The Viremic Phase and Humoral Immune Response Against African Horse Sickness Virus That Emerged in Thailand in 2020
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
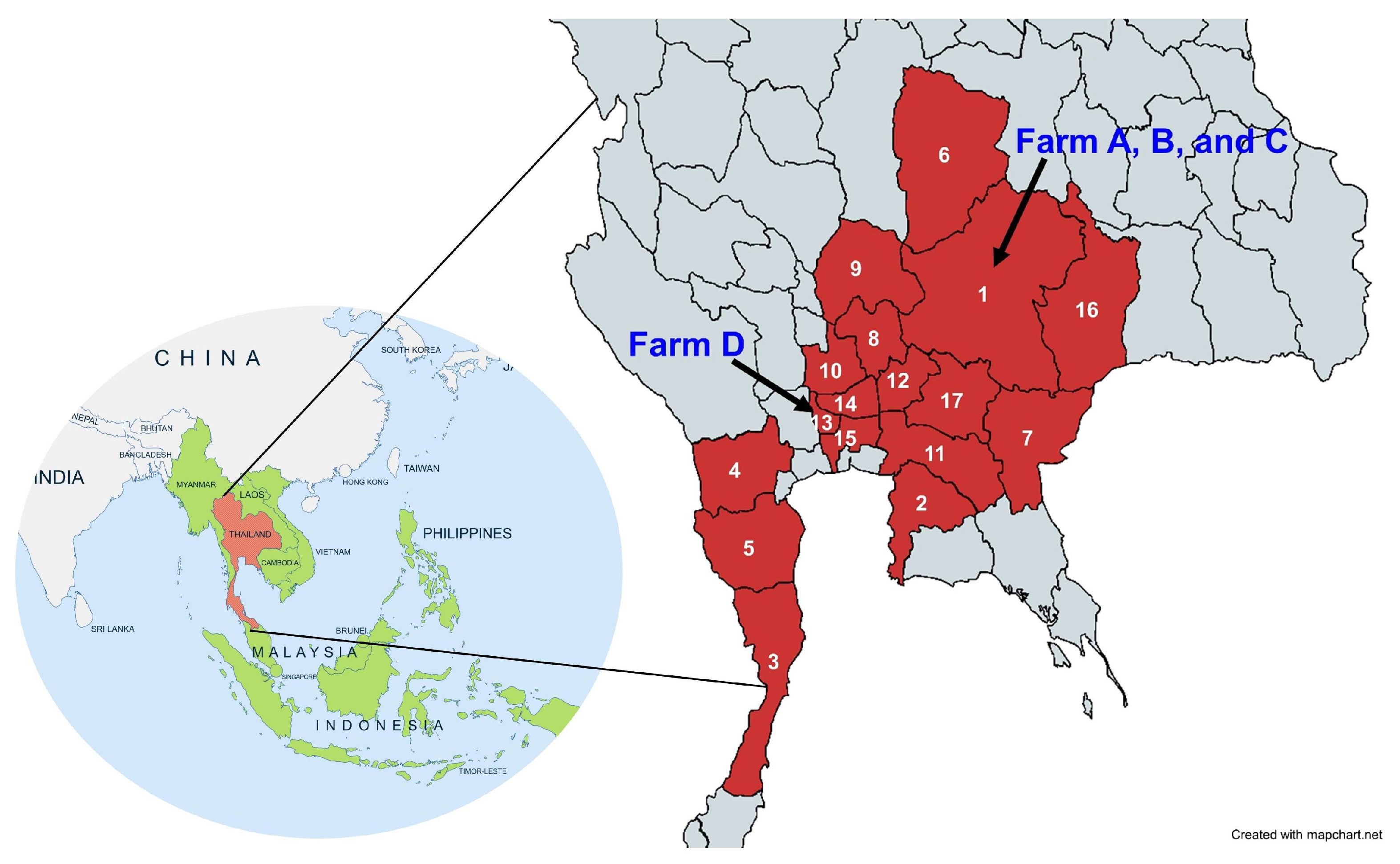
2.1. Study Design and Sample Collection
2.2. RNA Isolation and cDNA Synthesis
2.3. AHSV Detection Using Group-Specific (VP7) RT-PCR and RT Real-Time PCR
2.4. VP5 Gene Sequencing and Phylogenetic Analysis
2.5. Type-Specific (VP5) RT Nested PCR and RT Real-Time PCR for AHSV-1 Thai Field Strain and Vaccine Strain Differentiation
2.6. Immune Response Against AHSV by Blocking ELISA (bELISA)
2.7. Statistics
3. Results
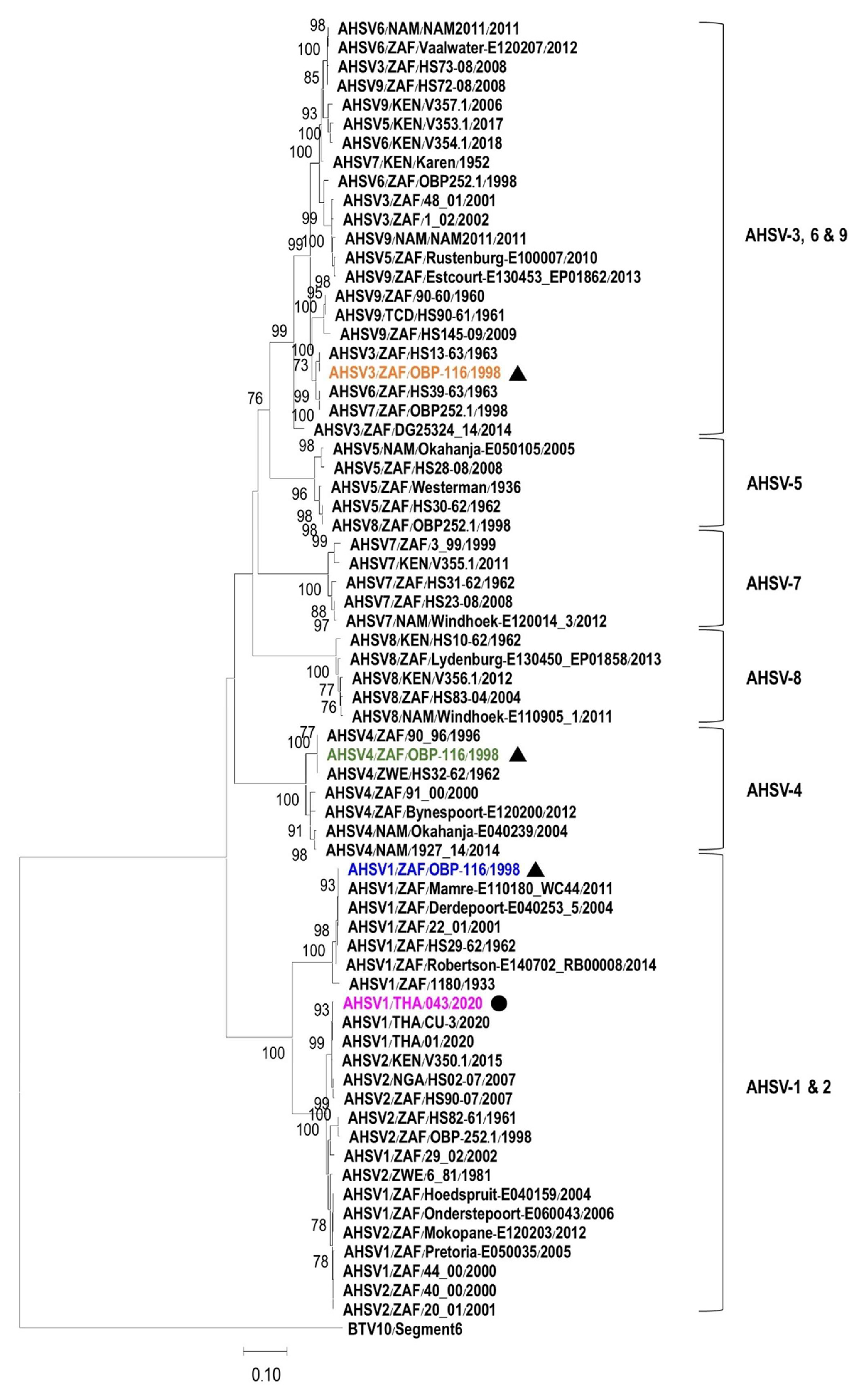
3.1. Characterization and Phylogenetic Analysis of VP5 Gene
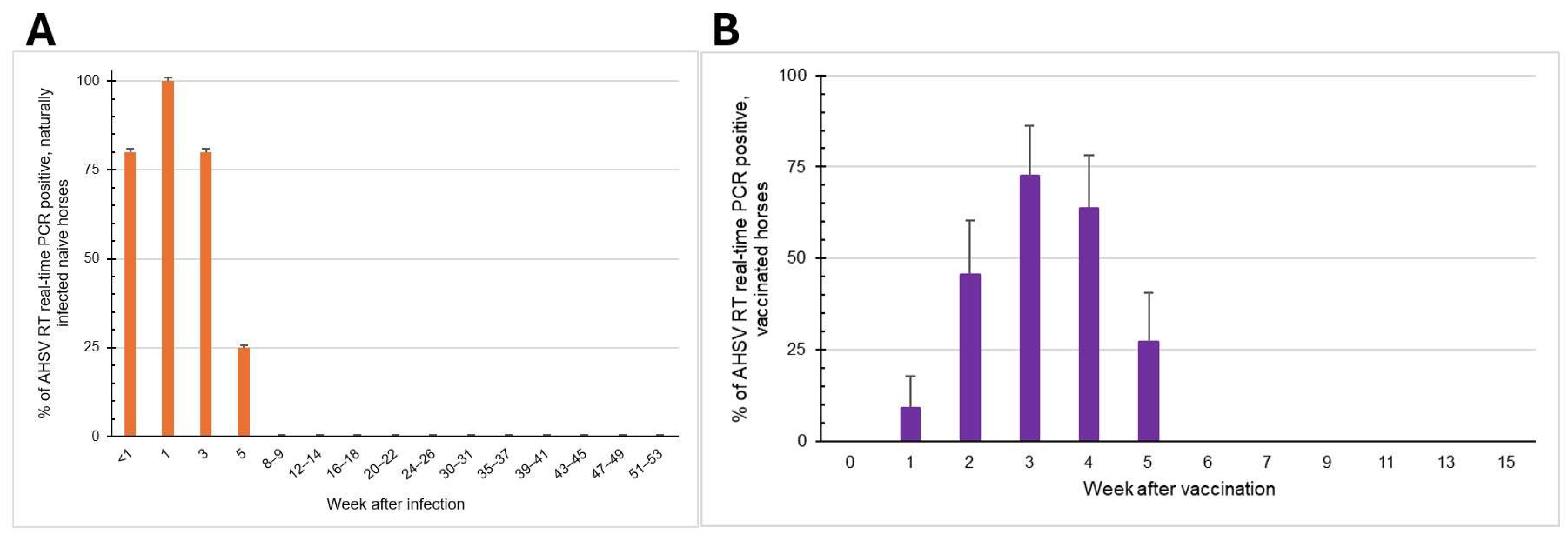
3.2. Viremic Phase and Clinical Signs of Naturally Infected Naïve Horses
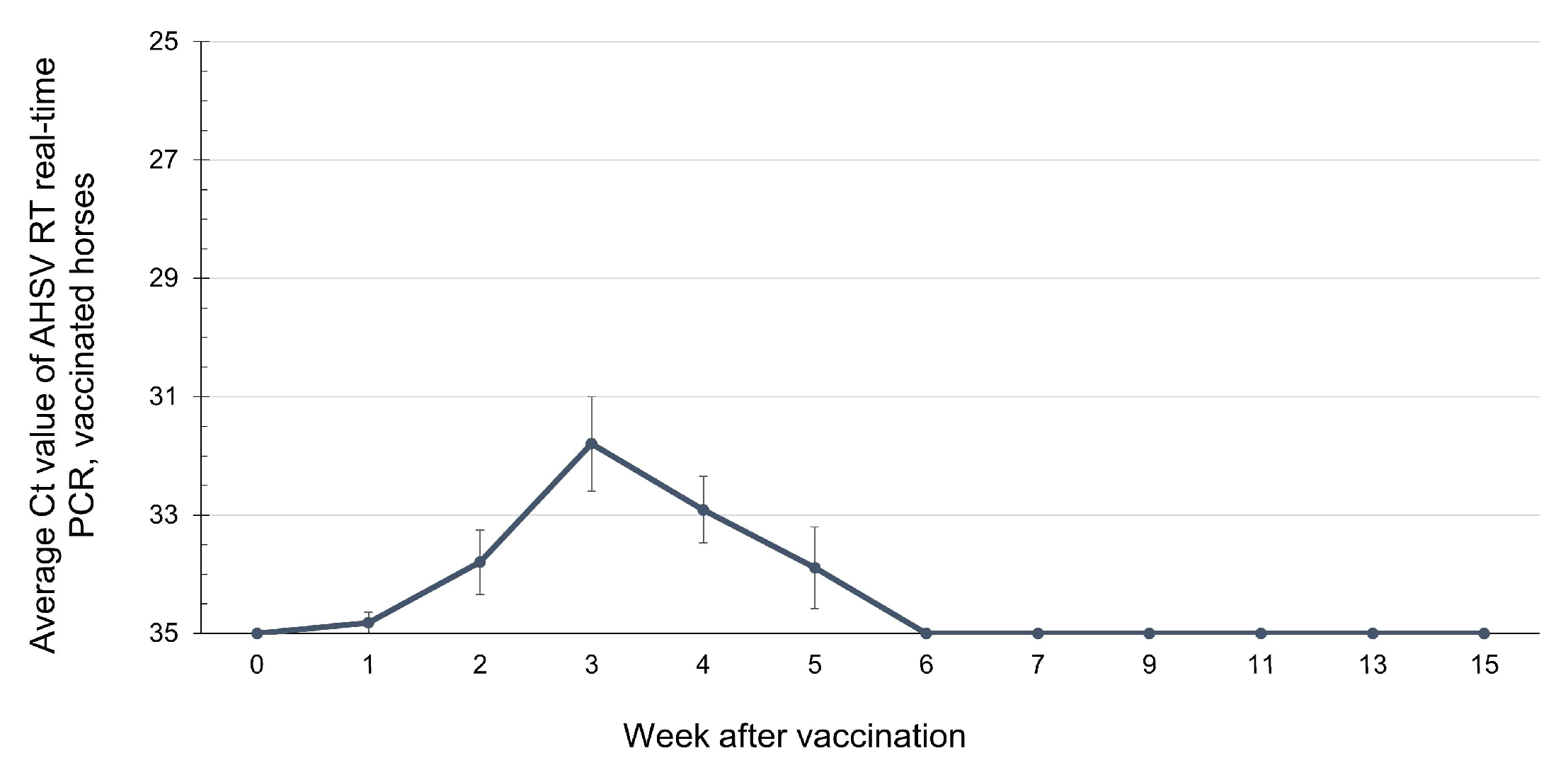
3.3. Viremic Phase of AHSV-LAV Combination-1-Vaccinated Horses
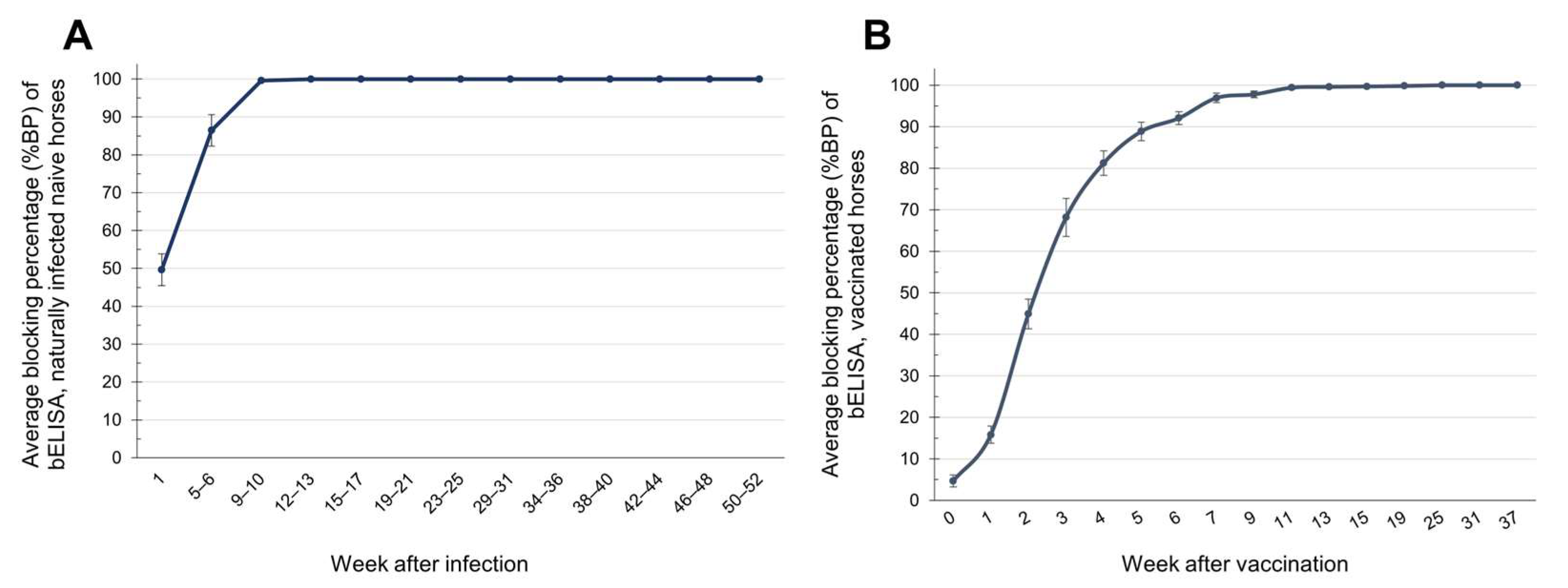
3.4. Detection of Immune Responses Against AHSV Using bELISA
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AHS | African Horse Sickness |
| AHSV | African Horse Sickness Virus |
| bELISA | Blocking ELISA |
| BLASTn | Nucleotide BLAST |
| bp | Base Pair |
| BP | Blocking Percentage |
| cDNA | Complementary Deoxyribonucleic Acid |
| CES | Capillary Electrophoresis Sequencing |
| Ct | Cycle Threshold |
| DLD | Department of Livestock Development |
| DNA | Deoxyribonucleic Acid |
| DNase | Deoxyribonuclease |
| dNTP | Deoxyribonucleotide Triphosphate |
| EDTA | Ethylenediaminetetraacetic Acid |
| ELISA | Enzyme-linked Immunosorbent Assay |
| KVDC | Kamphaeng Saen Veterinary Diagnostic Center |
| LAV | Live attenuated Vaccine |
| Mab | Monoclonal Antibody |
| NIAH | National Institute of Animal Health |
| NS | Viral Non-structural Protein |
| OBP | Onderstepoort Biological Products |
| OD | Optical Density |
| PCR | Polymerase Chain Reaction |
| RFU | Relative Fluorescence Unit |
| RNA | Ribonucleic Acid |
| RNase | Ribonuclease |
| Rpm | Revolutions Per Minute |
| RT | Reverse Transcription |
| RT-PCR | Reverse Transcription Polymerase Chain Reaction |
| SNT | Serum Neutralization Test |
| Tm | Melting Temperature |
| U | Unit |
| VP | Viral Structural Protein |
| WOAH | World Organisation for Animal Health |
| °C | Degree Celsius |
| mM | Millimolar |
| ml | Milliliter |
| µL | Microliter |
| μM | Micromolar |
References
- World Organisation for Animal Health. The OIE Technical Disease Cards: African Horse Sickness. Available online: https://www.woah.org/app/uploads/2021/09/african-horse-sickness-1.pdf (accessed on 30 March 2025).
- Spickler, A.R. African Horse Sickness. 2024. Available online: https://www.cfsph.iastate.edu/diseaseinfo/factsheets/ (accessed on 22 July 2025).
- Verhoef, F.A.; Venter, G.J.; Weldon, C.W. Thermal Limits of Two Biting Midges, Culicoides Imicola Kieffer and C. Bolitinos Meiswinkel (Diptera: Ceratopogonidae). Parasites Vectors 2014, 7, 384. [Google Scholar] [CrossRef] [PubMed]
- World Organisation for Animal Health. Terrestrial Animal Health Code: Chapter 1.3. Diseases, Infections and Infestations Listed by WOAH. Available online: https://www.woah.org/en/what-we-do/standards/codes-and-manuals/terrestrial-code-online-access/?id=169&L=1&htmfile=chapitre_oie_listed_disease.htm (accessed on 26 December 2024).
- World Organisation for Animal Health. WOAH Terrestrial Manual: Chapter 3.6.1.—African Horse Sickness (Infection with African Horse Sickness Virus). Available online: https://www.woah.org/fileadmin/Home/fr/Health_standards/tahm/3.06.01_AHS.pdf (accessed on 30 March 2025).
- King, S.; Rajko-Nenow, P.; Ashby, M.; Frost, L.; Carpenter, S.; Batten, C. Outbreak of African Horse Sickness in Thailand, 2020. Transbound. Emerg. Dis. 2020, 67, 1764–1767. [Google Scholar] [CrossRef]
- Bunpapong, N.; Charoenkul, K.; Nasamran, C.; Chamsai, E.; Udom, K.; Boonyapisitsopa, S.; Tantilertcharoen, R.; Kesdangsakonwut, S.; Techakriengkrai, N.; Suradhat, S.; et al. African Horse Sickness Virus Serotype 1 on Horse Farm, Thailand, 2020. Emerg. Infect. Dis. 2021, 27, 2208–2211. [Google Scholar] [CrossRef]
- Office of Agricultural Affairs. OIE (World Organisation for Animal Health) Has Announced Thailand’s Reinstatement of Status to a Member Recognised as Free from AHS (African Horse Sickness). Available online: https://agrithai.be/regulation/oie-world-organisation-for-animal-health-has-announced-thailands-reinstatement-of-status-to-a-member-recognised-as-free-from-ahs-african-horse-sickness/ (accessed on 18 April 2023).
- Long, M.T.; Guthrie, A.J. Equine Infectious Diseases. In Equine Infectious Diseases; Sellon, D.C., Long, M.T., Eds.; Saunders: Collingwood, ON, Canada, 2013; pp. 181–188.e4. [Google Scholar] [CrossRef]
- von Teichman, B.F.; Smit, T.K. Evaluation of the Pathogenicity of African Horsesickness (AHS) Isolates in Vaccinated Animals. Vaccine 2008, 26, 5014–5021. [Google Scholar] [CrossRef] [PubMed]
- Weyer, C.T.; Grewar, J.D.; Burger, P.; Joone, C.; Lourens, C.; MacLachlan, N.J.; Guthrie, A.J. Dynamics of African Horse Sickness Virus Nucleic Acid and Antibody in Horses Following Immunization with a Commercial Polyvalent Live Attenuated Vaccine. Vaccine 2017, 35, 2504–2510. [Google Scholar] [CrossRef] [PubMed]
- Kunanusont, N.; Taesuji, M.; Kulthonggate, U.; Rattanamas, K.; Mamom, T.; Thongsri, K.; Phannithi, T.; Ruenphet, S. Longitudinal Humoral Immune Response and Maternal Immunity in Horses after a Single Live-Attenuated Vaccination against African Horse Sickness During the Disease Outbreak in Thailand. Vet. World 2023, 16, 1690–1694. [Google Scholar] [CrossRef] [PubMed]
- Pipitpornsirikul, P.; Thangthamniyom, N.; Songkasupa, T.; Lekcharoensuk, P. Development of RT Nested-PCR Technique to Differentiate African Horse Sickness Virus Serotype 1 (AHSV-1) Circulating in Thailand from the Vaccine Strains. In Proceedings of the 63rd Kasetsart University Annual Conference, Bangkok, Thailand, 4–6 March 2025; p. 17. Available online: https://annualconference.ku.ac.th/v3/program/KUConf63-Full%20Abstracts1-1Mar2025.pdf (accessed on 30 March 2025).
- Agüero, M.; Gómez-Tejedor, C.; Cubillo, M.Á.; Rubio, C.; Romero, E.; Jiménez-Clavero, M.A. Real-Time Fluorogenic Reverse Transcription Polymerase Chain Reaction Assay for Detection of African Horse Sickness Virus. J. Vet. Diagn. Investig. 2008, 20, 325–328. [Google Scholar] [CrossRef] [PubMed]
- Guthrie, A.J.; Coetzee, P.; Martin, D.P.; Lourens, C.W.; Venter, E.H.; Weyer, C.T.; Joone, C.; le Grange, M.; Harper, C.K.; Howell, P.G.; et al. Complete Genome Sequences of the Three African Horse Sickness Virus Strains from a Commercial Trivalent Live Attenuated Vaccine. Genome Announc. 2015, 3, 814–829. [Google Scholar] [CrossRef] [PubMed]
- Toh, X.; Wang, Y.; Rajapakse, M.P.; Lee, B.; Songkasupa, T.; Suwankitwat, N.; Kamlangdee, A.; Judith Fernandez, C.; Huangfu, T. Use of Nanopore Sequencing to Characterize African Horse Sickness Virus (AHSV) from the African Horse Sickness Outbreak in Thailand in 2020. Transbound. Emerg. Dis. 2022, 69, 1010–1019. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Ong, J.; Ng, O.W.; Songkasupa, T.; Koh, E.Y.; Wong, J.P.S.; Puangjinda, K.; Fernandez, C.J.; Huangfu, T.; Ng, L.C.; et al. Development of Differentiating Infected from Vaccinated Animals (DIVA) Real-Time PCR for African Horse Sickness Virus Serotype 1. Emerg. Infect. Dis. 2022, 28, 2446–2454. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.H.; Xu, T.G.; Kannan Villalan, A.; Chi, T.Y.; Yu, X.J.; Jin, M.I.; Wu, R.N.; Ni, G.Y.; Sui, S.F.; Wang, Z.L.; et al. Environmental and Historical Determinants of African Horse Sickness: Insights from Predictive Modeling. Transbound. Emerg. Dis. 2024, 2024, 5586647. [Google Scholar] [CrossRef] [PubMed]
- Baker, T.; Carpenter, S.; Gubbins, S.; Newton, R.; Lo Iacono, G.; Wood, J.; Harrup, L.E. Can Insecticide-Treated Netting Provide Protection for Equids from Culicoides Biting Midges in the United Kingdom? Parasites Vectors 2015, 8, 604. [Google Scholar] [CrossRef] [PubMed]
- Durán-Ferrer, M.; Villalba, R.; Fernández-Pacheco, P.; Tena-Tomás, C.; Jiménez-Clavero, M.A.; Bouzada, J.A.; Ruano, M.J.; Fernández-Pinero, J.; Arias, M.; Castillo-Olivares, J.; et al. Clinical, Virological and Immunological Responses after Experimental Infection with African Horse Sickness Virus Serotype 9 in Immunologically Naïve and Vaccinated Horses. Viruses 2022, 14, 1545. [Google Scholar] [CrossRef] [PubMed]
- Crafford, J.E.; Lourens, C.W.; Smit, T.K.; Gardner, I.A.; MacLachlan, N.J.; Guthrie, A.J. Serological Response of Foals to Polyvalent and Monovalent Live-Attenuated African Horse Sickness Virus Vaccines. Vaccine 2014, 32, 3611–3616. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Molini, U.; Marucchella, G.; Maseke, A.; Ronchi, G.F.; Di Ventura, M.; Salini, R.; Scacchia, M.; Pini, A. Immunization of Horses with a Polyvalent Live-Attenuated African Horse Sickness Vaccine: Serological Response and Disease Occurrence under Field Conditions. Trials Vaccinol. 2015, 4, 24–28. [Google Scholar] [CrossRef]
- Genis, M.L.; Crafford, J.E.; Weyer, C.T.; Pollard, D.; Grewar, J.D.; Guthrie, A.J. African Horse Sickness Vaccination Status Correlated with Disease Outcome in South Africa. J. S. Afr. Vet. Assoc. 2023, 94, 99–106. [Google Scholar] [CrossRef]
| Primers | Sequences (5′ → 3′) | Purpose | Target | PCR Product Size (bp) |
|---|---|---|---|---|
| VP7_F | CGC-GAT-AGC-AGC-AAG-AGC-C | Group-specific amplification | VP7 | 256 |
| VP7_R | GTT-GCC-AAC-GCC-TGA-TCA-TA | |||
| VP5_F | GTT-TAT-TTT-TCC-AGA-AGC-CAT-GGG-TAA-ATT-C | Cloning | VP5 | 1564 |
| VP5_R | GTA-AGT-GTT-TTT-CCC-GCC-CAC-AGG-CTC-C | |||
| OP1_F | TCG-CAT-CTC-AAG-GTT-GC | First round of nested PCR (outer primers) | VP5 genes of AHSV-1 | 684 |
| OP1_R | AAG-CGC-GTT-CAT-TAT-CGT-CC | |||
| OP2_F | TAC-GTM-GAA-AAA-GCG-CT | First round of nested PCR (outer primers) | VP5 genes of AHSV-3 and -4 | 686 |
| OP2_R | TGA-TGA-TGC-GGY-GCA-ATG | |||
| ITH1_F | GCT-AGC-GGT-TGC-AAT-CAA-GTC-AAA-G | Second round of nested PCR (inner primers) | VP5 genes of Thai AHSV-1 | 547 |
| ITH1_R | CAG-ATC-TGT-GTT-ATG-CAC-CAG-CTG-TAG-T | |||
| IV1_F | ATA-TAA-TGC-ATG-GGG-GTG-CTG-TT | Second round of nested PCR (inner primers) | VP5 genes of AHSV-1 vaccine strain | 228 |
| IV1_R | GGA-GAT-CAG-TAT-TAT-GAA-CCA-ACT-GTA-AA | |||
| IV3_F | CCT-CCA-AAC-GGA-AGA-GGA-TTT-AAG-AAC-TTC | Second round of nested PCR (inner primers) | VP5 genes of AHSV-3 vaccine strain | 469 |
| IV3_R | TTC-GTA-TTC-CTT-CTT-CAC-TAG-AGG-CAT-G | |||
| IV4_F | GTT-ACA-AAC-AGA-GGA-AGA-TTT-GCG-GAC-ACG | Second round of nested PCR (inner primers) | VP5 genes of AHSV-4 vaccine strain | 437 |
| IV4_R | CAT-CGA-TTA-CGT-GCT-GCG-TTT-CTA-CG |
| Strain | Accession no. | Location | Year | %Similarity to AHSV2020/043 VP5 |
|---|---|---|---|---|
| Field AHSV-1 | MT586217 | Thailand | 2020 | 99.9 |
| Vaccine AHSV-1 | KT030334 | South Africa | 1998 | 85.5 |
| Vaccine AHSV-3 | KT030344 | South Africa | 1998 | 72.4 |
| Vaccine AHSV-4 | KT030354 | South Africa | 1998 | 71.1 |
| Field AHSV-2 | OM289934 | Kenya | 2015 | 99.5 |
| Field AHSV-2 | KP009636 | South Africa | 2007 | 99.1 |
| Field AHSV-2 | FJ196589 | Nigeria | 2008 | 99 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pipitpornsirikul, P.; Thangthamniyom, N.; Laikul, A.; Songkasupa, T.; Pathomsakulwong, W.; Apichaimongkonkun, T.; Kasemsuwan, S.; E-kobon, T.; Lekcharoensuk, P. The Viremic Phase and Humoral Immune Response Against African Horse Sickness Virus That Emerged in Thailand in 2020. Vet. Sci. 2025, 12, 878. https://doi.org/10.3390/vetsci12090878
Pipitpornsirikul P, Thangthamniyom N, Laikul A, Songkasupa T, Pathomsakulwong W, Apichaimongkonkun T, Kasemsuwan S, E-kobon T, Lekcharoensuk P. The Viremic Phase and Humoral Immune Response Against African Horse Sickness Virus That Emerged in Thailand in 2020. Veterinary Sciences. 2025; 12(9):878. https://doi.org/10.3390/vetsci12090878
Chicago/Turabian StylePipitpornsirikul, Paphavee, Nattarat Thangthamniyom, Aree Laikul, Tapanut Songkasupa, Watcharapol Pathomsakulwong, Tawanhathai Apichaimongkonkun, Suwicha Kasemsuwan, Teerasak E-kobon, and Porntippa Lekcharoensuk. 2025. "The Viremic Phase and Humoral Immune Response Against African Horse Sickness Virus That Emerged in Thailand in 2020" Veterinary Sciences 12, no. 9: 878. https://doi.org/10.3390/vetsci12090878
APA StylePipitpornsirikul, P., Thangthamniyom, N., Laikul, A., Songkasupa, T., Pathomsakulwong, W., Apichaimongkonkun, T., Kasemsuwan, S., E-kobon, T., & Lekcharoensuk, P. (2025). The Viremic Phase and Humoral Immune Response Against African Horse Sickness Virus That Emerged in Thailand in 2020. Veterinary Sciences, 12(9), 878. https://doi.org/10.3390/vetsci12090878





