A Novel Lightweight Dairy Cattle Body Condition Scoring Model for Edge Devices Based on Tail Features and Attention Mechanisms
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
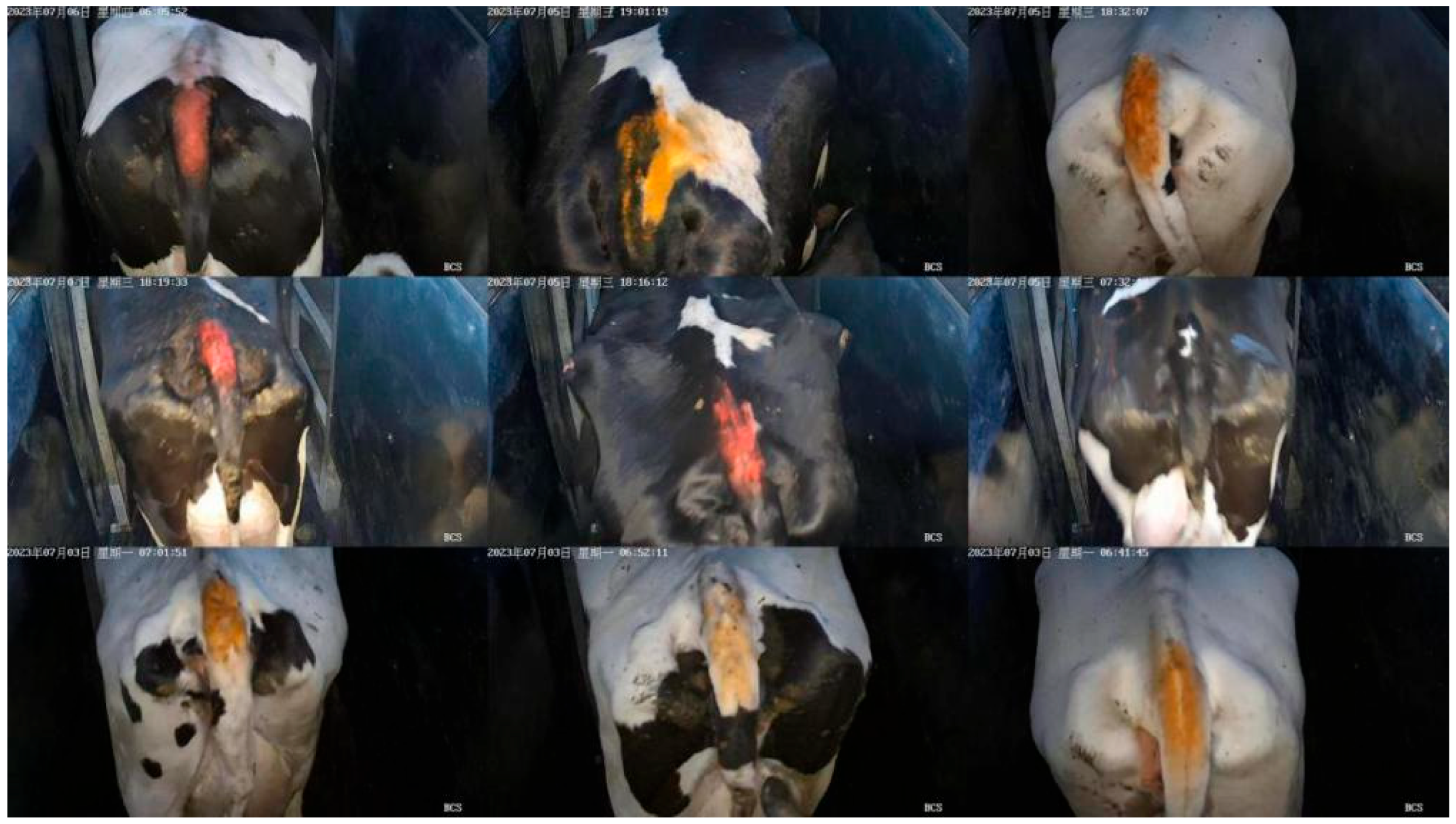
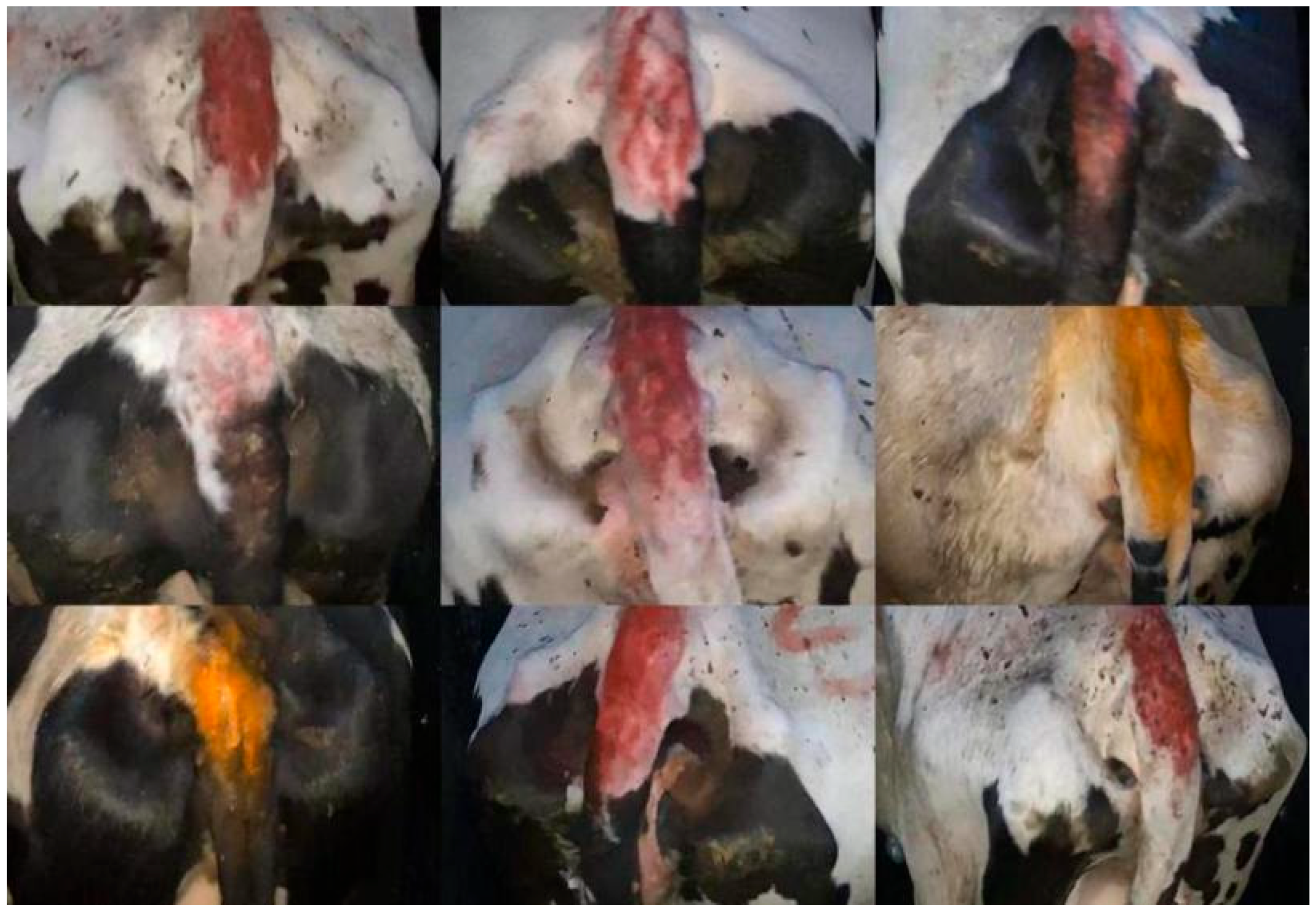
2.1. Selection of Dataset
2.2. Test Environment
3. Results
3.1. Selection of Benchmark Network Model
3.1.1. Confusion Matrix of Benchmark Network Model
3.1.2. Comparative Experiment of Basic Model
3.2. Improvement of Benchmark Network Model
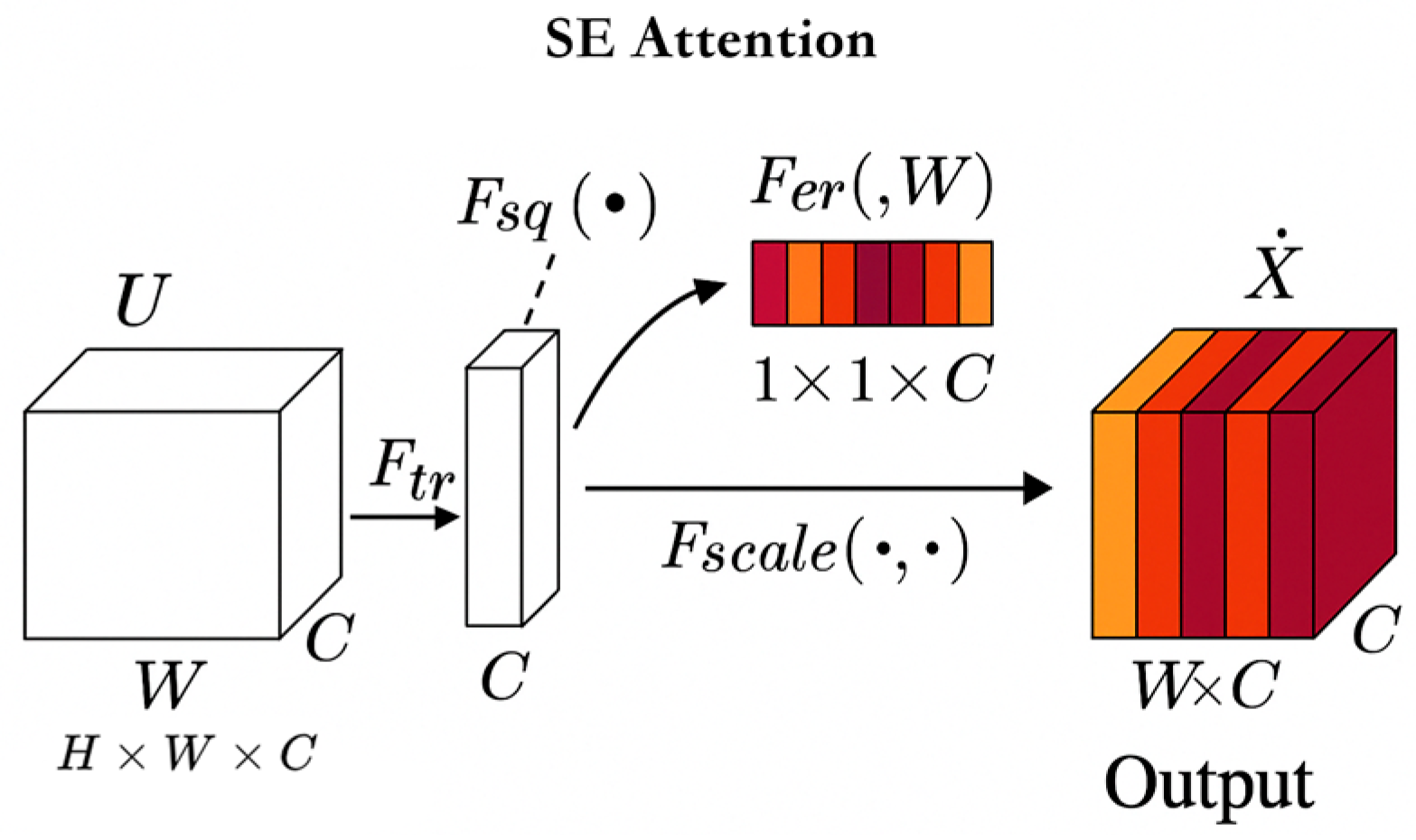
3.2.1. Introducing SE Attention Channel Attention Module
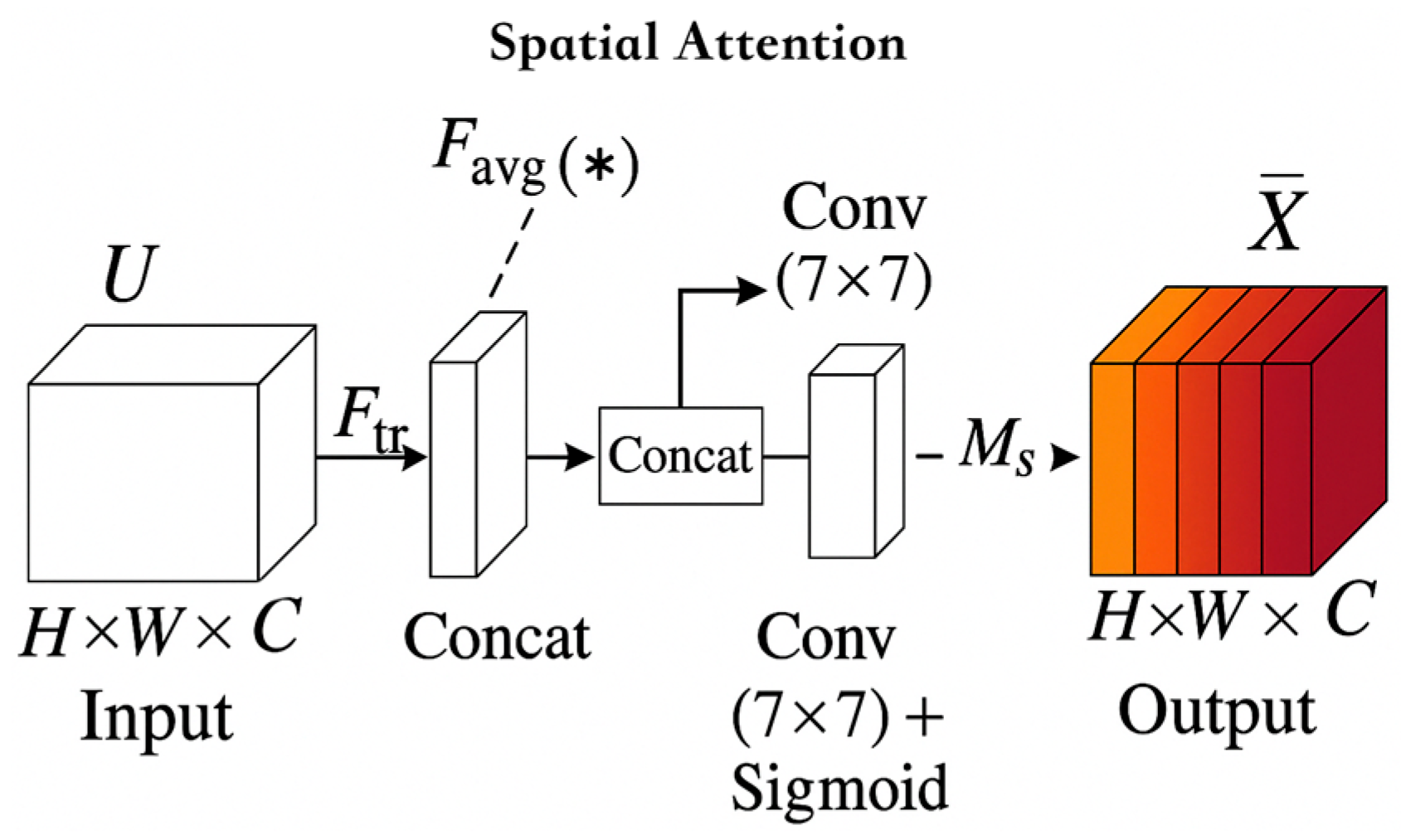
3.2.2. Introducing the Spatial Attention Module
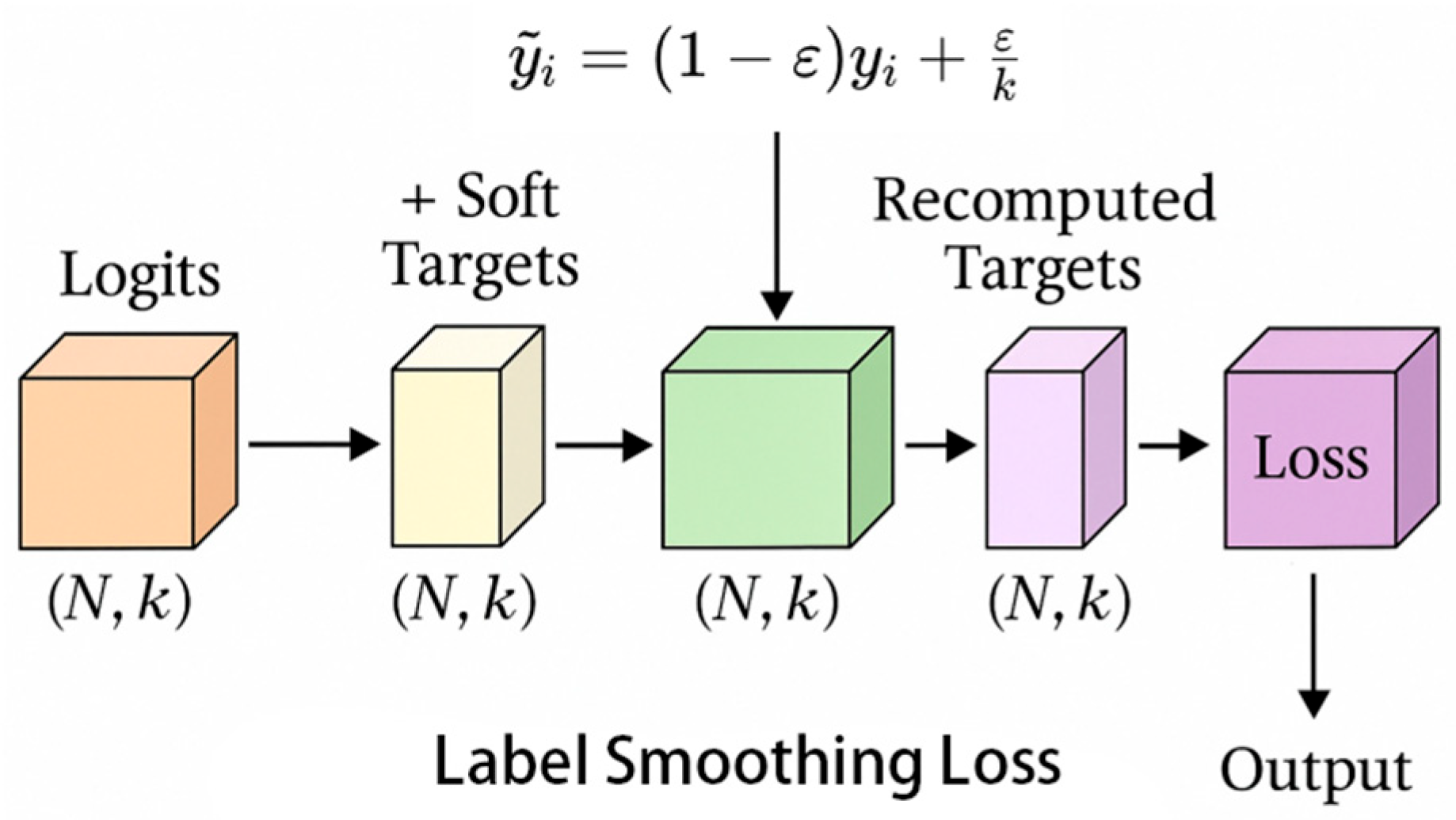
3.2.3. Introducing Label Smoothing Loss
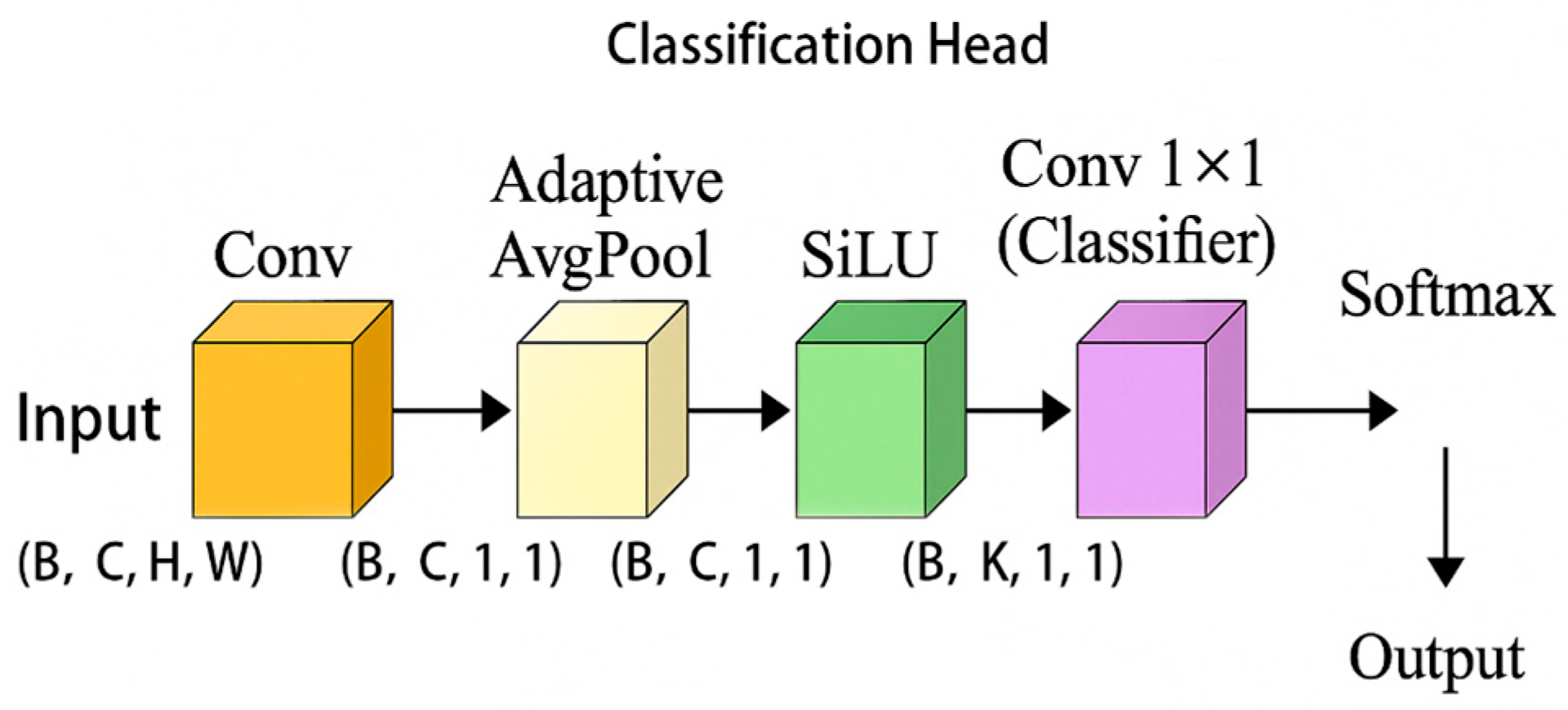
3.2.4. Introducing YOLOv5 Classification Header
3.3. Model Recognition Performance Test
3.3.1. The Confusion Matrix of the Model
3.3.2. Model Ablation Test
3.4. Model Lightweighting Processing
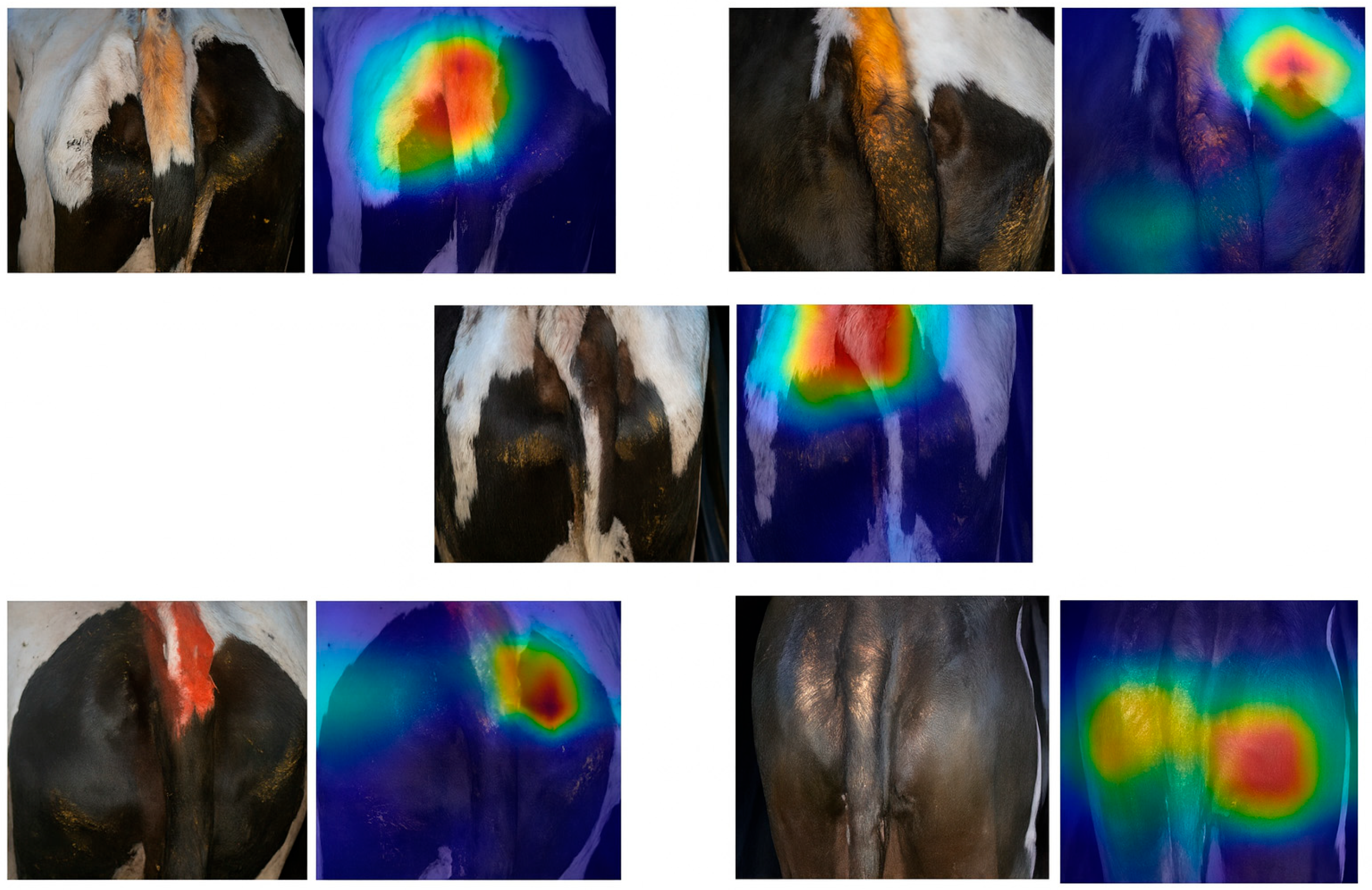
3.5. Model Interpretability Analysis (Grad CAM)
4. Discussion
4.1. Benchmark Model Selection
4.2. SE and Spatial Attention
4.3. Label Smoothing Loss
4.4. YOLOv5 Head and Ablation
4.5. Limitations and Future Research
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Edmonson, A.J.; Lean, I.J.; Weaver, L.D.; Farver, T.; Webster, G. A body condition scoring chart for holstein dairy cows. J. Dairy Sci. 1989, 72, 68–78. [Google Scholar] [CrossRef]
- Bewley, J.; Schutz, M. An interdisciplinary review of body condition scoring for dairy cattle. Prof. Anim. Sci. 2008, 24, 507–529. [Google Scholar] [CrossRef]
- Roche, J.; Macdonald, K.; Schütz, K.; Matthews, L.; Verkerk, G.; Meier, S.; Loor, J.; Rogers, A.; McGowan, J.; Morgan, S.; et al. Calving body condition score affects indicators of health in grazing dairy cows. J. Dairy Sci. 2013, 96, 5811–5825. [Google Scholar] [CrossRef]
- Roche, J.; Macdonald, K.; Burke, C.; Lee, J.; Berry, D. Associations among body condition score, body weight, and reproductive performance in seasonal-calving dairy cattle. J. Dairy Sci. 2007, 90, 376–391. [Google Scholar] [CrossRef]
- Roche, J.; Meier, S.; Heiser, A.; Mitchell, M.D.; Walker, C.; Crookenden, M.; Riboni, M.; Loor, J.; Kay, J. Effects of precalving body condition score and prepartum feeding level on production, reproduction, and health parameters in pasture-based transition dairy cows. J. Dairy Sci. 2015, 98, 7164–7182. [Google Scholar] [CrossRef] [PubMed]
- Pryce, J.; Coffey, M.; Simm, G. The relationship between body condition score and reproductive performance. J. Dairy Sci. 2001, 84, 1508–1515. [Google Scholar] [CrossRef] [PubMed]
- Wildman, E.; Jones, G.; Wagner, P.; Boman, R.; Troutt, H., Jr.; Lesch, T. A dairy cow body condition scoring system and its relationship to selected production characteristics. J. Dairy Sci. 1982, 65, 495–501. [Google Scholar] [CrossRef]
- Roche, J.R.; Friggens, N.C.; Kay, J.K.; Fisher, M.W.; Stafford, K.J.; Berry, D.P. Invited review: Body condition score and its association with dairy cow productivity, health, and welfare. J. Dairy Sci. 2009, 92, 5769–5801. [Google Scholar] [CrossRef]
- Waltner, S.; McNamara, J.; Hillers, J. Relationships of body condition score to production variables in high producing Holstein dairy cattle. J. Dairy Sci. 1993, 76, 3410–3419. [Google Scholar] [CrossRef]
- Paul, A.; Mondal, S.; Kumar, S.; Kumari, T. Body condition scoring in dairy cows-a conceptual and systematic review. Indian J. Anim. Res. 2020, 54, 929–935. [Google Scholar] [CrossRef]
- Goselink, R.M.; Schonewille, J.T.; van Duinkerken, G.; Hendriks, W.H. Physical exercise prepartum to support metabolic adaptation in the transition period of dairy cattle: A proof of concept. J. Anim. Physiol. Anim. Nutr. 2020, 104, 790–801. [Google Scholar] [CrossRef]
- Dandıl, E.; Çevik, K.K.; Boğa, M. Automated Classification System Based on YOLO Architecture for Body Condition Score in Dairy Cows. Vet. Sci. 2024, 11, 399. [Google Scholar] [CrossRef]
- Warden, P.; Situnayake, D. Tinyml: Machine Learning with Tensorflow Lite on Arduino and Ultra-Low-Power Microcontrollers; O’Reilly Media: Sebastopol, CA, USA, 2019. [Google Scholar]
- Zhou, Z.; Chen, X.; Li, E.; Zeng, L.; Luo, K.; Zhang, J. Edge intelligence: Paving the last mile of artificial intelligence with edge computing. Proc. IEEE 2019, 107, 1738–1762. [Google Scholar] [CrossRef]
- Deng, S.; Zhao, H.; Fang, W.; Yin, J.; Dustdar, S.; Zomaya, A.Y. Edge intelligence: The confluence of edge computing and artificial intelligence. IEEE Internet Things J. 2020, 7, 7457–7469. [Google Scholar] [CrossRef]
- Sharma, A.; Jain, A.; Gupta, P.; Chowdary, V. Machine learning applications for precision agriculture: A comprehensive review. IEEE Access 2020, 9, 4843–4873. [Google Scholar] [CrossRef]
- Zheng, Y.-Y.; Kong, J.-L.; Jin, X.-B.; Wang, X.-Y.; Su, T.-L.; Zuo, M. CropDeep: The crop vision dataset for deep-learning-based classification and detection in precision agriculture. Sensors 2019, 19, 1058. [Google Scholar] [CrossRef]
- Upadhyay, A.; Chandel, N.S.; Singh, K.P.; Chakraborty, S.K.; Nandede, B.M.; Kumar, M.; Subeesh, A.; Upendar, K.; Salem, A.; Elbeltagi, A. Deep learning and computer vision in plant disease detection: A comprehensive review of techniques, models, and trends in precision agriculture. Artif. Intell. Rev. 2025, 58, 92. [Google Scholar] [CrossRef]
- Wu, Z.; Chen, Y.; Zhao, B.; Kang, X.; Ding, Y. Review of weed detection methods based on computer vision. Sensors 2021, 21, 3647. [Google Scholar] [CrossRef]
- Fernandes, A.F.A.; Dórea, J.R.R.; Rosa, G.J.d.M. Image analysis and computer vision applications in animal sciences: An overview. Front. Vet. Sci. 2020, 7, 551269. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Huang, Y.; Chen, Z.; Chesser, G.D., Jr.; Purswell, J.L.; Linhoss, J.; Zhao, Y. Practices and applications of convolutional neural network-based computer vision systems in animal farming: A review. Sensors 2021, 21, 1492. [Google Scholar] [CrossRef] [PubMed]
- Antognoli, V.; Presutti, L.; Bovo, M.; Torreggiani, D.; Tassinari, P. Computer Vision in Dairy Farm Management: A Literature Review of Current Applications and Future Perspectives. Animals 2025, 15, 2508. [Google Scholar] [CrossRef]
- Min, X.; Ye, Y.; Xiong, S.; Chen, X. Computer Vision Meets Generative Models in Agriculture: Technological Advances, Challenges and Opportunities. Appl. Sci. 2025, 15, 7663. [Google Scholar] [CrossRef]
- Shao, D.; He, Z.; Fan, H.; Sun, K. Detection of cattle key parts based on the improved Yolov5 algorithm. Agriculture 2023, 13, 1110. [Google Scholar] [CrossRef]
- Lewis, R.; Kostermans, T.; Brovold, J.W.; Laique, T.; Ocepek, M. Automated Body Condition Scoring in Dairy Cows Using 2D Imaging and Deep Learning. Agriengineering 2025, 7, 241. [Google Scholar] [CrossRef]
- Feng, T.; Guo, Y.; Huang, X.; Wu, J.; Cheng, C. A Method of Body Condition Scoring for Dairy Cows Based on Lightweight Convolution Neural Network. J. ASABE 2024, 67, 409–420. [Google Scholar] [CrossRef]
- Li, J.; Zeng, P.; Yue, S.; Zheng, Z.; Qin, L.; Song, H. Automatic body condition scoring system for dairy cows in group state based on improved YOLOv5 and video analysis. Artif. Intell. Agric. 2025, 15, 350–362. [Google Scholar] [CrossRef]
- Li, W.-Y.; Shen, Y.; Wang, D.-J.; Yang, Z.-K.; Yang, X.-T. Automatic dairy cow body condition scoring using depth images and 3D surface fitting. In Proceedings of the 2019 IEEE International Conference on Unmanned Systems and Artificial Intelligence (ICUSAI), Xi’an, China, 22–24 November 2019; pp. 155–159. [Google Scholar]
- Summerfield, G.I.; De Freitas, A.; van Marle-Koster, E.; Myburgh, H.C. Automated cow body condition scoring using multiple 3D cameras and convolutional neural networks. Sensors 2023, 23, 9051. [Google Scholar] [CrossRef]
- Lee, J.G.; Lee, S.S.; Alam, M.; Lee, S.M.; Seong, H.-S.; Park, M.N.; Nguyen, H.-P.; Nguyen, D.T.; Baek, M.K.; Dang, C.G. Body Condition Score Assessment of Dairy Cows Using Deep Neural Network and 3D Imaging. Preprint 2024, 202401. [Google Scholar] [CrossRef]
- Luo, Y.; Zeng, Z.; Lu, H.; Lv, E. Posture detection of individual pigs based on lightweight convolution neural networks and efficient channel-wise attention. Sensors 2021, 21, 8369. [Google Scholar] [CrossRef]
- Yukun, S.; Pengju, H.; Yujie, W.; Ziqi, C.; Yang, L.; Baisheng, D.; Runze, L.; Yonggen, Z. Automatic monitoring system for individual dairy cows based on a deep learning framework that provides identification via body parts and estimation of body condition score. J. Dairy Sci. 2019, 102, 10140–10151. [Google Scholar] [CrossRef]
- Nagy, S.Á.; Kilim, O.; Csabai, I.; Gábor, G.; Solymosi, N. Impact evaluation of score classes and annotation regions in deep learning-based dairy cow body condition prediction. Animals 2023, 13, 194. [Google Scholar] [CrossRef]
- Giannone, C.; Sahraeibelverdy, M.; Lamanna, M.; Cavallini, D.; Formigoni, A.; Tassinari, P.; Torreggiani, D.; Bovo, M.J.S.A.T. Automated dairy cow identification and feeding behaviour analysis using a computer vision model based on YOLOv8. Smart Agric. Technol. 2025, 12, 101304. [Google Scholar] [CrossRef]
- Huang, X.; Li, X.; Hu, Z. Cow tail detection method for body condition score using Faster R-CNN. In Proceedings of the 2019 IEEE International Conference on Unmanned Systems and Artificial Intelligence (ICUSAI), Xi’an, China, 22–24 November 2019; pp. 347–351. [Google Scholar]
- Tan, M.; Le, Q. Efficientnet: Rethinking model scaling for convolutional neural networks. In Proceedings of the International Conference on Machine Learning, Long Beach, CA, USA, 9–15 June 2019; pp. 6105–6114. [Google Scholar]
- Tan, M.; Le, Q. Efficientnetv2: Smaller models and faster training. In Proceedings of the International Conference on Machine Learning, Virtual, 18–24 July 2021; pp. 10096–10106. [Google Scholar]
- Yin, X.; Wu, D.; Shang, Y.; Jiang, B.; Song, H. Using an EfficientNet-LSTM for the recognition of single Cow’s motion behaviours in a complicated environment. Comput. Electron. Agric. 2020, 177, 105707. [Google Scholar] [CrossRef]
- Hu, J.; Shen, L.; Sun, G. Squeeze-and-excitation networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–22 June 2018; pp. 7132–7141. [Google Scholar]
- Jaderberg, M.; Simonyan, K.; Zisserman, A. Spatial transformer networks. In Proceedings of the Advances in Neural Information Processing Systems (NeurIPS 2015), Montreal, QC, Canada, 7–12 December 2015; Volume 28. [Google Scholar]
- Müller, R.; Kornblith, S.; Hinton, G.E. When does label smoothing help? In Proceedings of the Advances in Neural Information Processing Systems 32 (NeurIPS 2019), Vancouver, BC, Canada, 8–14 December 2019; Volume 32. [Google Scholar]
- Bochkovskiy, A.; Wang, C.-Y.; Liao, H.-Y.M. Yolov4: Optimal speed and accuracy of object detection. arXiv 2020, arXiv:2004.10934. [Google Scholar] [CrossRef]
- Khanam, R.; Hussain, M. What is YOLOv5: A deep look into the internal features of the popular object detector. arXiv 2024, arXiv:2407.20892. [Google Scholar] [CrossRef]
- Redmon, J.; Farhadi, A. YOLO9000: Better, faster, stronger. In Proceedings of the 30th IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 7263–7271. [Google Scholar] [CrossRef]
- Huang, X.; Dou, Z.; Huang, F.; Zheng, H.; Hou, X.; Wang, C.; Feng, T.; Rao, Y. Dairy Cow Body Condition Score Target Detection Data Set [DS/OL], Version 3. Science Data Bank. 2025. Available online: https://cstr.cn/31253.11.sciencedb.16704 (accessed on 7 September 2025).
- Howard, A.; Sandler, M.; Chen, B.; Wang, W.; Chen, L.-C.; Tan, M.; Chu, G.; Vasudevan, V.; Zhu, Y.; Pang, R.; et al. Searching for mobilenetv3. In Proceedings of the 2019 IEEE/CVF International Conference on Computer Vision (ICCV), Seoul, Republic of Korea, 27 October–2 November 2019; pp. 1314–1324. [Google Scholar] [CrossRef]
- Sandler, M.; Howard, A.; Zhu, M.; Zhmoginov, A.; Chen, L. Mobilenetv2: Inverted residuals and linear bottlenecks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–22 June 2018; pp. 4510–4520. [Google Scholar] [CrossRef]
- Iandola, F.N.; Han, S.; Moskewicz, M.W.; Ashraf, K.; Dally, W.J.; Keutzer, K. SqueezeNet: AlexNet-level accuracy with 50× fewer parameters and <0.5 MB model size. arXiv 2016, arXiv:1602.07360. [Google Scholar]
- Zhang, X.; Zhou, X.; Lin, M.; Sun, J. Shufflenet: An extremely efficient convolutional neural network for mobile devices. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–22 June 2018; pp. 6848–6856. [Google Scholar]
- Hinton, G.; Vinyals, O.; Dean, J. Distilling the knowledge in a neural network. arXiv 2015, arXiv:1503.02531. [Google Scholar] [CrossRef]
- Romero, J.A.; Sanchis, R.; Arrebola, E. Experimental study of event based PID controllers with different sampling strategies. Application to brushless DC motor networked control system. In Proceedings of the 2015 XXV International Conference on Information, Communication and Automation Technologies (ICAT), Sarajevo, Bosnia and Herzegovina, 29–31 October 2015; pp. 1–6. [Google Scholar]
- Sanh, V.; Debut, L.; Chaumond, J.; Wolf, T. DistilBERT, a distilled version of BERT: Smaller, faster, cheaper and lighter. arXiv 2019, arXiv:1910.01108. [Google Scholar]
- Jiao, X.; Yin, Y.; Shang, L.; Jiang, X.; Chen, X.; Li, L.; Wang, F.; Liu, Q. Tinybert: Distilling bert for natural language understanding. In Proceedings of theFindings of the Association for Computational Linguistics: EMNLP 2020, Online Event, 16–20 November 2020; pp. 4163–4174. [Google Scholar]
- Wang, W.; Wei, F.; Dong, L.; Bao, H.; Yang, N.; Zhou, M. Minilm: Deep self-attention distillation for task-agnostic compression of pre-trained transformers. In Proceedings of the Advances in Neural Information Processing Systems 33 (NeurIPS 2020), Virtual, 6–12 December 2020; Volume 33, pp. 5776–5788. [Google Scholar]
- Selvaraju, R.R.; Cogswell, M.; Das, A.; Vedantam, R.; Parikh, D.; Batra, D. Grad-cam: Visual explanations from deep networks via gradient-based localization. In Proceedings of the 2017 IEEE International Conference on Computer Vision (ICCV), Venice, Italy, 22–29 October 2017; pp. 618–626. [Google Scholar] [CrossRef]
- Chattopadhay, A.; Sarkar, A.; Howlader, P.; Balasubramanian, V.N. Grad-cam++: Generalized gradient-based visual explanations for deep convolutional networks. In Proceedings of the 2018 IEEE Winter Conference on Applications of Computer Vision (WACV), Lake Tahoe, NV, USA, 12–15 March 2018; pp. 839–847. [Google Scholar]
- Kim, J.; Canny, J. Interpretable learning for self-driving cars by visualizing causal attention. In Proceedings of the 2017 IEEE International Conference on Computer Vision (ICCV), Venice, Italy, 22–29 October 2017; pp. 2942–2950. [Google Scholar]
- Ngiam, J.; Khosla, A.; Kim, M.; Nam, J.; Lee, H.; Ng, A.Y. Multimodal deep learning. In Proceedings of the ICML, Bellevue, WA, USA, 28 June–2 July 2011; pp. 689–696. [Google Scholar]
- Reed, R. Pruning algorithms-a survey. IEEE Trans. Neural Netw. 1993, 4, 740–747. [Google Scholar] [CrossRef]
- Liang, T.; Glossner, J.; Wang, L.; Shi, S.; Zhang, X. Pruning and quantization for deep neural network acceleration: A survey. Neurocomputing 2021, 461, 370–403. [Google Scholar] [CrossRef]
- Raj, E.F.I.; Appadurai, M.; Athiappan, K. Precision farming in modern agriculture. In Smart Agriculture Automation Using Advanced Technologies: Data Analytics and Machine Learning, Cloud Architecture, Automation and IoT; Springer: Berlin/Heidelberg, Germany, 2022; pp. 61–87. [Google Scholar]
- Marios, S.; Georgiou, J. Precision agriculture: Challenges in sensors and electronics for real-time soil and plant monitoring. In Proceedings of the2017 IEEE Biomedical Circuits and Systems Conference (BioCAS), Turin, Italy, 19–21 October 2017; pp. 1–4. [Google Scholar]
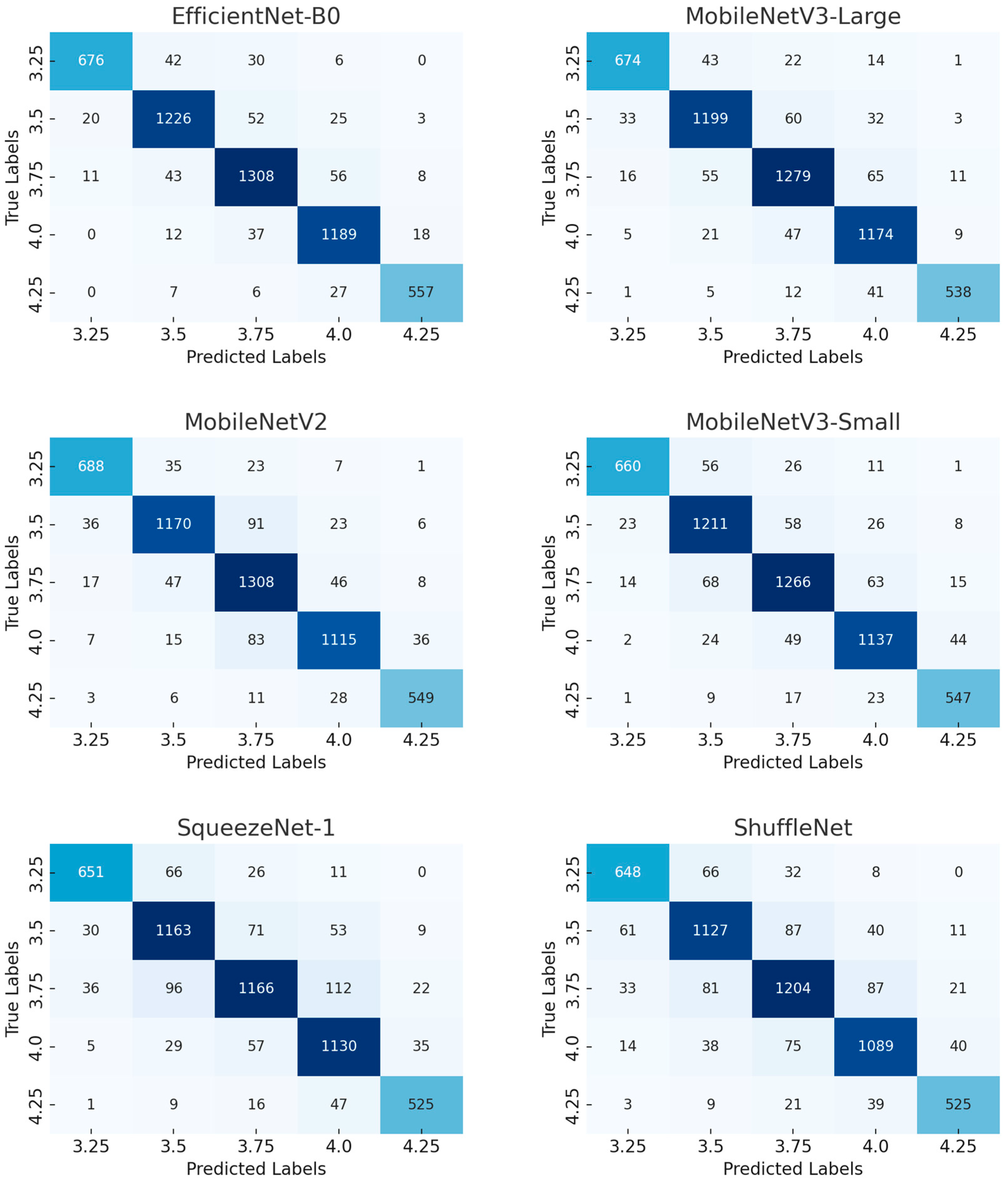
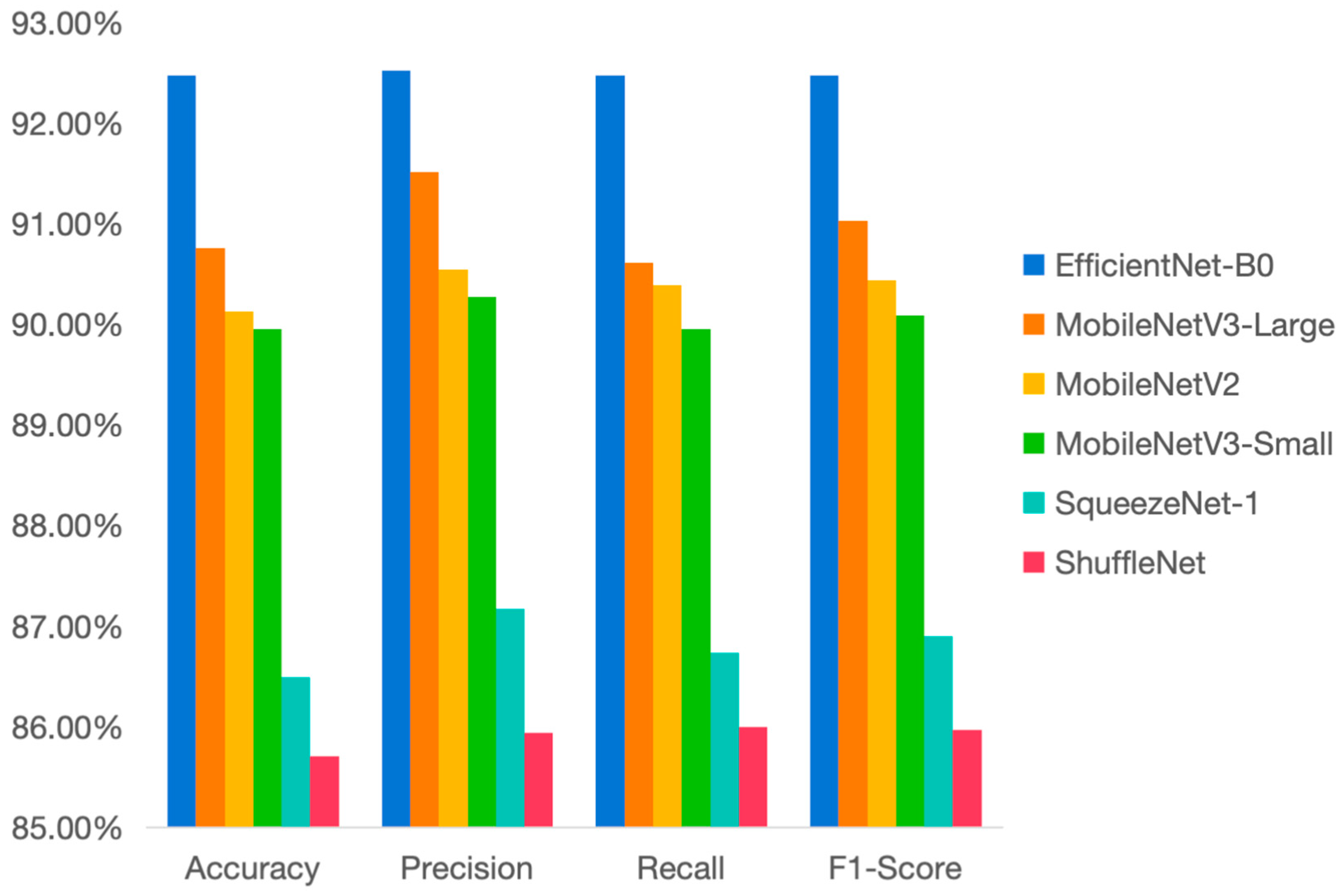


| Metric | EfficientNet-B0 | MobileNetV3-Large | MobileNetV2 | MobileNetV3-Small | SqueezeNet-1 | ShuffleNet |
|---|---|---|---|---|---|---|
| Accuracy | 92.48 | 90.76 | 90.13 | 89.96 | 86.49 | 85.71 |
| Precision | 92.53 | 91.52 | 90.55 | 90.28 | 87.17 | 85.94 |
| Recall | 92.48 | 90.62 | 90.39 | 89.96 | 86.74 | 86.00 |
| F1-Score | 92.48 | 91.03 | 90.44 | 90.09 | 86.90 | 85.97 |
| Model Size | 15.31 | 16.63 | 8.7 | 5.9 | 2.8 | 5.0 |
| FLOPS | 400.39 | 224.76 | 312.92 | 58.63 | 263.06 | 147.79 |
| Parameters | 4.01 | 4.21 | 2.23 | 1.52 | 0.73 | 1.26 |
| Per-sample latency | 0.6829 | 0.9772 | 0.9386 | 0.8761 | 0.8928 | 0.8834 |
| BCS ±0.25 | 97.98 | 97.35 | 97.63 | 97.13 | 96.08 | 95.71 |
| BCS ±0.5 | 99.70 | 99.48 | 99.44 | 99.40 | 99.37 | 99.16 |
| Evaluation Indicators | No SE | Have SE | Increase Amplitude |
|---|---|---|---|
| Accuracy | 92.48 | 92.55 | +0.07 |
| Precision | 92.53 | 92.58 | +0.05 |
| Recall | 92.48 | 92.55 | +0.07 |
| F1-Score | 92.48 | 92.56 | +0.08 |
| FLOPS | 400.39 | 414.14 | −13.75 |
| Parameters | 4.01 | 4.22 | +0.21 |
| Per-sample latency | 0.6829 | 0.6423 | −0.0406 |
| Model Size | 15.31 | 16.38 | +1.07 |
| BCS ± 0.25 | 97.98 | 98.02 | +0.04 |
| BCS ± 0.5 | 99.70 | 99.68 | −0.02 |
| Evaluation Indicators | No Spatial | Have Spatial | Increase Amplitude |
|---|---|---|---|
| Accuracy | 92.48 | 92.82 | +0.34 |
| Precision | 92.53 | 92.83 | +0.30 |
| Recall | 92.48 | 92.82 | +0.34 |
| F1-Score | 92.48 | 92.82 | +0.34 |
| FLOPS | 400.39 | 403.46 | +3.07 |
| Parameters | 4.01 | 4.08 | +0.07 |
| Per-sample latency | 0.6829 | 0.7063 | +0.0234 |
| Model Size | 15.31 | 15.84 | +0.53 |
| BCS ± 0.25 | 97.98 | 98.04 | +0.06 |
| BCS ± 0.5 | 99.70 | 99.70 | +0 |
| Evaluation Indicators | No Label Smoothing | Have Label Smoothing | Increase Amplitude |
|---|---|---|---|
| Accuracy | 92.48 | 92.70 | +0.22 |
| Precision | 92.53 | 92.74 | +0.21 |
| Recall | 92.48 | 92.70 | +0.22 |
| F1-Score | 92.48 | 92.71 | +0.23 |
| FLOPS | 400.39 | 413.87 | +13.48 |
| Parameters | 4.01 | 4.01 | +0 |
| Per-sample latency | 0.6829 | 0.7124 | +0.0295 |
| Model Size | 15.31 | 16.4 | +1.09 |
| BCS ± 0.25 | 97.98 | 98.28 | +0.30 |
| BCS ± 0.5 | 99.70 | 99.78 | +0.08 |
| Evaluation Indicators | No YOLO Classification Head | Have YOLO Classification Head | Increase Amplitude |
|---|---|---|---|
| Accuracy | 92.48 | 92.89 | +0.41 |
| Precision | 92.53 | 92.92 | +0.39 |
| Recall | 92.48 | 92.89 | +0.41 |
| F1-Score | 92.48 | 92.89 | +0.41 |
| FLOPS | 400.39 | 415.52 | +15.13 |
| Parameters | 4.01 | 5.65 | +1.64 |
| Per-sample latency | 0.6829 | 1.3398 | +0.6569 |
| Model Size | 15.31 | 21.9 | +6.59 |
| BCS ± 0.25 | 97.98 | 98.10 | +0.12 |
| BCS ± 0.5 | 99.70 | 99.70 | +0 |
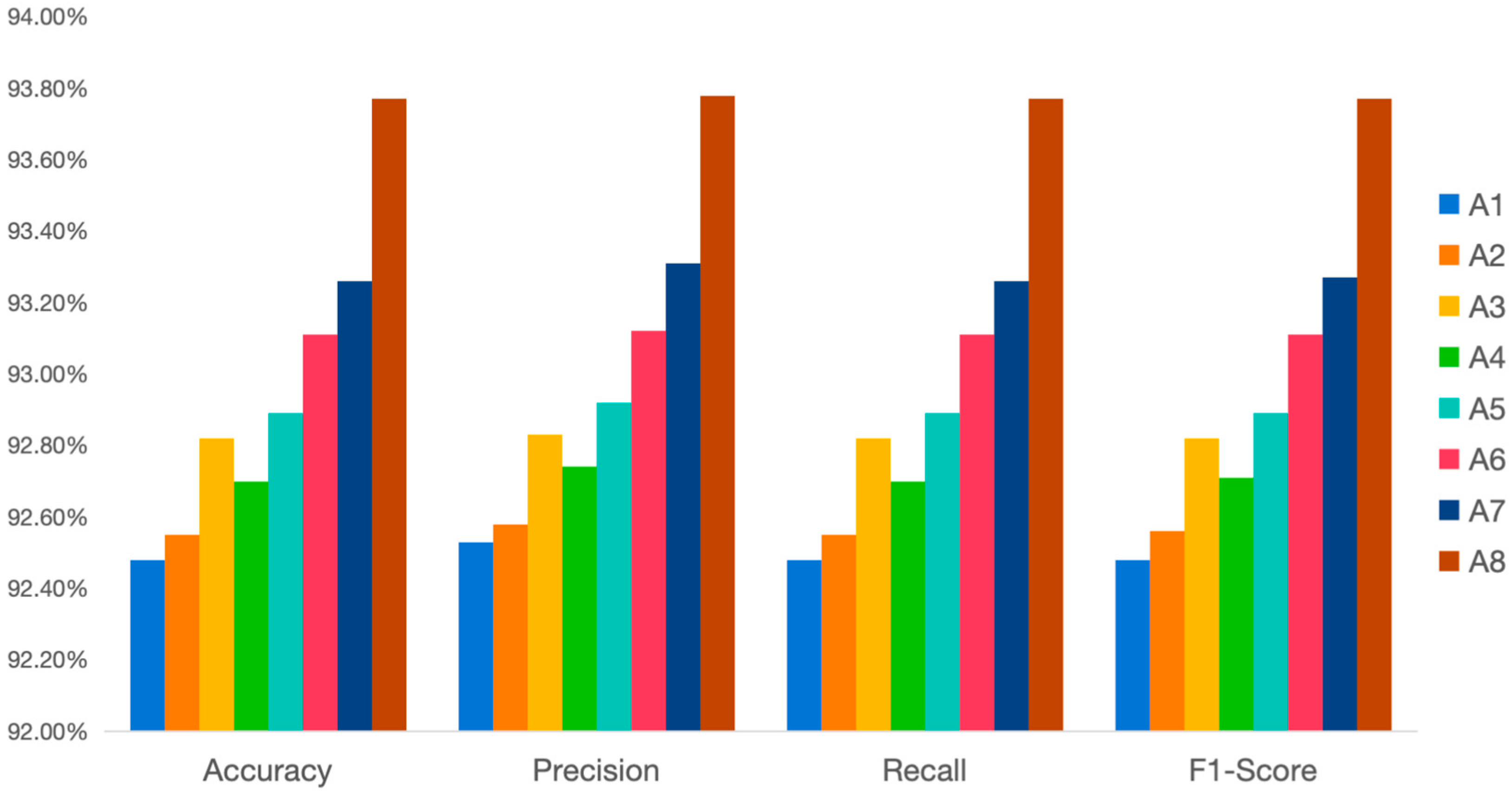
| Metric | A1 | A2 | A3 | A4 | A5 | A6 | A7 | A8 |
|---|---|---|---|---|---|---|---|---|
| Attention module | none | SE | Spatial | none | none | SE + Spatial | SE + Spatial | SE + Spatial |
| Loss function | CrossEntropy | CrossEntropy | CrossEntropy | CrossEntropy | CrossEntropy | CrossEntropy | LabelSmoothing | LabelSmoothing |
| Describe | base model | Add SE attention module | Add Spatial Attention Module | Add label smoothing loss | Add YOLO classification header | Add combined attention | Add label smoothing loss function | Add YOLO classification header |
| Accuracy | 92.48 | 92.55 | 92.82 | 92.70 | 92.89 | 93.11 | 93.26 | 93.77 |
| Precision | 92.53 | 92.58 | 92.83 | 92.74 | 92.92 | 93.12 | 93.31 | 93.78 |
| Recall | 92.48 | 92.55 | 92.82 | 92.70 | 92.89 | 93.11 | 93.26 | 93.77 |
| F1-Score | 92.48 | 92.56 | 92.82 | 92.71 | 92.89 | 93.11 | 93.27 | 93.77 |
| FLOPS | 400.39 | 414.14 | 403.46 | 413.87 | 415.52 | 403.72 | 403.73 | 481.07 |
| Parameters | 4.01 | 4.22 | 4.08 | 4.01 | 5.65 | 4.28 | 4.28 | 5.86 |
| Per-sample latency | 0.6829 | 0.6423 | 0.7063 | 0.7124 | 1.3398 | 0.6723 | 0.6872 | 0.9752 |
| Model Size | 15.31 | 16.38 | 15.84 | 16.4 | 21.9 | 16.6 | 17.4 | 23.8 |
| BCS ± 0.25 | 97.98 | 98.02 | 98.04 | 98.28 | 98.10 | 98.28 | 98.06 | 98.19 |
| BCS ± 0.5 | 99.70 | 99.68 | 99.70 | 99.78 | 99.70 | 99.76 | 99.72 | 99.63 |
| Evaluation Indicators | Before Distillation | After Distillation |
|---|---|---|
| Accuracy | 93.77 | 91.10 |
| Precision | 93.78 | 91.14 |
| Recall | 93.77 | 91.10 |
| F1-Score | 93.77 | 91.10 |
| FLOPS | 481.07 | 312.92 |
| Parameters | 5.86 | 2.23 |
| Per-sample latency | 0.9752 | 0.8711 |
| Model Size | 23.8 | 8.7 |
| BCS ± 0.25 | 98.19 | 97.57 |
| BCS ± 0.5 | 99.63 | 99.72 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, F.; Zhang, Y.; Liu, Y.; Li, J.; Li, M.; Yao, J. A Novel Lightweight Dairy Cattle Body Condition Scoring Model for Edge Devices Based on Tail Features and Attention Mechanisms. Vet. Sci. 2025, 12, 906. https://doi.org/10.3390/vetsci12090906
Liu F, Zhang Y, Liu Y, Li J, Li M, Yao J. A Novel Lightweight Dairy Cattle Body Condition Scoring Model for Edge Devices Based on Tail Features and Attention Mechanisms. Veterinary Sciences. 2025; 12(9):906. https://doi.org/10.3390/vetsci12090906
Chicago/Turabian StyleLiu, Fan, Yongan Zhang, Yanqiu Liu, Jia Li, Meian Li, and Jianping Yao. 2025. "A Novel Lightweight Dairy Cattle Body Condition Scoring Model for Edge Devices Based on Tail Features and Attention Mechanisms" Veterinary Sciences 12, no. 9: 906. https://doi.org/10.3390/vetsci12090906
APA StyleLiu, F., Zhang, Y., Liu, Y., Li, J., Li, M., & Yao, J. (2025). A Novel Lightweight Dairy Cattle Body Condition Scoring Model for Edge Devices Based on Tail Features and Attention Mechanisms. Veterinary Sciences, 12(9), 906. https://doi.org/10.3390/vetsci12090906






