional domains in the nonstructural polyprotein of hepatitis E virus: Delineation of an additional group of positive-strand RNA plant and animal viruses. Proc. Natl. Acad. Sci. USA 1992, 89, 8259–8263. [Google Scholar] [CrossRef]
- Kenney, S.P.; Pudupakam, R.S.; Huang, Y.W.; Pierson, F.W.; LeRoith, T.; Meng, X.J. The PSAP motif within the ORF3 protein of an avian strain of the hepatitis E virus is not critical for viral infectivity in vivo but plays a role in virus release. J. Virol. 2012, 86, 5637–5646. [Google Scholar] [CrossRef]
- Tian, D.; Yugo, D.M.; Kenney, S.P.; Lynn Heffron, C.; Opriessnig, T.; Karuppannan, A.K.; Bayne, J.; Halbur, P.G.; Meng, X.J. Dissecting the potential role of hepatitis E virus ORF1 nonstructural gene in cross-species infection by using intergenotypic chimeric viruses. J. Med. Virol. 2020, 92, 3563–3571. [Google Scholar] [CrossRef]
- Ding, Q.; Heller, B.; Capuccino, J.M.; Song, B.; Nimgaonkar, I.; Hrebikova, G.; Contreras, J.E.; Ploss, A. Hepatitis E virus ORF3 is a functional ion channel required for release of infectious particles. Proc. Natl. Acad. Sci. USA 2017, 114, 1147–1152. [Google Scholar] [CrossRef] [PubMed]
- McNulty, H.; Pentieva, K.; Ward, M. Causes and Clinical Sequelae of Riboflavin Deficiency. Annu. Rev. Nutr. 2023, 43, 101–122. [Google Scholar] [CrossRef] [PubMed]
- Da’dara, A.A.; Nation, C.S.; Skelly, P.J. Metabolism of FAD, FMN and riboflavin (vitamin B2) in the human parasitic blood fluke Schistosoma mansoni. BMC Infect. Dis. 2024, 24, 636. [Google Scholar] [CrossRef] [PubMed]
- Xu, N.; Wei, L.; Liu, J. Biotechnological advances and perspectives of gamma-aminobutyric acid production. World J. Microbiol. Biotechnol. 2017, 33, 64. [Google Scholar] [CrossRef]
- Jiao, H.; Zhao, Y.; Zhou, Z.; Li, W.; Li, B.; Gu, G.; Luo, Y.; Shuai, X.; Fan, C.; Wu, L.; et al. Identifying Circular RNAs in HepG2 Expressing Genotype IV Swine Hepatitis E Virus ORF3 Via Whole Genome Sequencing. Cell Transplant. 2021, 30, 9636897211055042. [Google Scholar] [CrossRef]
- Jiao, H.; Shuai, X.; Luo, Y.; Zhou, Z.; Zhao, Y.; Li, B.; Gu, G.; Li, W.; Li, M.; Zeng, H.; et al. Deep Insight Into Long Non-coding RNA and mRNA Transcriptome Profiling in HepG2 Cells Expressing Genotype IV Swine Hepatitis E Virus ORF3. Front. Vet. Sci. 2021, 8, 625609. [Google Scholar] [CrossRef]
- Jiao, H.; Meng, C.; Jiao, F.; Zhou, G.; Wang, L.; Wu, S.; Fan, C.; Li, J.; Cao, L.; Zhao, Y.; et al. Descriptive Comparative Transcriptomic Analysis of Genotype IV SHEV ORF3-Expressing HepG2 Cells. Microorganisms 2025, 13, 412. [Google Scholar] [CrossRef]
- Zang, J.; Lu, D.; Xu, A. The interaction of circRNAs and RNA binding proteins: An important part of circRNA maintenance and function. J. Neurosci. Res. 2020, 98, 87–97. [Google Scholar]
- Rong, D.; Sun, H.; Li, Z.; Liu, S.; Dong, C.; Fu, K.; Tang, W.; Cao, H. An emerging function of circRNA-miRNAs-mRNA axis in human diseases. Oncotarget 2017, 8, 73271–73281. [Google Scholar] [CrossRef]
- Nguyen, H.T.; Shukla, P.; Torian, U.; Faulk, K.; Emerson, S.U. Hepatitis E virus genotype 1 infection of swine kidney cells in vitro is inhibited at multiple levels. J. Virol. 2014, 88, 868–877. [Google Scholar] [CrossRef] [PubMed]
- Shukla, P.; Nguyen, H.T.; Faulk, K.; Mather, K.; Torian, U.; Engle, R.E.; Emerson, S.U. Adaptation of a genotype 3 hepatitis E virus to efficient growth in cell culture depends on an inserted human gene segment acquired by recombination. J. Virol. 2012, 86, 5697–5707. [Google Scholar] [CrossRef] [PubMed]
- Panda, S.K.; Ansari, I.H.; Durgapal, H.; Agrawal, S.; Jameel, S. The in vitro-synthesized RNA from a cDNA clone of hepatitis E virus is infectious. J. Virol. 2000, 74, 2430–2437. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.-C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar]
- Kovaka, S.; Zimin, A.V.; Pertea, G.M.; Razaghi, R.; Salzberg, S.L.; Pertea, M. Transcriptome assembly from long-read RNA-seq alignments with StringTie2. Genome Biol. 2019, 20, 278. [Google Scholar]
- Blaising, J.; Pécheur, E.I. Lipids: A key for hepatitis C virus entry and a potential target for antiviral strategies. Biochimie 2013, 95, 96–102. [Google Scholar] [CrossRef]
- Douam, F.; Lavillette, D.; Cosset, F.L. The mechanism of HCV entry into host cells. Prog. Mol. Biol. Transl. Sci. 2015, 129, 63–107. [Google Scholar]
- Meng, X.J. Swine hepatitis E virus: Cross-species infection and risk in xenotransplantation. Curr. Top. Microbiol. Immunol. 2003, 278, 185–216. [Google Scholar]
- Himmelsbach, K.; Bender, D.; Hildt, E. Life cycle and morphogenesis of the hepatitis E virus. Emerg. Microbes Infect. 2018, 7, 196. [Google Scholar] [CrossRef]
- Nan, Y.; Wu, C.; Zhao, Q.; Sun, Y.; Zhang, Y.J.; Zhou, E.M. Vaccine Development against Zoonotic Hepatitis E Virus: Open Questions and Remaining Challenges. Front. Microbiol. 2018, 9, 266. [Google Scholar] [CrossRef]
- Nan, Y.; Wu, C.; Zhao, Q.; Zhou, E.M. Zoonotic Hepatitis E Virus: An Ignored Risk for Public Health. Front. Microbiol. 2017, 8, 2396. [Google Scholar] [CrossRef]
- Zhou, J.H.; Shang, Y.; Cao, X.A.; Wang, Y.N.; Liu, Y.; Hu, Y.; Lan, X. Potential effects of hepatitis E virus infection in swine on public health in China. Infect. Genet. Evol. J. Mol. Epidemiol. Evol. Genet. Infect. Dis. 2019, 68, 113–118. [Google Scholar] [CrossRef]
- Bian, X.; Gao, W.; Wang, Y.; Yao, Z.; Xu, Q.; Guo, C.; Li, B. Riboflavin deficiency affects lipid metabolism partly by reducing apolipoprotein B100 synthesis in rats. J. Nutr. Biochem. 2019, 70, 75–81. [Google Scholar] [CrossRef]
- Duerden, J.M.; Bates, C.J. Effect of riboflavin deficiency on lipid metabolism of liver and brown adipose tissue of sucking rat pups. Br. J. Nutr. 1985, 53, 107–115. [Google Scholar] [CrossRef]
- Olpin, S.E.; Bates, C.J. Lipid metabolism in riboflavin-deficient rats. 1. Effect of dietary lipids on riboflavin status and fatty acid profiles. Br. J. Nutr. 1982, 47, 577–596. [Google Scholar] [CrossRef]
- Manthey, K.C.; Chew, Y.C.; Zempleni, J. Riboflavin deficiency impairs oxidative folding and secretion of apolipoprotein B-100 in HepG2 cells, triggering stress response systems. J. Nutr. 2005, 135, 978–982. [Google Scholar] [CrossRef]
- Zhang, Z.; He, G.; Han, G.S.; Zhang, J.; Catanzaro, N.; Diaz, A.; Wu, Z.; Carman, G.M.; Xie, L.; Wang, X. Host Pah1p phosphatidate phosphatase limits viral replication by regulating phospholipid synthesis. PLoS Pathog. 2018, 14, e1006988. [Google Scholar] [CrossRef]
- Zhou, Z.Y.; Bi, X.X. Experimental studies on the inactivation of HBV in blood via riboflavin photochemical treatment. Exp. Ther. Med. 2017, 13, 222–224. [Google Scholar] [CrossRef]
- Yang, Y.L.; Nan, Y.C. Open reading frame 3 protein of hepatitis E virus: Multi-function protein with endless potential. World J. Gastroenterol. 2021, 27, 2458–2473. [Google Scholar] [CrossRef]
- Solomon, K. The host immune response to Clostridium difficile infection. Ther. Adv. Infect. Dis. 2013, 1, 19–35. [Google Scholar] [CrossRef]
- Zhao, X.; Zhong, Y.; Wang, X.; Shen, J.; An, W. Advances in Circular RNA and Its Applications. Int. J. Med. Sci. 2022, 19, 975–985. [Google Scholar] [CrossRef]
- Yu, C.Y.; Kuo, H.C. The emerging roles and functions of circular RNAs and their generation. J. Biomed. Sci. 2019, 26, 29. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Yang, L.; Chen, L.L. The Biogenesis, Functions, and Challenges of Circular RNAs. Mol. Cell 2018, 71, 428–442. [Google Scholar] [CrossRef] [PubMed]
- Panda, A.C. Circular RNAs Act as miRNA Sponges. Adv. Exp. Med. Biol. 2018, 1087, 67–79. [Google Scholar] [PubMed]
- Li, Y.; Ji, M.; Huang, X.; Yang, F.; Yang, Z.; Shen, L. CircRNA_0040705 promotes the progression of hepatocellular carcinoma. IUBMB Life 2022, 74, 408–418. [Google Scholar] [CrossRef]
- Zhang, M.; Bai, X.; Zeng, X.; Liu, J.; Liu, F.; Zhang, Z. circRNA-miRNA-mRNA in breast cancer. Clin. Chim. Acta Int. J. Clin. Chem. 2021, 523, 120–130. [Google Scholar] [CrossRef]
- Chen, S.; Zhang, Y.; Ding, X.; Li, W. Identification of lncRNA/circRNA-miRNA-mRNA ceRNA Network as Biomarkers for Hepatocellular Carcinoma. Front. Genet. 2022, 13, 838869. [Google Scholar] [CrossRef]
- Jia, S.; Yu, L.; Wang, L.; Peng, L. The functional significance of circRNA/miRNA/mRNA interactions as a regulatory network in lung cancer biology. Int. J. Biochem. Cell Biol. 2024, 169, 106548. [Google Scholar] [CrossRef]
| GO_ID | GO_Term | p-Value |
|---|---|---|
| GO:0031100 | animal organ regeneration | 0.00 |
| GO:0060412 | ventricular septum morphogenesis | 0.00 |
| GO:0008420 | RNA polymerase II CTD heptapeptide repeat phosphatase activity | 0.00 |
| GO:0007406 | negative regulation of neuroblast proliferation | 0.00 |
| GO:0006886 | intracellular protein transport | 0.00 |
| GO:0046976 | histone methyltransferase activity (H3-K27 specific) | 0.00 |
| GO:0098609 | cell–cell adhesion | 0.00 |
| GO:0070971 | endoplasmic reticulum exit site | 0.00 |
| GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator | 0.00 |
| GO:0003714 | transcription corepressor activity | 0.00 |
| GO:0046872 | metal ion binding | 0.00 |
| GO:0047485 | protein N-terminus binding | 0.00 |
| GO:0005114 | type II transforming growth factor beta receptor binding | 0.00 |
| GO:0046826 | negative regulation of protein export from nucleus | 0.00 |
| GO:0001763 | morphogenesis of a branching structure | 0.00 |
| GO:0008328 | ionotropic glutamate receptor complex | 0.00 |
| GO:0043175 | RNA polymerase core enzyme binding | 0.00 |
| GO:0006642 | triglyceride mobilization | 0.00 |
| GO:0010587 | miRNA catabolic process | 0.01 |
| GO:0090114 | COPII-coated vesicle budding | 0.01 |
| Pathway_ID | Pathway_Name |
|---|---|
| ko05225 | Hepatocellular carcinoma |
| ko03018 | RNA degradation |
| ko04961 | Endocrine and other factor-regulated calcium reabsorption |
| ko04390 | Hippo signaling pathway |
| ko04520 | Adherens junction |
| ko04960 | Aldosterone-regulated sodium reabsorption |
| ko04150 | mTOR signaling pathway |
| ko04120 | Ubiquitin mediated proteolysis |
| ko05212 | Pancreatic cancer |
| ko00740 | Riboflavin metabolism |
| ko05161 | Hepatitis B |
| ko04744 | Phototransduction |
| ko04914 | Progesterone-mediated oocyte maturation |
| ko05146 | Amoebiasis |
| ko05226 | Gastric cancer |
| ko04933 | AGE-RAGE signaling pathway in diabetic complications |
| ko05202 | Transcriptional misregulation in cancer |
| ko01524 | Platinum drug resistance |
| ko05210 | Colorectal cancer |
| ko04024 | cAMP signaling pathway |
| circRNA Gene ID | circRNA Gene Name | Log2 (Fold Change) | p-Value | q-Value | Significant * |
|---|---|---|---|---|---|
| circRNA13511 | ENPP3 | Inf a | 0.12 | 1 | yes |
| ciRNA140 | ENSG00000154269 | −inf a | 0.19 | 1 | yes |
| hsa_circ_0130711 | ENPP1 | −inf a | 0.26 | 1 | yes |
| hsa_circ_0130715 | ENPP1 | −1.21 | 0.79 | 1 | yes |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tu, J.; Wu, S.; Wang, L.; Meng, C.; Zhou, G.; Guo, J.; Li, J.; Cao, L.; Song, Z.; Jiao, H. Identification of Riboflavin Metabolism Pathway in HepG2 Cells Expressing Genotype IV Swine Hepatitis E Virus ORF3 Protein. Vet. Sci. 2025, 12, 912. https://doi.org/10.3390/vetsci12090912
Tu J, Wu S, Wang L, Meng C, Zhou G, Guo J, Li J, Cao L, Song Z, Jiao H. Identification of Riboflavin Metabolism Pathway in HepG2 Cells Expressing Genotype IV Swine Hepatitis E Virus ORF3 Protein. Veterinary Sciences. 2025; 12(9):912. https://doi.org/10.3390/vetsci12090912
Chicago/Turabian StyleTu, Jing, Shengping Wu, Lingjie Wang, Chi Meng, Gengxu Zhou, Jianhua Guo, Jixiang Li, Liting Cao, Zhenhui Song, and Hanwei Jiao. 2025. "Identification of Riboflavin Metabolism Pathway in HepG2 Cells Expressing Genotype IV Swine Hepatitis E Virus ORF3 Protein" Veterinary Sciences 12, no. 9: 912. https://doi.org/10.3390/vetsci12090912
APA StyleTu, J., Wu, S., Wang, L., Meng, C., Zhou, G., Guo, J., Li, J., Cao, L., Song, Z., & Jiao, H. (2025). Identification of Riboflavin Metabolism Pathway in HepG2 Cells Expressing Genotype IV Swine Hepatitis E Virus ORF3 Protein. Veterinary Sciences, 12(9), 912. https://doi.org/10.3390/vetsci12090912







