Isolation and Characterization of Equine Lymph Node Endothelial Cells
Simple Summary
Abstract
1. Introduction
2. Methodology
2.1. Primary Cell Extraction
2.2. Cell Monitoring and Contamination Control
2.3. Trypsin Differentiation
2.4. Growth Curve Assay
2.5. Growth Media Curve Assay
2.6. Cell Characterization Using Immunofluorescence Assay
2.7. Cell Contamination Assay
2.8. Whole-Genome Sequencing
2.8.1. DNA Isolation
2.8.2. Library Preparation and Sequencing
3. Results
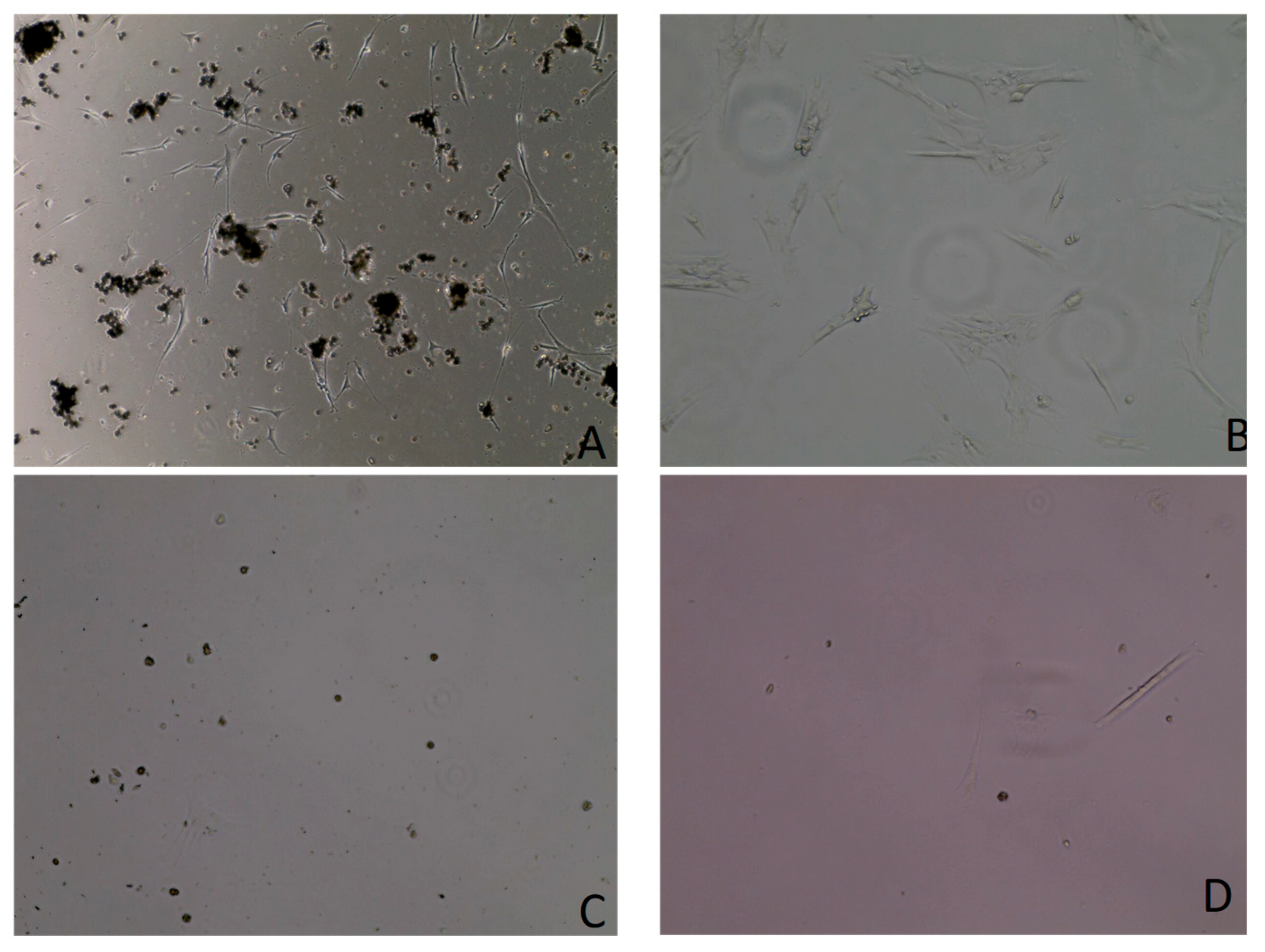
3.1. Cell Culture
3.2. Growth Curve Assay Results
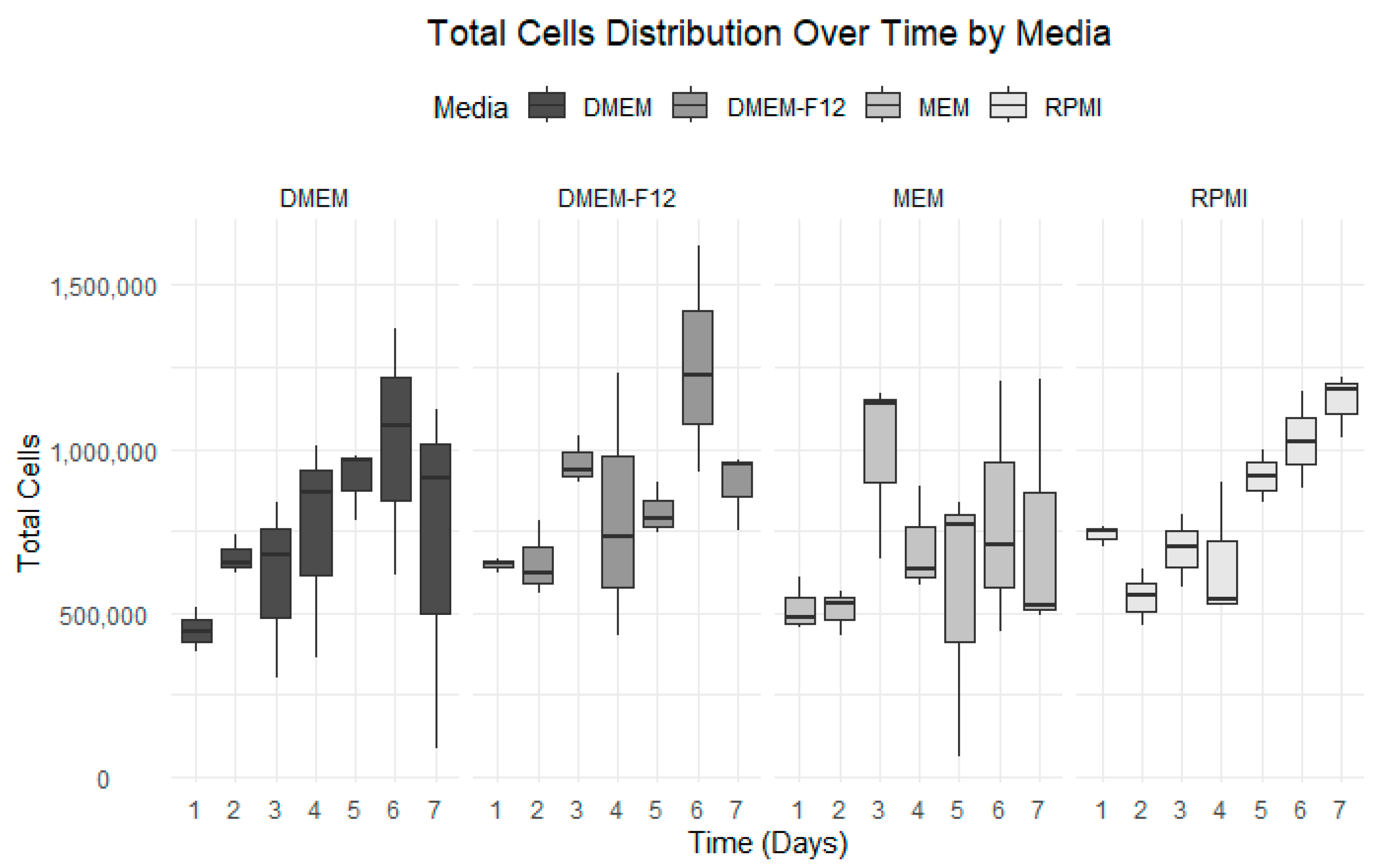
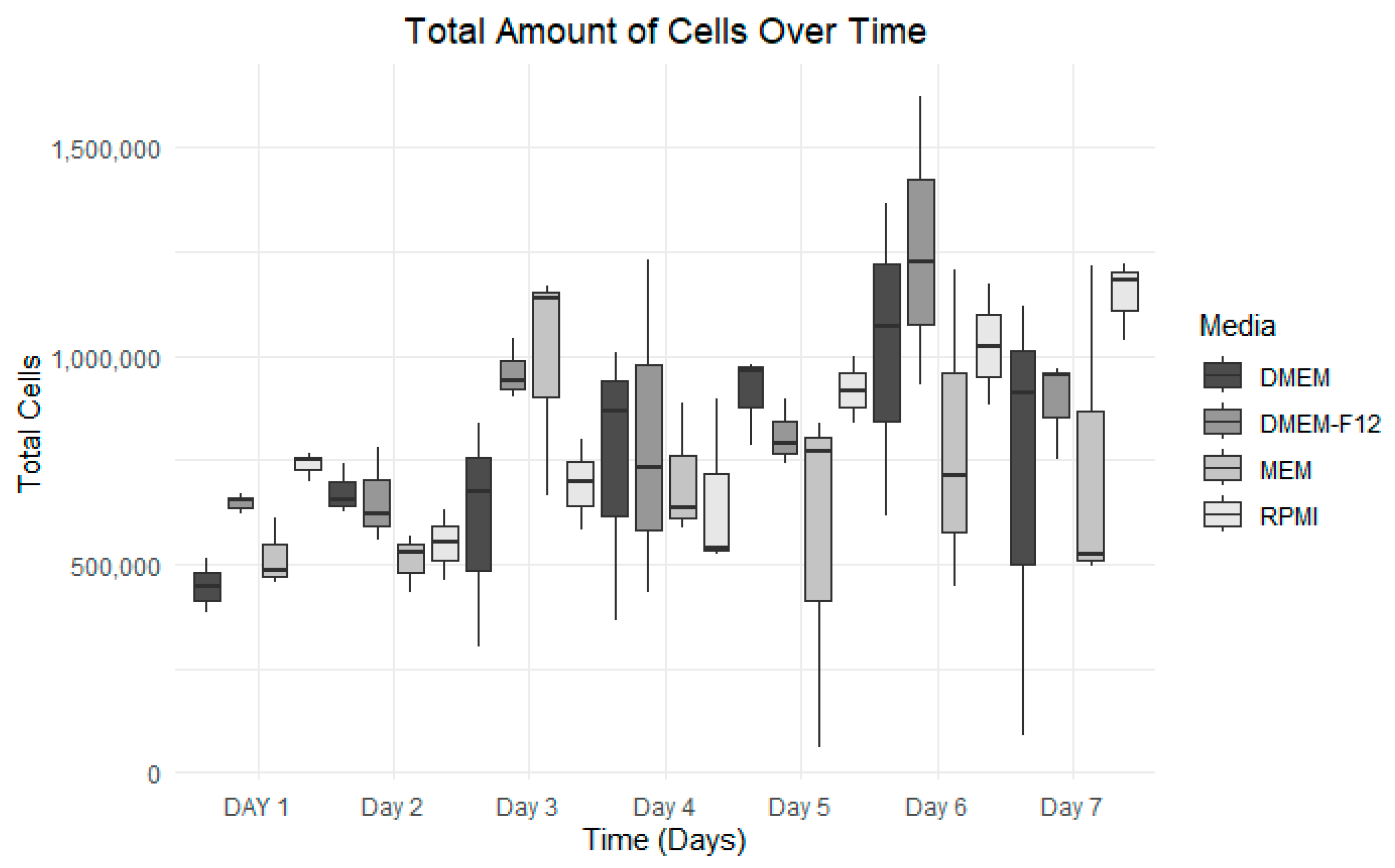
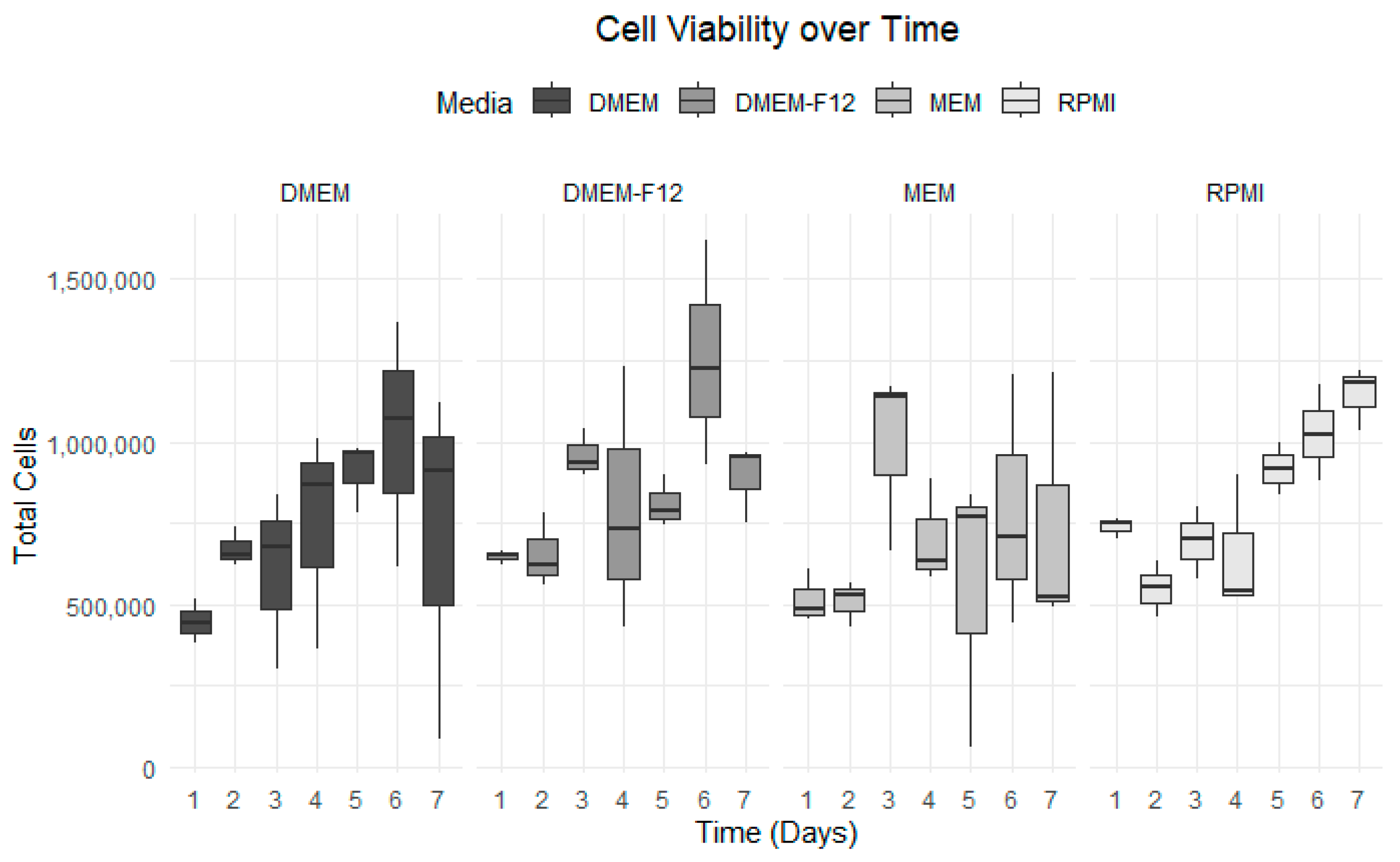
3.3. Growth Curve Media Assay
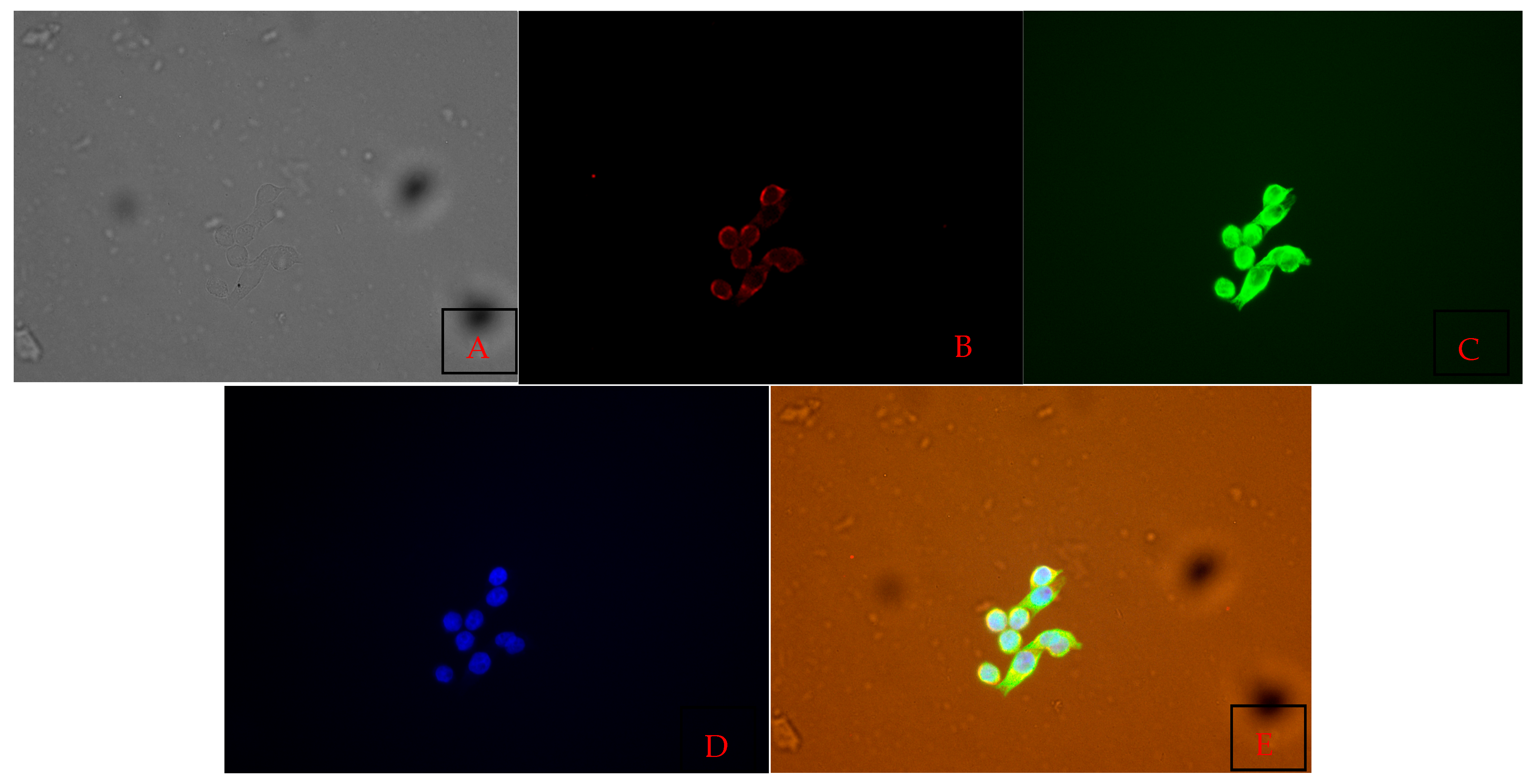
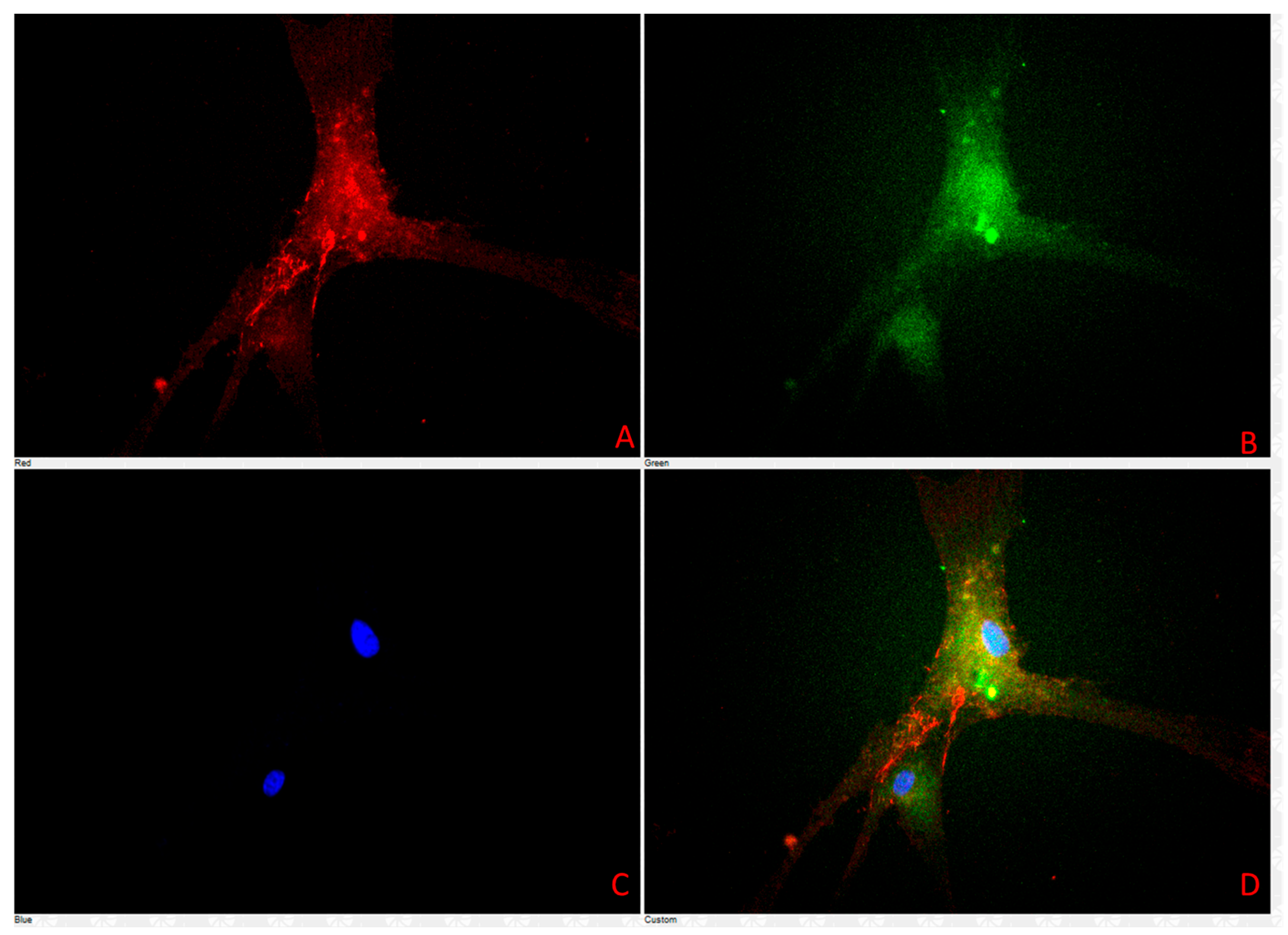
3.4. Cell Characterization Using Immunofluorescence Assay Results
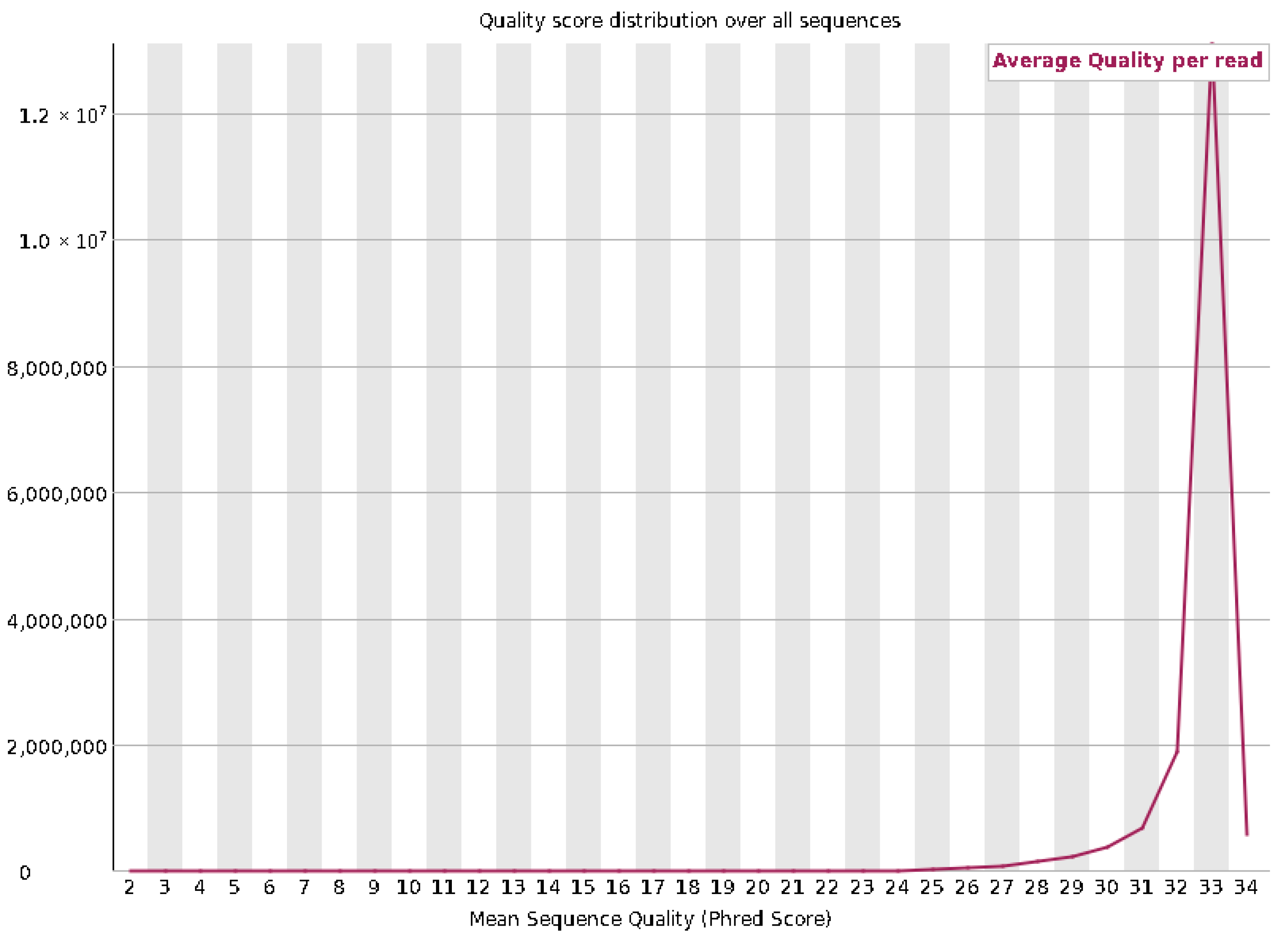
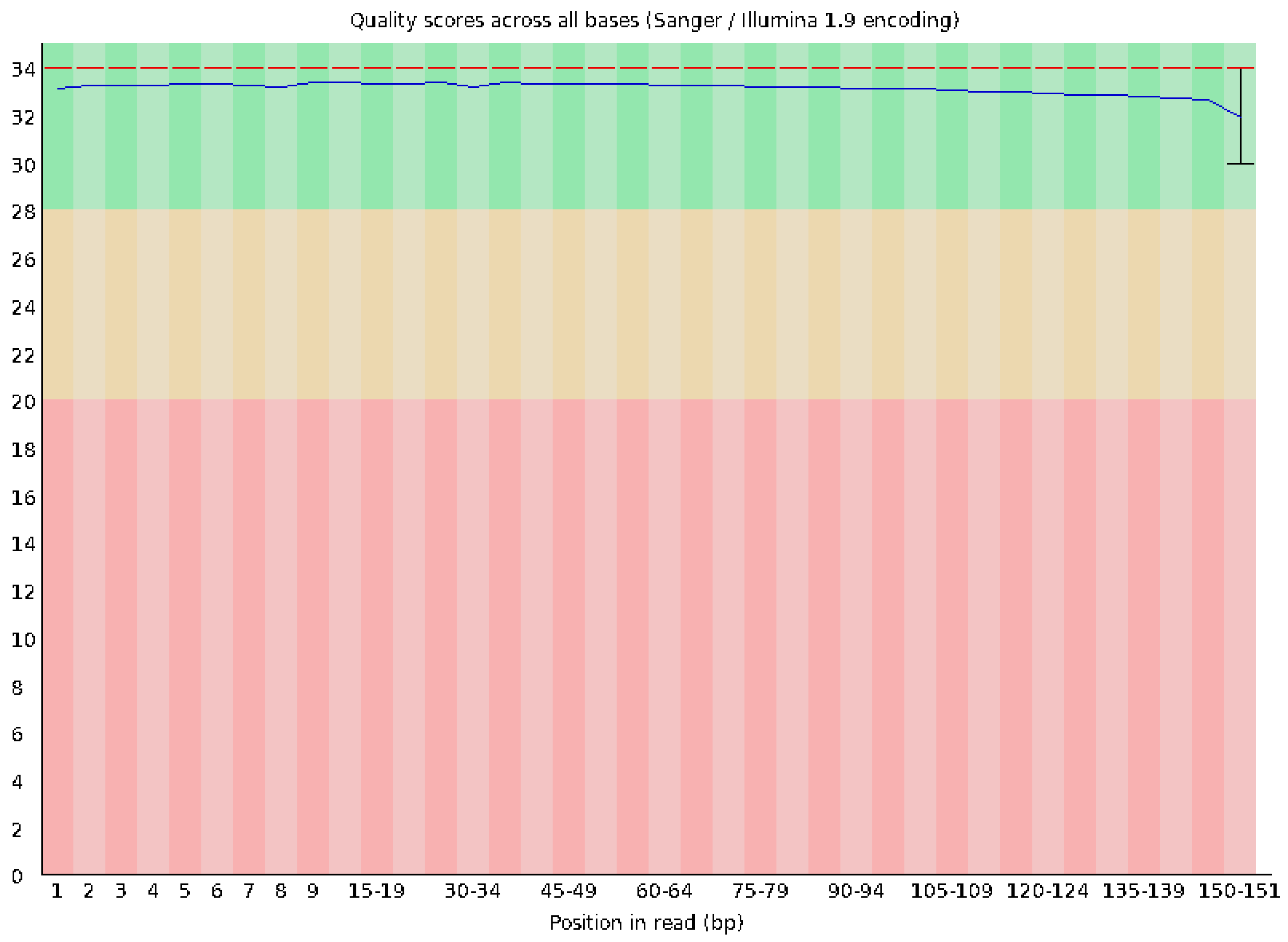
3.5. Whole-Genome Sequencing Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Willard-Mack, C.L. Normal Structure, Function, and Histology of Lymph Nodes. Toxicol. Pathol. 2006, 34, 409–424. [Google Scholar] [CrossRef]
- Grant, S.M.; Lou, M.; Yao, L.; Germain, R.N.; Radtke, A.J. The lymph node at a glance–how spatial organization optimizes the immune response. J. Cell Sci. 2020, 133, jcs241828. [Google Scholar] [CrossRef] [PubMed]
- Ozulumba, T.; Montalbine, A.N.; Ortiz-Cárdenas, J.E.; Pompano, R.R. New tools for immunologists: Models of lymph node function from cells to tissues. Front. Immunol. 2023, 14, 1183286. [Google Scholar] [CrossRef] [PubMed]
- Morgan, E.A. Surgical Pathology of Hematopoietic Neoplasms. Pathobiol. Hum. Dis. A Dyn. Encycl. Dis. Mech. 2014, 3607–3627. [Google Scholar] [CrossRef]
- Pereira, E.R.; Jones, D.; Jung, K.; Padera, T.P. The lymph node microenvironment and its role in the progression of metastatic cancer. Semin. Cell Dev. Biol. 2015, 38, 98–105. [Google Scholar] [CrossRef]
- Liao, S.; Padera, T.P. Lymphatic function and immune regulation in health and disease. Lymphat. Res. Biol. 2013, 11, 136–143. [Google Scholar] [CrossRef]
- Hammel, J.H.; Cook, S.R.; Belanger, M.C.; Munson, J.M.; Pompano, R.R. Modeling Immunity in Vitro: Slices, Chips, and Engineered Tissues. Annu. Rev. Biomed. Eng. 2021, 23, 461–491. [Google Scholar] [CrossRef]
- Hughes, J.P.; Rees, S.S.; Kalindjian, S.B.; Philpott, K.L. Principles of early drug discovery. Br. J. Pharmacol. 2011, 162, 1239–1249. [Google Scholar] [CrossRef]
- Ramaiahgari, S.C.; den Braver, M.W.; Herpers, B.; Terpstra, V.; Commandeur, J.N.M.; Van De Water, B.; Price, L.S. A 3D in vitro model of differentiated HepG2 cell spheroids with improved liver-like properties for repeated dose high-throughput toxicity studies. Arch. Toxicol. 2014, 88, 1083–1095. [Google Scholar] [CrossRef]
- Roh, T.T.; Chen, Y.; Rudolph, S.; Gee, M.; Kaplan, D.L. In Vitro Models of Intestine Innate Immunity. Trends Biotechnol. 2021, 39, 274–285. [Google Scholar] [CrossRef]
- Wang, L.; Hu, D.; Xu, J.; Hu, J.; Wang, Y. Complex in vitro model: A transformative model in drug development and precision medicine. Clin. Transl. Sci. 2024, 17, e13695. [Google Scholar] [CrossRef] [PubMed]
- Richter, M.; Piwocka, O.; Musielak, M.; Piotrowski, I.; Suchorska, W.M.; Trzeciak, T. From Donor to the Lab: A Fascinating Journey of Primary Cell Lines. Front. Cell Dev. Biol. 2021, 9, 711381. [Google Scholar] [CrossRef]
- Dietze, K.; Slosarek, I.; Fuhrmann-Selter, T.; Hopperdietzel, C.; Plendl, J.; Kaessmeyer, S. Isolation of equine endothelial cells and life cell angiogenesis assay. Clin. Hemorheol. Microcirc. 2014, 58, 127–146. [Google Scholar] [CrossRef]
- Fidalgo-Carvalho, I.; Craigo, J.K.; Barnes, S.; Costa-Ramos, C.; Montelaro, R.C. Characterization of an equine macrophage cell line: Application to studies of EIAV infection. Vet. Microbiol. 2009, 136, 8–19. [Google Scholar] [CrossRef]
- Lessiak, U.; Pratscher, B.; Tichy, A.; Nell, B. Bevacizumab Efficiently Inhibits VEGF-Associated Cellular Processes in Equine Umbilical Vein Endothelial Cells: An In Vitro Characterization. Vet. Sci. 2023, 10, 632. [Google Scholar] [CrossRef] [PubMed]
- Bussche, L.; Van de Walle, G.R. Peripheral Blood-Derived Mesenchymal Stromal Cells Promote Angiogenesis via Paracrine Stimulation of Vascular Endothelial Growth Factor Secretion in the Equine Model. Stem Cells Transl. Med. 2014, 3, 1514–1525. [Google Scholar] [CrossRef]
- Bosseler, L.; Verryken, K.; Bauwens, C.; de Vries, C.; Deprez, P.; Ducatelle, R.; Vandenabeele, S. Equine multisystemic eosinophilic epitheliotropic disease: A case report and review of literature Equine multisystemic eosinophilic epitheliotropic disease: A case report and review of literature. N. Z. Vet. J. 2013, 61, 177–182. [Google Scholar] [CrossRef]
- Schumacher, J.; Edwards, J.F.; Cohen, N.D. Chronic Idiopathic Inflammatory Bowel Diseases of the Horse. J. Vet. Intern. Med. 2000, 14, 258–265. [Google Scholar] [CrossRef]
- Horan, E.M.; Metcalfe, L.V.A.; de Swarte, M.; Cahalan, S.D.; Katz, L.M. Pulmonary and hepatic eosinophilic granulomas and epistaxis in a horse suggestive of multi-systemic eosinophilic epitheliotropic disease. Equine Vet. Educ. 2013, 25, 607–613. [Google Scholar] [CrossRef]
- Villagrán, C.C.; Vogt, D.; Gupta, A.; Fernández, E.A. Case Report Rapport de cas Inflammatory bowel disease characterized by multisystemic eosinophilic epitheliotropic disease (MEED) in a horse in Saskatchewan, Canada. Can. Vet. J. 2021, 62, 1190. [Google Scholar] [PubMed]
- Paraschou, G.; Vogel, P.E.; Lee, A.M.; Trawford, R.F.; Priestnall, S.L. Multisystemic eosinophilic epitheliotropic disease in three donkeys. J. Comp. Pathol. 2023, 201, 105–108. [Google Scholar] [CrossRef]
- La Perle, K.M.D.; Piercy, R.J.; Long, J.F.; Blomme, E.A.G. Multisystemic, eosinophilic, epitheliotropic disease with intestinal lymphosarcoma in a horse. Vet. Pathol. 1998, 35, 144–146. [Google Scholar] [CrossRef]
- Simon-Assmann, P.; Turck, N.; Sidhoum-Jenny, M.; Gradwohl, G.; Kedinger, M. In vitro models of intestinal epithelial cell differentiation. Cell Biol. Toxicol. 2007, 23, 241–256. [Google Scholar] [PubMed]
- Wang, Y.; Gunasekara, D.B.; Attayek, P.J.; Reed, M.I.; DiSalvo, M.; Nguyen, D.L.; Dutton, J.S.; Lebhar, M.S.; Bultman, S.J.; Sims, C.E.; et al. In Vitro Generation of Mouse Colon Crypts. ACS Biomater. Sci. Eng. 2017, 3, 2502–2513. [Google Scholar] [CrossRef] [PubMed]
- Rooney, M.F.; Neto, N.G.B.; Monaghan, M.G.; Hill, E.W.; Porter, R.K. Conditionally immortalised equine skeletal muscle cell lines for in vitro analysis. Biochem. Biophys. Rep. 2023, 33, 101391. [Google Scholar] [CrossRef]
- National Research Council. Summary of Advantages and Disadvantages of In Vitro and In Vivo Methods. In Monoclonal Antibody Production; National Academies Press: Washington, DC, USA, 1999. [Google Scholar]
- Marycz, K.; Bourebaba, N.; Serwotka-Suszczak, A.; Mularczyk, M.; Galuppo, L.; Bourebaba, L. In Vitro Generated Equine Hepatic-Like Progenitor Cells as a Novel Potent Cell Pool for Equine Metabolic Syndrome (EMS) Treatment. Stem Cell Rev. Rep. 2023, 19, 1124–1134. [Google Scholar] [CrossRef]
- Yuan, Z.Q.; Gault, E.; Gobeil, P.; Nixon, C.; Campo, M.; Nasir, L. Establishment and characterization of equine fibroblast cell lines transformed in vivo and in vitro by BPV-1: Model systems for equine sarcoids. Virology 2008, 373, 352–361. [Google Scholar] [CrossRef] [PubMed]
- Evans, E.; Paillot, R.; López-Álvarez, M.R. A comprehensive analysis of e-CAS cell line reveals they are mouse macrophages. Sci. Rep. 2018, 8, 8237. [Google Scholar] [CrossRef]
- Yu, K.; Chen, B.; Aran, D.; Charalel, J.; Yau, C.; Wolf, D.M.; van ‘t Veer, L.J.; Butte, A.J.; Goldstein, T.; Sirota, M. Comprehensive transcriptomic analysis of cell lines as models of primary tumors across 22 tumor types. Nat. Commun. 2019, 10, 3574. [Google Scholar] [CrossRef]
- Kaur, G.; Dufour, J.M. Cell lines: Valuable tools or useless artifacts. Spermatogenesis 2012, 2, 1–5. [Google Scholar] [CrossRef]
- Lertkiatmongkol, P.; Liao, D.; Mei, H.; Hu, Y.; Newman, P.J. Endothelial functions of PECAM-1 (CD31). Curr. Opin. Hematol. 2016, 23, 253–259. [Google Scholar]
- Dave, J.M.; Bayless, K.J. Vimentin as an integral regulator of cell adhesion and endothelial sprouting. Microcirculation 2014, 21, 333–344. [Google Scholar] [CrossRef]
- Tornavaca, O.; Chia, M.; Dufton, N.; Almagro, L.O.; Conway, D.E.; Randi, A.M.; Schwartz, M.A.; Matter, K.; Balda, M.S. ZO-1 controls endothelial adherens junctions, cell-cell tension, angiogenesis, and barrier formation. J. Cell Biol. 2015, 208, 821–838. [Google Scholar] [CrossRef] [PubMed]
- Van Roy, F.; Berx, G. The cell-cell adhesion molecule E-cadherin. Cell. Mol. Life Sci. 2008, 65, 3756–3788. [Google Scholar] [CrossRef] [PubMed]
- Johnson, L.A.; Prevo, R.; Clasper, S.; Jackson, D.G. Inflammation-induced Uptake and Degradation of the Lymphatic Endothelial Hyaluronan Receptor LYVE-1. J. Biol. Chem. 2007, 282, 33671–33680. [Google Scholar] [CrossRef] [PubMed]
- Gordon, E.J.; Gale, N.W.; Harvey, N.L. Expression of the hyaluronan receptor LYVE-1 is not restricted to the lymphatic vasculature; LYVE-1 is also expressed on embryonic blood vessels. Dev. Dyn. 2008, 237, 1901–1909. [Google Scholar] [CrossRef]
- Banerji, S.; Ni, J.; Wang, S.X.; Clasper, S.; Su, J.; Tammi, R.; Jones, M.; Jackson, D.G. LYVE-1, a new homologue of the CD44 glycoprotein, is a lymph-specific receptor for hyaluronan. J. Cell Biol. 1999, 144, 789–801. [Google Scholar] [CrossRef]
- Jackson, D.G. Hyaluronan in the lymphatics: The key role of the hyaluronan receptor LYVE-1 in leucocyte trafficking. Matrix Biol. 2019, 78–79, 219–235. [Google Scholar] [CrossRef] [PubMed]
- Jackson, D.G. Biology of the lymphatic marker LYVE-1 and applications in research into lymphatic trafficking and lymphangiogenesis. APMIS 2004, 112, 526–538. [Google Scholar] [CrossRef]
- Wilting, J.; Papoutsi, M.; Christ, B.; Nicolaides, K.H.; Kaisenberg, C.S.; Borges, J.; Stark, G.B.; Alitalo, K.; Tomarev, S.I.; Niemeyer, C.; et al. The transcription factor Prox1 is a marker for lymphatic endothelial cells in normal and diseased human tissues. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2002, 16, 1271–1273. [Google Scholar]
- Sawa, Y. New trends in the study of podoplanin as a cell morphological regulator. Jpn. Dent. Sci. Rev. 2010, 46, 165–172. [Google Scholar] [CrossRef][Green Version]
- Quintanilla, M.; Montero, L.M.; Renart, J.; Villar, E.M. Podoplanin in inflammation and cancer. Int. J. Mol. Sci. 2019, 20, 707. [Google Scholar] [CrossRef]
- Astarita, J.L.; Acton, S.E.; Turley, S.J. Podoplanin: Emerging functions in development, the immune system, and cancer. Front. Immunol. 2012, 3, 283. [Google Scholar] [CrossRef] [PubMed]
- Vigl, B.; Aebischer, D.; Nitschké, M.; Iolyeva, M.; Röthlin, T.; Antsiferova, O.; Halin, C. Tissue inflammation modulates gene expression of lymphatic endothelial cells and dendritic cell migration in a stimulus-dependent manner. Blood 2011, 118, 205–215. [Google Scholar] [CrossRef]
- Chu, P.G.; Arber, D.A. CD79: A review. Appl. Immunohistochem. Mol. Morphol. 2001, 9, 97–106. [Google Scholar] [CrossRef] [PubMed]
- Cyster, J.G.; Allen, C.D.C. B Cell Responses: Cell Interaction Dynamics and Decisions. Cell 2019, 177, 524–540. [Google Scholar] [CrossRef]
- Ager, A. High endothelial venules and other blood vessels: Critical regulators of lymphoid organ development and function. Front. Immunol. 2017, 8, 45. [Google Scholar] [CrossRef]
- Hussain, B.; Kasinath, V.; Ashton-Rickardt, G.P.; Clancy, T.; Uchimura, K.; Tsokos, G.; Abdi, R. High endothelial venules as potential gateways for therapeutics. Trends Immunol. 2022, 43, 728–740. [Google Scholar] [CrossRef]
- Vella, G.; Guelfi, S.; Bergers, G. High Endothelial Venules: A Vascular Perspective on Tertiary Lymphoid Structures in Cancer. Front. Immunol. 2021, 12, 736670. [Google Scholar] [CrossRef]
- Blanchard, L.; Girard, J.P. High endothelial venules (HEVs) in immunity, inflammation and cancer. Angiogenesis 2021, 24, 719–753. [Google Scholar] [CrossRef]
- Zhu, W.; Zheng, D.; Wang, D.; Yang, L.; Zhao, C.; Huang, X. Emerging Roles of Ubiquitin-Specific Protease 25 in Diseases. Front. Cell Dev. Biol. 2021, 9, 698751. [Google Scholar] [CrossRef] [PubMed]
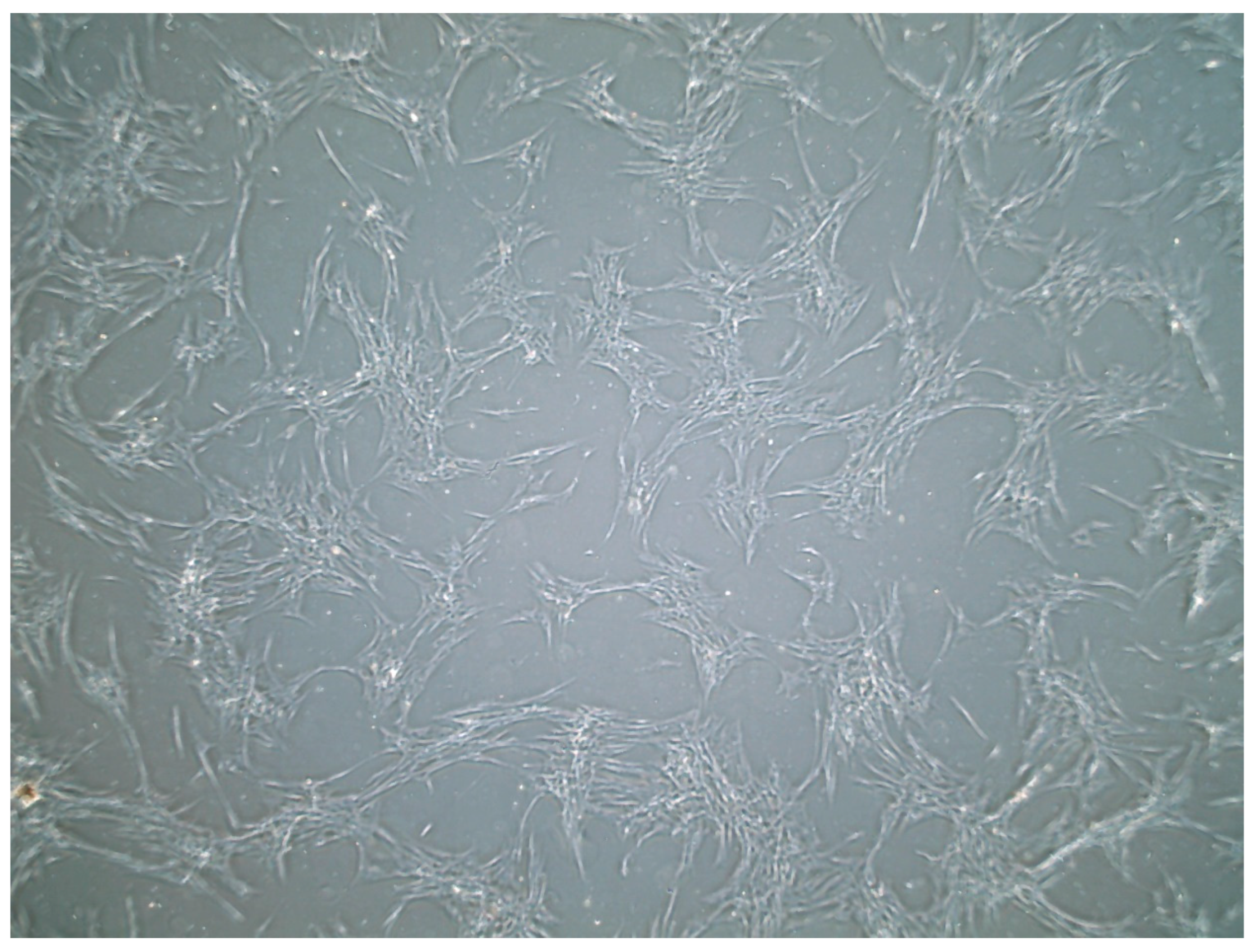
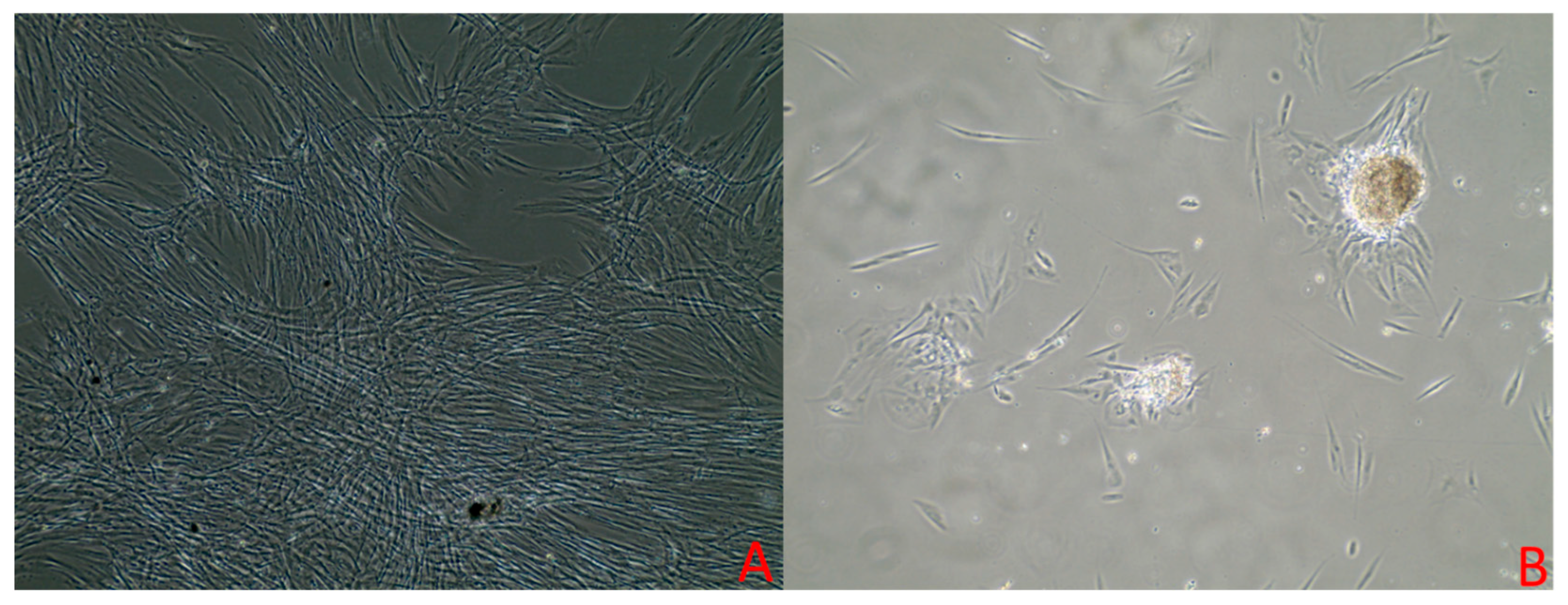
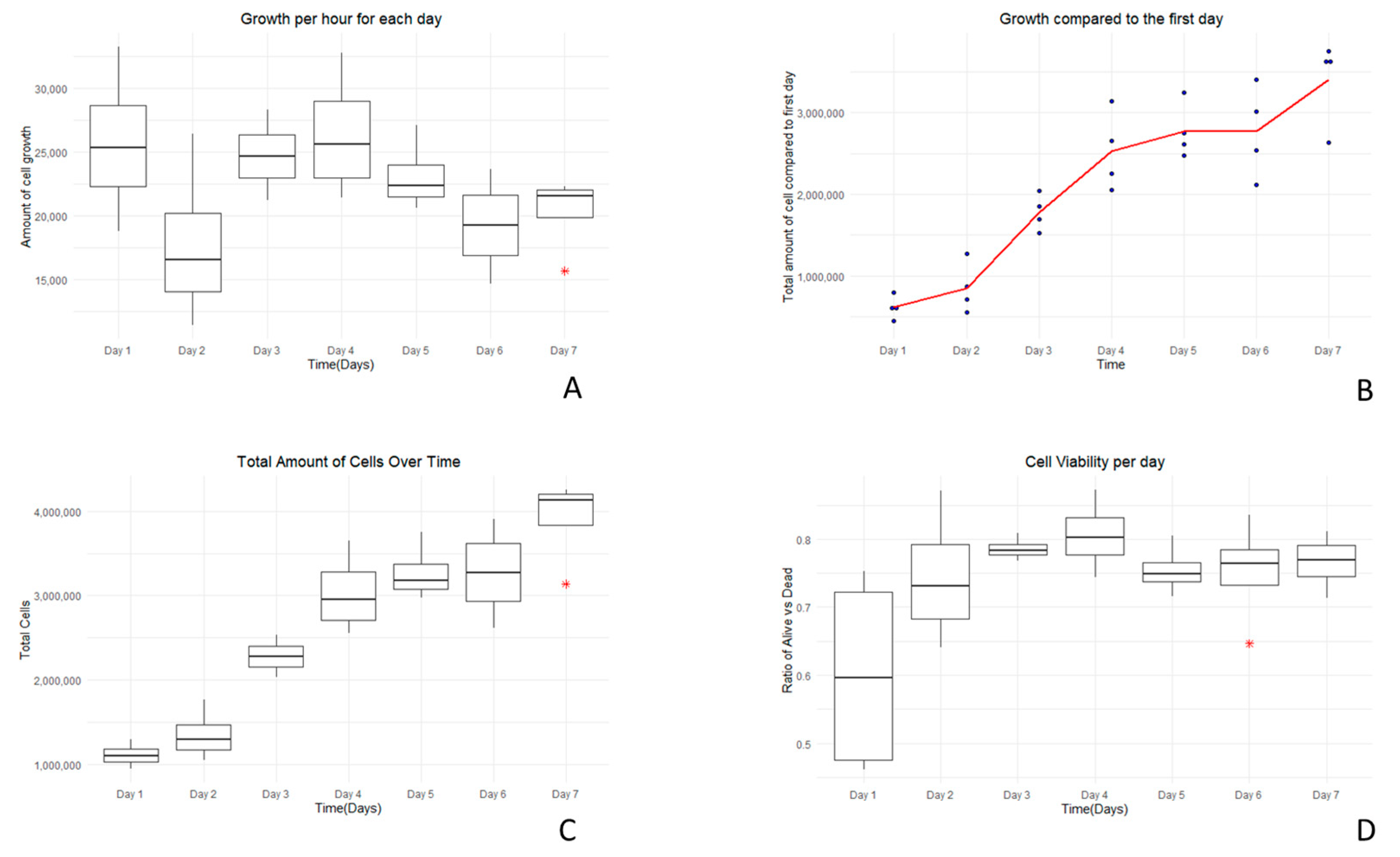
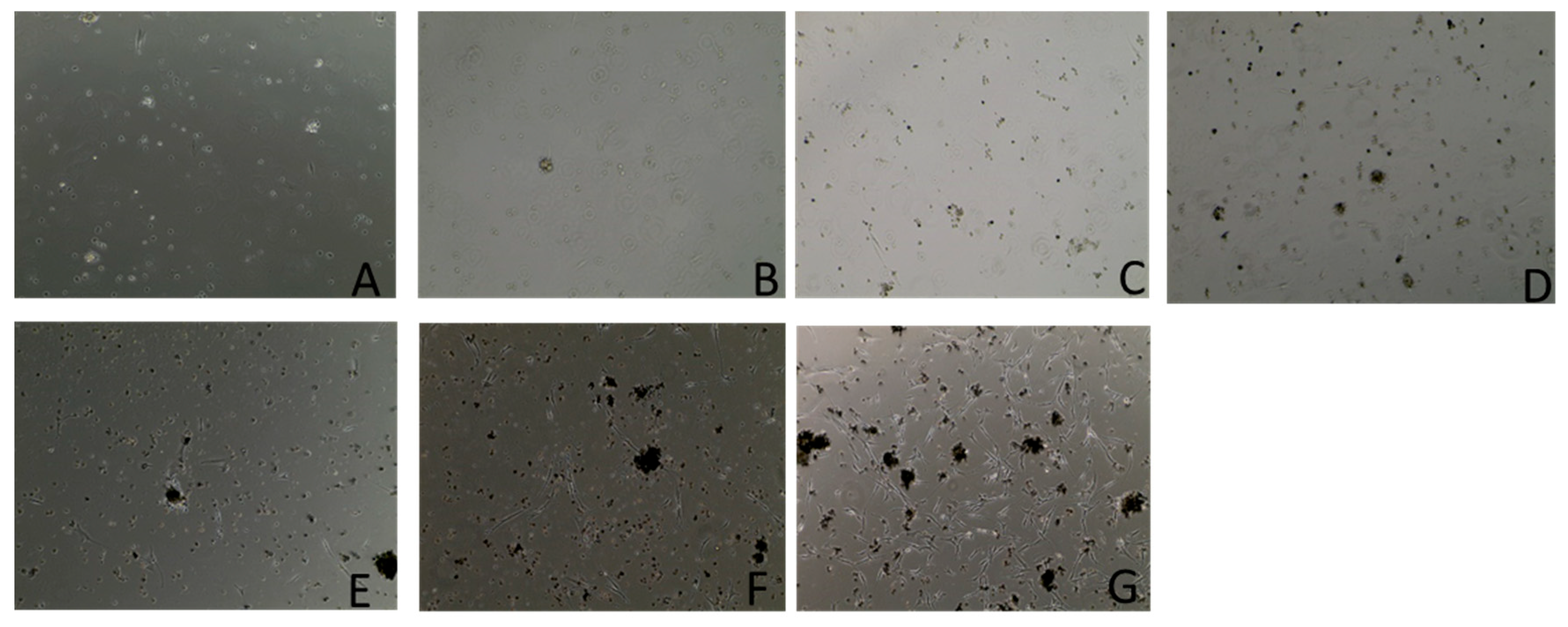

| Growth Curve Assay in RPMI Media | ||||
|---|---|---|---|---|
| Number of Days | Average Amount of Cells | Average Amount of Dead Cells | Average Amount of Live Cells | Average Amount of Total Cells |
| Day 1 | 1.12 × 106 | 5.56 × 105 | 8.39 × 105 | 1.40 × 106 |
| Day 2 | 1.35 × 106 | 4.86 × 105 | 1.20 × 106 | 1.69 × 106 |
| Day 3 | 2.28 × 106 | 4.44 × 105 | 1.63 × 106 | 2.07 × 106 |
| Day 4 | 3.03 × 106 | 5.04 × 105 | 2.25 × 106 | 2.75 × 106 |
| Day 5 | 3.27 × 106 | 5.78 × 105 | 1.76 × 106 | 2.34 × 106 |
| Day 6 | 3.27 × 106 | 5.55 × 105 | 1.78 × 106 | 2.34 × 106 |
| Day 7 | 3.91 × 106 | 6.49 × 105 | 2.14 × 106 | 2.80 × 106 |
| Number of Days | DMEM | DMEM-F12 | MEM | RPMI |
|---|---|---|---|---|
| Day 1 | 4.48 × 105 | 6.48 × 105 | 5.18 × 105 | 7.39 × 105 |
| Day 2 | 6.73 × 105 | 6.55 × 105 | 5.10 × 105 | 5.49 × 105 |
| Day 3 | 6.04 × 105 | 9.59 × 105 | 9.91 × 105 | 6.93 × 105 |
| Day 4 | 7.47 × 105 | 7.97 × 105 | 7.05 × 105 | 6.53 × 105 |
| Day 5 | 9.10 × 105 | 8.10 × 105 | 5.56 × 105 | 9.17 × 105 |
| Day 6 | 1.02 × 106 | 1.26 × 106 | 7.88 × 105 | 1.03 × 106 |
| Day 7 | 7.07 × 105 | 8.91 × 105 | 7.44 × 105 | 1.15 × 106 |
| Biomarker | Positive | Negative |
|---|---|---|
| CD-3 | Negative | |
| CD-31 | Positive | |
| CD-79 | Positive | |
| E-cadherin | Positive | |
| Vimentin | Positive | |
| Beta-Catenin | Positive | |
| IBA1 | Negative | |
| LY6G | Negative | |
| LYE1 | Positive |
| Category | Count | Percentage |
|---|---|---|
| Total reads | 16,855,223 | 100.00% |
| Paired reads | 16,855,223 | 100.00% |
| Concordant alignments | ||
| Aligned concordantly 0 times | 3,758,073 | 22.30% |
| Aligned concordantly exactly 1 time | 10,345,915 | 61.38% |
| Aligned concordantly >1 time | 2,751,235 | 16.32% |
| Discordant alignments | ||
| Pairs aligned concordantly 0 times | 3,758,073 | |
| Aligned discordantly 1 time | 2,748,824 | 73.14% |
| Unaligned pairs | ||
| Pairs aligned neither concordantly nor discordantly | 1,009,249 | |
| Mates making up these pairs | 2,018,498 | |
| Aligned 0 times | 379,206 | 18.79% |
| Aligned exactly 1 time | 323,314 | 16.02% |
| Aligned >1 time | 1,315,978 | 65.20% |
| Overall alignment rate | 98.88% |
| Category | Biallelic SNVs | Biallelic Indels | Biallelic STRs | Other Biallelic | Multiallelic SNVs | Multiallelic Indels | Multiallelic STRs | Other Multiallelic |
|---|---|---|---|---|---|---|---|---|
| Variants | 1,202,674 | 264,537 | 1366 | 0 | 220 | 692 | 942 | 0 |
| Genotype calls | 4859 | 316 | 23 | 0 | 25 | 14 | 10 | 0 |
| Coding variants | 107,385 | 23,754 | 123 | 0 | 15 | 55 | 93 | 0 |
| Category | Count |
|---|---|
| Total Biallelic SNVs | 1,202,674 |
| Coding Variants | 107,385 |
| Synonymous Variants | 5499 |
| Missense Variants | 4891 |
| Stop-Lost Variants | 13 |
| Stop-Gained Variants | 45 |
| Start-Lost Variants | 3 |
| Splice Donor Variants | 24 |
| Splice Acceptor Variants | 17 |
| Exonic Splice Region Variants | 154 |
| Splice Region Variants | 1062 |
| 5′ UTR Variants | 80,899 |
| 3′ UTR Variants | 14,778 |
| Upstream Transcript Variants | 10,368 |
| Downstream Transcript Variants | 3117 |
| Intron Variants | 357,493 |
| Intergenic Variants | 724,311 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lugo, T.; Myers, S.; Nguyen, T.A. Isolation and Characterization of Equine Lymph Node Endothelial Cells. Vet. Sci. 2025, 12, 905. https://doi.org/10.3390/vetsci12090905
Lugo T, Myers S, Nguyen TA. Isolation and Characterization of Equine Lymph Node Endothelial Cells. Veterinary Sciences. 2025; 12(9):905. https://doi.org/10.3390/vetsci12090905
Chicago/Turabian StyleLugo, Tomas, Stephanie Myers, and Thu Annelise Nguyen. 2025. "Isolation and Characterization of Equine Lymph Node Endothelial Cells" Veterinary Sciences 12, no. 9: 905. https://doi.org/10.3390/vetsci12090905
APA StyleLugo, T., Myers, S., & Nguyen, T. A. (2025). Isolation and Characterization of Equine Lymph Node Endothelial Cells. Veterinary Sciences, 12(9), 905. https://doi.org/10.3390/vetsci12090905







