Comparative Genetics of Canine and Human Cancers
Simple Summary
Abstract
1. Introduction
2. Genetics of Canine Cancer
2.1. How Similar Are Domestic Dogs to Humans in Terms of Cancer Genetics?
2.2. How Similar Are Domestic and Wild Canids in Terms of Cancer Genetics?
2.3. Are There Associations Between Individual Breeds and Individual Cancer Types in Domestic Dogs?
3. Canine Cancer Types and Their Genetic Defects in Comparison to Human Disease
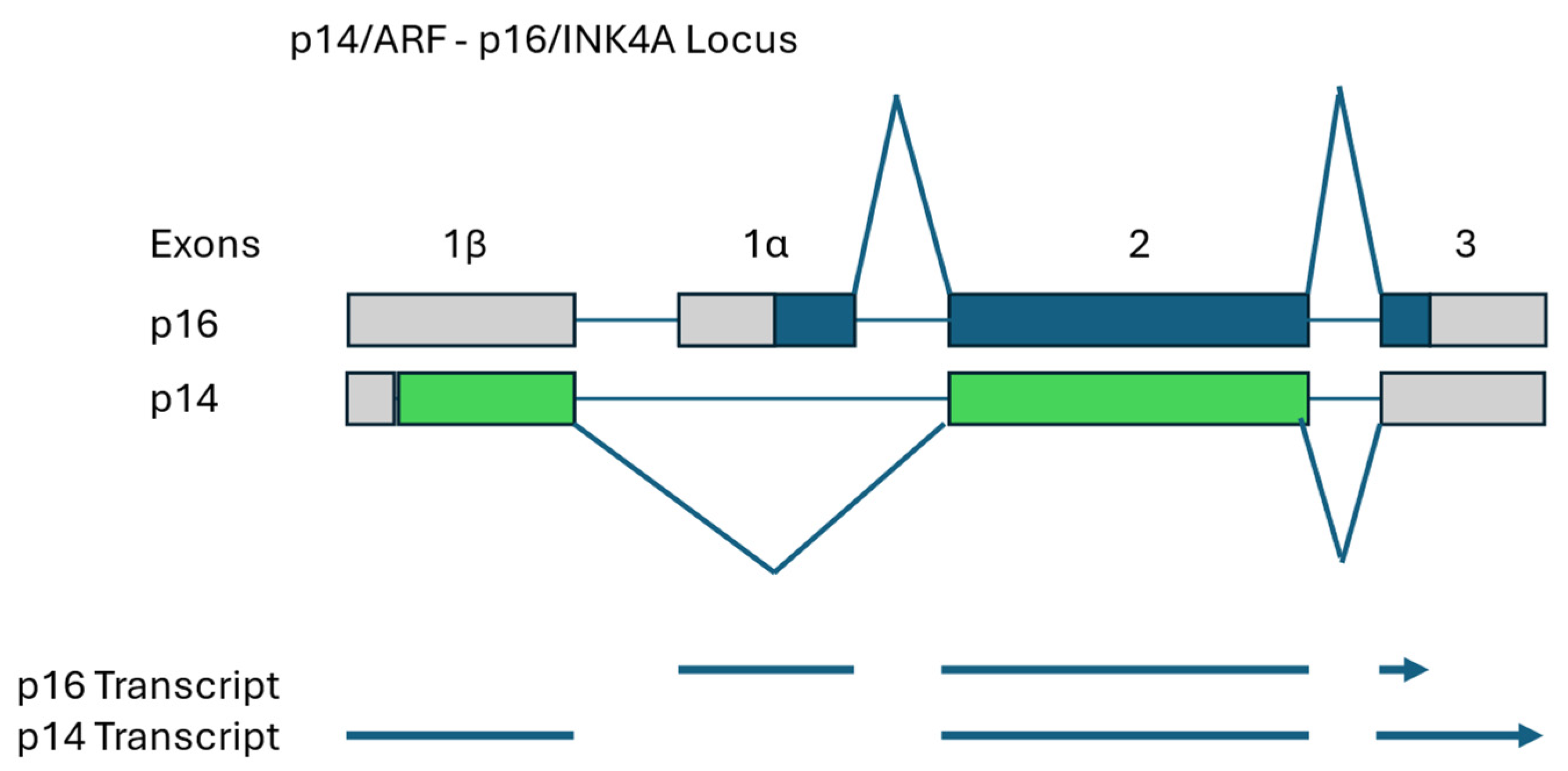
3.1. Canine Breast/Mammary Cancer
3.2. Canine Malignant Melanoma
3.3. Canine Osteosarcoma
3.4. Canine Lymphoma
3.5. Canine Transmissible Venereal Tumor—cTVT
3.6. Prostate Cancer
3.7. Can Cancer Incidence Be Modified in Individual Dogs?
4. Concluding Comments
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| SKA2 | Spindle and kinetochore-associated complex subunit 2 gene |
| NEU1 | Neuraminidase 1 gene |
| ATAC | Assay for transposase-accessible chromatin |
| TIL | Tumor infiltrating lymphocyte |
| KIT | Receptor tyrosine kinase |
| Flt3 | fms-like tyrosine kinase 3 |
| SYK | Spleen-associated tyrosine kinase |
| TIMP1 | TIMP metallopeptidase inhibitor 1 |
| RPS/L | Ribosomal protein small/large subunit |
| EDEM1 | ER degradation enhancing alpha-mannosidase-like protein 1 |
| PTK2B | Protein tyrosine kinase 2 beta |
| MMP9 | Matrix metalloproteinase 9 |
| JAK1 | Janus kinase 1 |
| PCTL | Peripheral T-cell lymphomas |
| GATA3 | Trans-Acting T-Cell-Specific Transcription Factor GATA-3 |
| PI3K | Phosphoinositol 3 kinase |
| Akt | AKT Serine/Threonine Kinase 1 |
| mTOR | Mechanistic Target Of Rapamycin Kinase |
| CCL | Chemokine |
| ITK | Interleukin-2-inducible T-cell kinase |
| LAT | Linker for activation of T-cells |
| CREBBP | cAMP response element-binding protein–binding protein |
| PD | Programmed death receptor/ligand 1 or 2 |
| Myc | Myc oncogene |
| Erg | Erythroblast transformation-specific/ETS-related gene |
| Bcl-2 | BCL2 Apoptosis Regulator gene |
| BRAF | Member of the RAS/MAPK pathway |
| CAR | Coxsackie and Adenovirus Receptor |
| MHC | Major histocompatibility gene |
| TLR | Toll-like receptor |
| DLBCL | Diffuse large B-cell lymphoma |
| BRCA | Breast cancer gene 1 and 2 |
| PAF1 | Polymerase-associated factor 1 complex subunit gene |
| PSMG2 | Proteasome assembly chaperone 2 |
| NOB1 | NIN1/RPN12 binding protein 1 homolog gene |
| MDM2 | Homolog of mouse double minute 2 p53-binding protein |
| PTEN | Phosphatase and tensin homolog gene |
| STAT3 | Signal transducer and activator of transcription 3 gene |
| ER | Estrogen receptor |
| PR | Progesterone receptor |
| erb | EGF receptor B (epidermal-like growth factor) |
| HER-2 | Human epidermal growth factor receptor 2 |
| INK | Inhibitor of cyclin-dependent kinase |
| CDK | Cyclin-dependent kinase |
| ARF | Alternative reading frame |
| Q-RT-PCR | Quantitative reverse-transcriptase polymerase chain reaction |
| FISH | Fluorescence in situ hybridization |
| FHIT | Fragile histidine triad |
| CAR | Coxsakie and adenovirus receptor |
| CHOP | Cyclophosphamide, hydroxydaunorubicin, oncovin and prednisone therapy |
| cTVT | Canine transmissible venereal tumor |
| NK | Natural killer |
References
- Iasi, L.N.M.; Chintalapati, M.; Skov, L.; Bossoms Mesa, A.; Hajdinjak, M.; Peter, B.M.; Moorjani, P. Neanderthal Ancestry Through Time: Insights From Genomes of Ancient and Present-Day Humans. Science 2024, 384, 1170–1172. [Google Scholar] [CrossRef]
- Martinón-Torres, M.; Lalueza-Fox, C. Ancient Human Genomes Offer Clues About the Earliest Migrations Out of Africa. Nature 2025, 38, 620–621. [Google Scholar] [CrossRef] [PubMed]
- Pennisi, E.A. Shaggy Dog History. Science 2002, 298, 1540–1542. [Google Scholar] [CrossRef]
- Frantz, L.A.F.; Bradley, D.; Larson, G.; Orlando, L. Animal Domestication in the Era of Ancient Genomics. Nat. Rev. Genet. 2020, 21, 449–460. [Google Scholar] [CrossRef]
- Kornegay, J.N. The Golden Retriever Model of Duchenne Muscular Dystrophy. Skelet. Muscle 2017, 7, 9. [Google Scholar] [CrossRef]
- Grimm, D. Dogs May Have Been Domesticated More than Once But All Living Dogs Have Asian Roots. Science 2016, 352, 1153–1154. [Google Scholar] [CrossRef]
- Sexton, C.L.; Ruple, A. How Can We Achieve More Accurate Reporting of Average Dog Lifespan? J. Am. Vet. Med. Assoc. 2024, 262, 1–5. [Google Scholar] [CrossRef]
- Julve, M.; Lythgoe, M.P.; Larkin, J.; Furness, A.J.S. Lifileucel: The First Cellular Therapy Approved for Solid Tumours. Trends Cancer 2024, 10, 475–477. [Google Scholar] [CrossRef]
- Bhutta, Z.A.; Choi, K.C. Canine Mammary Tumors as a Promising Adjunct Preclinical Model for Human Breast Cancer Research: Similarities, Opportunities, and Challenges. Arch. Pharm. Res. 2025, 48, 43–61. [Google Scholar] [CrossRef] [PubMed]
- London, C.A.; Gardner, H.; Zhao, S.; Knapp, D.W.; Utturkar, S.M.; Duval, D.L.; Chambers, M.L.; Ostrander, E.; Trent, J.F.; Kuffel, G. Leading the Pack: Best Practices in Comparative Canine Cancer. Genomics to Inform Human Oncology. Vet. Comp. Oncol. 2023, 21, 565–577. [Google Scholar] [CrossRef] [PubMed]
- Bryn Nelson, B.; Faquin, W. Retrieving New Clues About a Dog Breed’s “Insane” Cancer Risk Cancer. Cytopathology 2024, 132, 541–542. [Google Scholar]
- Bird, R.C. Defects in Genes Regulating the Cell Cycle in Spontaneous Canine Models of Cancer. In Trends in Cell Cycle Research; Yoshida, K., Ed.; Research Sign Post: Kerala, India, 2009; pp. 209–236. [Google Scholar]
- Lutful Kabir, F.M.; DeInnocentes, P.; Bird, R.C. Altered miRNA Expression Profiles Identify miR-141 as the Regulator of INK4A (p16 and p14ARF) Tumor Suppressor Genes in Canine Breast Cancer Models. J. Cell. Biochem. 2015, 116, 2956–2969. [Google Scholar] [CrossRef]
- Lutful Kabir, F.M.; DeInnocentes, P.; Agarwal, P.; Mill, C.P.; Riese, D.J., II; Bird, R.C. Estrogen Receptor-α, Progesterone Receptor and c-erbB/HER-Family Receptor mRNA Detection and Phenotype Analysis in Spontaneous Canine Models of Breast Cancer. J. Vet. Sci. 2017, 18, 149–158. [Google Scholar] [CrossRef] [PubMed]
- Osaki, T.; Sunden, Y.; Sugiyama, A.; Azuma, K.; Murahata, Y.; Tsuka, T.; Ito, N.; Imagawa, T.; Okamoto, Y. Establishment of a Canine Mammary Gland Tumor Cell Line and Characterization of its miRNA Expression. J. Vet. Sci. 2016, 17, 385–390. [Google Scholar] [CrossRef]
- Fish, E.J.; Irizarry, K.J.; DeInnocentes, P.; Ellis, C.; Prasad, N.; Moss, A.G.; Bird, R.C. Malignant Canine Mammary Epithelial Cells Shed Exosomes Containing Differentially Expressed MicroRNA That Regulate Oncogenic Networks. BMC Cancer 2018, 18, 832–851. [Google Scholar] [CrossRef]
- Fish, E.J.; Martinez Romero, E.G.; DeInnocentes, P.; Kohler, J.; Smith, A.N.; Prasad, N.; Bird, R.C. Circulating MicroRNA as Biomarkers of Canine Mammary Carcinoma. J. Vet. Intern. Med. 2020, 34, 1282–1290. [Google Scholar] [CrossRef]
- Gojobori, J.; Arakawa, N.; Xiaokaiti, X.; Matsumoto, Y.; Matsumura, S.; Hongo, H.; Ishiguro, N.; Terai, Y. Japanese Wolves are Most Closely Related to Dogs and Share DNA With East Eurasian Dogs. Nat. Commun. 2024, 15, 1680–1691. [Google Scholar] [CrossRef] [PubMed]
- Bergström, A.; Frantz, L.; Schmidt, R.; Ersmark, E.; Lebrasseur, O.; Flink, L.G.; Lin, A.T.; Storå, J.; Sjögren, K.-G.; Anthony, D.; et al. Origins and Genetic Legacy of Prehistoric Dogs. Science 2020, 370, 557–564. [Google Scholar] [CrossRef] [PubMed]
- Morell, V. Rethinking the North American Wolf. Science 2016, 353, 434–435. [Google Scholar] [CrossRef]
- Freedman, A.H.; Wayne, R.K. Deciphering the Origin of Dogs: From Fossils to Genomes. Annu. Rev. Anim. Biosci. 2017, 5, 281–307. [Google Scholar] [CrossRef]
- Kopaliani, N.; Shakarashvili, M.; Gurielidze, Z.; Qurkhuli, T.; Tarkhnishvili, D. Gene Flow Between Wolf and Shepherd Dog Populations in Georgia (Caucasus). J. Hered. 2014, 105, 345–353. [Google Scholar] [CrossRef]
- Stronen, A.V.; Mattucci, F.; Fabbri, E.; Galaverni, M.; Cocchiararo, B.; Nowak, C.; Godinho, R.; González, A.R.; Kusak, J.; Skrbinšek, T.; et al. A Reduced SNP Panel to Trace Gene Flow Across Southern European Wolf Populations and Detect Hybridization with Other Canis Taxa. Sci. Rep. 2022, 12, 4195–4208. [Google Scholar] [CrossRef] [PubMed]
- Seeley, K.E.; Garner, M.M.; Waddell, W.T.; Wolf, K.N. A Survey of Diseases in Captive Red Wolves (Canis rufus), 1997–2012. J. Zoo Wildl. Med. 2016, 47, 83–90. [Google Scholar] [CrossRef]
- Grimm, D. Siberia May be Long-Sought Site of Dog Domestication. Science 2021, 371, 451–452. [Google Scholar] [CrossRef]
- Spatola, G.J.; Buckley, R.M.; Dillon, M.; Dutrow, E.V.; Betz, J.A.; Pilot, M.; Parker, H.G.; Bogdanowicz, W.; Thomas, R.; Chyzhevskyi, I.; et al. The Dogs of Chernobyl: Demographic Insights Into Populations Inhabiting the Nuclear Exclusion Zone. Sci. Adv. 2023, 9, eade2537. [Google Scholar] [CrossRef]
- Huskey, A.L.W.; McNeely, I.; Merner, N.D. CEACAM Gene Family Mutations Associated with Inherited Breast Cancer Risk—A Comparative Oncology Approach to Discovery. Front. Genet. 2021, 12, 702889. [Google Scholar] [CrossRef]
- Goebel, K.; Merner, N.D. A Monograph Proposing the Use of Canine Mammary Tumours as a Model for the Study of Hereditary Breast Cancer Susceptibility Genes in Humans. Vet. Med. Sci. 2017, 3, 51–62. [Google Scholar] [CrossRef]
- Pinho, S.S.; Carvalho, S.; Cabral, J.; Reis, C.A.; Gärtner, F. Canine Tumors: A Spontaneous Animal Model of Human Carcinogenesis. Transl. Res. 2012, 159, 165–172. [Google Scholar] [CrossRef] [PubMed]
- Scott, M.C.; Sarver, A.L.; Gavin, K.J.; Thayanithy, V.; Getzy, D.M.; Newman, R.A.; Cutter, G.R.; Lindblad-Toh, K.; Kisseberth, W.C.; Hunter, L.E.; et al. Molecular Subtypes of Osteosarcoma Identified by Reducing Tumor Heterogeneity Through an Interspecies Comparative Approach. Bone 2011, 49, 356–367. [Google Scholar] [CrossRef] [PubMed]
- Dhein, E.S.; Heikkila, U.; Oevermann, A.; Blatter, S.; Meier, D.; Hartnack, S.; Guscetti, F. Incidence Rates of the Most Common Canine Tumors Based on Data From the Swiss Canine Cancer Registry (2008 to 2020). PLoS ONE 2024, 19, e0302231. [Google Scholar] [CrossRef]
- Vail, D.M.; Thamm, D.H.; Liptak, J.M. Withrow and MacEwen’s Small Animal Clinical Oncology, 6th ed.; Saunders: Philadelphia, PA, USA, 2019. [Google Scholar]
- Kumar, V.; Abbas, A.K.; Aster, J.C.; Deyrup, A.T. Robbins and Kumar’s Basic Pathology, 11th ed.; Elsevier: Cambridge, MA, USA, 2025. [Google Scholar]
- Paoloni, M.; Khanna, C. Translation of new cancer treatments from pet dogs to humans. Nat. Rev. Cancer 2008, 8, 147–150. [Google Scholar] [CrossRef] [PubMed]
- LeBlanc, A.K.; Breen, M.; Choyke, P.; Dewhirst, M.; Fan, T.M.; Gustafson, D.L.; Helman, L.J.; Kastan, M.B.; Knapp, D.W.; Levin, W.J.; et al. Perspectives from man’s best friend: National Academy of Medicine’s Workshop on Comparative Oncology. Sci. Transl. Med. 2016, 8, 324ps5. [Google Scholar] [CrossRef]
- Carvalho, P.T.; Niza-Ribeiro, J.; Amorim, I.; Queiroga, F.; Severo, M.; Ribeiro, A.I.; Pinello, K. Comparative Epidemiological Study of Breast Cancer in Humans and Canine Mammary Tumors: Insights From Portugal. Front. Vet. Sci. 2023, 10, 1271097. [Google Scholar] [CrossRef]
- Dobson, J.M. Breed Predispositions to Cancer in Pedigree Dogs. ISRN Vet. Sci. 2013, 2013, 941275. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.-W.; Lim, H.-Y.; Shin, J.-I.; Seung, B.-J.; Ju, J.-H.; Sur, J.-H. Breed- and Age-Related Differences in Canine Mammary Tumors. Can. J. Vet. Res. 2016, 80, 146–155. [Google Scholar] [PubMed]
- Munson, L.; Moresco, A. Comparative Pathology of Mammary Gland Cancers in Domestic and Wild Animals. Breast. Dis. 2007, 28, 7–21. [Google Scholar] [CrossRef]
- Vazquez, E.; Lipovka, Y.; Cervantes-Arias, A.; Garibay-Escobar, A.; Haby, M.M.; Queiroga, F.L.; Velazquez, C. Canine Mammary Cancer: State of the Art and Future Perspectives. Animals 2023, 13, 3147. [Google Scholar] [CrossRef]
- Pasaol, J.C.; Smieszek, A.; Pawlak, A. Exploring the Therapeutic Potential of BRCA1 and BRCA2 as Targets in Canine Oncology: A Comprehensive Review of Their Role in Cancer Development and Treatment. Int. J. Mol. Sci. 2025, 26, 1768. [Google Scholar] [CrossRef]
- Cassali, G.D.; Nakagaki, K.Y.R.; Salvi, M.; Possa dos Reys, M.; Rocha, M.A.N.; Bonolo de Campos, C.; Ferreira, E.; Rodrigues, A.C.B.; Carlos dos Reis, D.; Damasceno, K.A.; et al. Canine, Feline, and Murine Mammary Tumors as a Model for Translational Research in Breast Cancer. Vet. Sci. 2025, 12, 189. [Google Scholar] [CrossRef]
- Ahern, T.E.; Bird, R.C.; Church Bird, A.E.; Wolfe, L.G. Overexpression of the Oncogene c-erbB-2 in Canine Mammary Carcinomas and Tumor-Derived Cell Lines. Am. J. Vet. Res. 1996, 57, 693–696. [Google Scholar]
- Lutful Kabir, F.M.; Alvarez, C.E.; Bird, R.C. Canine Mammary Carcinomas: A Comparative Analysis of Altered Gene Expression. Vet. Sci. 2015, 3, 1. [Google Scholar] [CrossRef] [PubMed]
- Bird, R.C.; DeInnocentes, P.; Church Bird, A.E.; Lutful Kabir, F.M.; Smith, A.; Smith, B.F. Autologous Hybrid Cell Fusion Vaccine in a Spontaneous Intermediate Model of Breast Carcinoma. J. Vet. Sci. 2019, 20, e48. [Google Scholar] [CrossRef] [PubMed]
- Bird, R.C.; DeInnocentes, P.; Church Bird, A.E.; van Ginkel, F.W.; Smith, B.F. An Autologous Dendritic Cell-Canine Mammary Tumor Hybrid-Cell Fusion Vaccine. Cancer Immunol. Immunother. 2011, 60, 87–97. [Google Scholar] [CrossRef] [PubMed]
- Murakami, A.; Mori, T.; Sakai, H.; Murakami, M.; Yanai, T.; Hoshino, Y.; Maruo, K. Analysis of KIT Expression and KIT Exon 11 Mutations in Canine Oral Malignant Melanomas. Vet. Comp. Oncol. 2011, 9, 219–224. [Google Scholar] [CrossRef]
- Dankort, D.; Curley, D.P.; Cartlidge, R.A.; Nelson, B.; Karnezis, A.N.; Damsky, W.E., Jr.; You, M.J.; DePinho, R.A.; McMahon, M.; Bosenberg, M. Braf(V600E) Cooperates With PTEN Loss to Induce Metastatic Melanoma. Nat. Genet. 2009, 41, 544–552. [Google Scholar] [CrossRef]
- Fowles, J.S.; Denton, C.L.; Gustafson, D.L. Comparative Analysis of MAPK and PI3K/AKT Pathway Activation and Inhibition in Human and Canine Melanoma. Vet. Comp. Oncol. 2015, 13, 288–304. [Google Scholar] [CrossRef]
- Smedley, R.C.; Thaiwong, T.; Deeth, L.E.; Kiupel, M. Correlation Between KIT Expression and c-Kit Mutations in 2 Subtypes of Canine Oral Melanocytic Neoplasms. Vet. Pathol. 2021, 58, 683–691. [Google Scholar] [CrossRef]
- Tani, H.; Miyamoto, R.; Noguchi, S.; Kurita, S.; Nagashima, T.; Michishita, M.; Yayoshi, N.; Tamura, K.; Bonkobara, M. A Canine Case of Malignant Melanoma Carrying a KIT c.1725_1733del Mutation Treated with Toceranib: A Case Report and In Vitro Analysis. BMC Vet. Res. 2021, 17, 147–153. [Google Scholar] [CrossRef]
- Ahern, T.E.; Bird, R.C.; Church Bird, A.E.; Wolfe, L.G. Overexpression of c-erbB-2 and c-myc but Not c-ras, in Canine Melanoma Cell Lines, is Associated With Metastatic Potential in Nude Mice. Anticancer Res. 1993, 13, 1365–1372. [Google Scholar]
- Stevenson, V.B.; Perry, S.N.; Todd, M.; Huckle, W.R.; LeRoith, T. PD-1, PD-L1, and PD-L2 Gene Expression and Tumor Infiltrating Lymphocytes in Canine Melanoma. Vet. Pathol. 2021, 58, 692–698. [Google Scholar] [CrossRef]
- Inanaga, S.; Igase, M.; Sakai, Y.; Tanabe, M.; Shimonohara, N.; Itamoto, K.; Nakaichi, M.; Mizuno, T. Mismatch Repair Deficiency in Canine Neoplasms. Vet. Pathol. 2021, 58, 1058–1063. [Google Scholar] [CrossRef] [PubMed]
- Hiddemann, W.; Roessner, A.; Wörmann, B.; Mellin, W.; Klockenkemper, B.; Bösing, T.; Büchner, T.; Grundmann, E. Tumor Heterogeneity in Osteosarcoma as Identified by Flow Cytometry. Cancer 1987, 59, 324–328. [Google Scholar] [CrossRef] [PubMed]
- Nance, R.L.; Sajib, A.M.; Smith, B.F. Canine Models of Human Cancer: Bridging the Gap to Improve Precision Medicine. Prog. Mol. Biol. Transl. Sci. 2022, 189, 67–99. [Google Scholar]
- Levine, R.A.; Forest, T.; Smith, C. Tumor Suppressor PTEN is Mutated in Canine Osteosarcoma Cell Lines and Tumors. Vet. Pathol. 2002, 39, 372–378. [Google Scholar] [CrossRef]
- Sarver, A.L.; Mills, L.J.; Makielski, K.M.; Temiz, N.A.; Wang, J.; Spector, L.G.; Subramanian, S.; Modiano, J.F. Distinct Mechanisms of PTEN Inactivation in Dogs and Humans Highlight Convergent Molecular Events That Drive Cell Division in the Pathogenesis of Osteosarcoma. Cancer Genet. 2023, 276–277, 1–11. [Google Scholar] [CrossRef]
- Tawa, G.J.; Braisted, J.; Gerhold, D.; Grewal, G.; Mazcko, C.; Breen, M.; Sittampalam, G.; LeBlanc, A.K. Transcriptomic Profiling in Canines and Humans Reveals Cancer Specific Gene Modules and Biological Mechanisms Common to Both Species. PLoS Comput. Biol. 2021, 17, e1009450. [Google Scholar] [CrossRef]
- Karlsson, E.K.; Sigurdsson, S.; Ivansson, E.; Thomas, R.; Elvers, I.; Wright, J.; Howald, C.; Tonomura, N.; Perloski, M.; Swofford, R.; et al. Genome-Wide Analyses Implicate 33 Loci in Heritable Dog Osteosarcoma, Including Regulatory Variants Near CDKN2A/B. Genome Biol. 2013, 14, R132. [Google Scholar] [CrossRef]
- Letko, A.; Minor, K.M.; Norton, E.M.; Marinescu, V.D.; Drögemüller, M.; Ivansson, E.; Megquier, K.; Noh, H.J.; Starkey, M.; Friedenberg, S.G.; et al. Genome-Wide Analyses for Osteosarcoma in Leonberger Dogs Reveal the CDKN2A/B Gene Locus as a Major Risk Locus. Genes 2021, 12, 1964. [Google Scholar] [CrossRef] [PubMed]
- Lucas, S.E.; Yang, T.; Wimberly, C.E.; Parmar, K.V.; Hansen, H.M.; de Smith, A.J.; Morimoto, L.M.; Metayer, C.; Ostrom, Q.T.; Eward, W.C.; et al. Genetic Variation Near GRB10 Associated With Bone Growth and Osteosarcoma Risk in Canine and Human Populations. Cancer Epidemiol. 2024, 12, 102599–102606. [Google Scholar] [CrossRef]
- Makielski, K.M.; Donnelly, A.J.; Khammanivong, A.; Scott, M.C.; Ortiz, A.R.; Galvan, D.C.; Tomiyasu, H.; Amaya, C.; Ward, K.A.; Montoya, A.; et al. Development of an Exosomal Gene Signature to Detect Residual Disease in Dogs with Osteosarcoma Using a Novel Xenograft Platform and Machine Learning. Lab. Investig. 2021, 101, 1585–1596. [Google Scholar] [CrossRef]
- Nance, R.L.; Wang, X.; Sandey, M.; Matz, B.M.; Thomas, A.; Smith, B.F. Single-Nuclei Multiome (ATAC + Gene Expression) Sequencing of a Primary Canine Osteosarcoma Elucidates Intra-Tumoral Heterogeneity and Characterizes the Tumor Microenvironment. Int. J. Mol. Sci. 2023, 24, 16365. [Google Scholar] [CrossRef]
- Ammons, D.; Hopkins, L.; Cronise, K.; Kurihara, J.; Regan, D.; Dow, S. Single-Cell RNA Sequencing Reveals the Cellular and Molecular Heterogeneity of Treatment-Naïve Primary Osteosarcoma in Dogs. 2024. Commun. Biol. 2024, 7, 496–513. [Google Scholar] [CrossRef]
- Mannheimer, J.D.; Tawa, G.; Gerhold, D.; Braisted, J.; Sayers, C.M.; McEachron, T.A.; Meltzer, P.; Mazcko, C.; Beck, J.A.; LeBlanc, A.K. Transcriptional Profiling of Canine Osteosarcoma Identifies Prognostic Gene Expression Signatures With Translational Value for Humans. Commun. Biol. 2023, 6, 856–873. [Google Scholar] [CrossRef]
- Zandvliet, M. Canine lymphoma: A review. Vet. Q. 2016, 36, 76–104. [Google Scholar] [CrossRef]
- Breen, M.; Modiano, J.F. Evolutionarily Conserved Cytogenetic Changes in Hematological Malignancies of Dogs and Humans--Man and His Best Friend Share More Than Companionship. Chromosome Res. 2008, 16, 145–154. [Google Scholar] [CrossRef] [PubMed]
- Tonomura, N.; Elvers, I.; Thomas, R.; Megquier, K.; Turner-Maier, J.; Howald, C.; Sarver, A.L.; Swofford, R.; Frantz, A.M.; Ito, D.; et al. Genome-Wide Association Study Identifies Shared Risk Loci Common to Two Malignancies in Golden Retrievers. PLoS Genet. 2015, 11, e1004922. [Google Scholar] [CrossRef] [PubMed]
- Hiraoka, H.; Minami, K.; Kaneko, N.; Shimokawa Miyama, T.; Okamura, Y.; Mizuno, T.; Okuda, M. Aberrations of the FHIT Gene and Fhit Protein in Canine Lymphoma Cell Lines. J. Vet. Med. Sci. 2009, 71, 769–777. [Google Scholar] [CrossRef]
- Musser, M.L.; Viall, A.K.; Phillips, R.L.; Fasina, O.; Johannes, C.M. Prostaglandin EP4 Receptor mRNA Expression in Canine Lymphoma. Vet. Comp. Oncol. 2022, 20, 127–133. [Google Scholar] [CrossRef]
- Suter, S.E.; Small, G.W.; Seiser, E.L.; Thomas, R.; Breen, M.; Richards, K.L. FLT3 Mutations in Canine Acute Lymphocytic Leukemia. BMC Cancer 2011, 11, 38–46. [Google Scholar] [CrossRef]
- Aricò, A.; Giantin, M.; Gelain, M.E.; Riondato, F.; Comazzi, S.; Rütgen, B.C.; Essler, S.E.; Dacasto, M.; Castagnaro, M.; Aresu, L. The Role of Vascular Endothelial Growth Factor and Matrix Metalloproteinases in Canine Lymphoma: In Vivo and In Vitro Study. BMC Vet. Res. 2013, 9, 94–103. [Google Scholar] [CrossRef] [PubMed]
- Mortarino, M.; Gelain, M.E.; Gioia, G.; Ciusani, E.; Bazzocchi, C.; Comazzi, S. ZAP-70 and Syk Expression in Canine Lymphoid Cells and Preliminary Results on Leukaemia Cases. Vet. Immunol. Immunopathol. 2009, 128, 395–401. [Google Scholar] [CrossRef] [PubMed]
- Aricò, A.; Giantin, M.; Gelain, M.; Riondato, F.; Mortarino, M.; Comazzi, S.; Dacasto, M.; Castagnaro, M.; Aresu, L. Matrix Metalloproteinases and Vascular Endothelial Growth Factor Expression in Canine Leukaemias. Vet. J. 2013, 196, 260–262. [Google Scholar] [CrossRef]
- Owens, E.; Harris, L.; Harris, A.; Yoshimoto, J.; Burnett, R.; Avery, A. The Gene Expression Profile and Cell of Origin of Canine Peripheral T-Cell Lymphoma. BMC Cancer 2024, 24, 18–36. [Google Scholar] [CrossRef]
- Toyoda, H.; Tani, A.; Goto-Koshino, Y.; Motegi, T.; Sakamoto, M.; Mochizuki, T.; Harada, K.; Kobayashi, T.; Setoguchi, A.; Shizuta, Y.; et al. Gene Expression Profiles Associated With Early Relapse During First Remission Induction in Canine Multicentric High-Grade B-Cell Lymphoma. J. Vet. Med. Sci. 2024, 86, 18–27. [Google Scholar] [CrossRef]
- Dittrich, K.; Yıldız-Altay, Ü.; Qutab, F.; Kwong, D.A.; Rao, Z.; Nievez-Lozano, S.A.; Gardner, H.L.; Richmond, J.M.; London, C.A. Baseline Tumor Gene Expression Signatures Correlate With Chemoimmunotherapy Treatment Responsiveness in Canine B Cell Lymphoma. PLoS ONE 2023, 18, e0290428. [Google Scholar] [CrossRef]
- Agarwal, P.; Gammon, E.A.; Sajib, A.M.; Sandey, M.; Smith, B.F. Cell-Surface Integrins and CAR Are Both Essential for Adenovirus Type 5 Transduction of Canine Cells of Lymphocytic Origin. PLoS ONE 2017, 12, e0169532. [Google Scholar] [CrossRef] [PubMed]
- VonHoldt, B.M.; Ostrander, E.A. The Singular History of a Canine Transmissible Tumor. Cell 2006, 126, 445–447. [Google Scholar] [CrossRef]
- Ostrander, E.A.; Davis, B.W.; Ostrander, G.K. Transmissible Tumors: Breaking the Cancer Paradigm. Trends Genet. 2016, 32, 1–15. [Google Scholar] [CrossRef]
- Baez-Ortega, A.; Gori, K.; Strakova, A.; Allen, J.L.; Allum, K.M.; Bansse-Issa, L.; Bhutia, T.N.; Bisson, J.L.; Briceño, C.; Castillo Domracheva, A.; et al. Somatic Evolution and Global Expansion of an Ancient Transmissible Cancer Lineage. Science 2019, 365, 440–446. [Google Scholar] [CrossRef]
- Strakova, A.; Ní Leathlobhair, M.; Wang, G.D.; Yin, T.T.; Airikkala-Otter, I.; Allen, J.L.; Allum, K.M.; Bansse-Issa, L.; Bisson, J.L.; Castillo Domracheva, A.; et al. Mitochondrial Genetic Diversity, Selection and Recombination in a Canine Transmissible Cancer. Elife 2016, 5, e14552. [Google Scholar] [CrossRef] [PubMed]
- Strakova, A.; Murchison, E.P. The Cancer Which Survived: Insights From the Genome of an 11,000 Year-Old Cancer. Curr. Opin. Genet. Dev. 2015, 30, 49–55. [Google Scholar] [CrossRef]
- Murgia, C.; Pritchard, J.K.; Kim, S.Y.; Fassati, A.; Weiss, R.A. Clonal Origin and Evolution of a Transmissible Cancer. Cell 2006, 126, 477–487. [Google Scholar] [CrossRef]
- Stockmann, D.; Ferrari, H.F.; Andrade, A.L.; Cardoso, T.C.; Luvizotto, M.C.R. Detection of the Tumour Suppressor Gene TP53 and Expression of p53, Bcl-2 and p63 Proteins in Canine Transmissible Venereal Tumour. Vet. Comp. Oncol. 2011, 9, 251–259. [Google Scholar] [CrossRef]
- Decker, B.; Davis, B.W.; Rimbault, M.; Long, A.H.; Karlins, E.; Jagannathan, V.; Reiman, R.; Parker, H.G.; Drögemüller, C.; Corneveaux, J.J.; et al. Comparison Against 186 Canid Whole-Genome Sequences Reveals Survival Strategies of an Ancient Clonally Transmissible Canine Tumor. Genome Res. 2015, 25, 1646–1655. [Google Scholar] [CrossRef] [PubMed]
- Chiang, H.C.; Liao, A.T.; Jan, T.R.; Wang, Y.S.; Lei, H.J.; Tsai, M.H.; Chen, M.F.; Lee, C.Y.; Lin, Y.C.; Chu, R.M.; et al. Gene-Expression Profiling to Identify Genes Related to Spontaneous Tumor Regression in a Canine Cancer Model. Vet. Immunol. Immunopathol. 2013, 151, 207–216. [Google Scholar] [CrossRef]
- Pai, C.; Kuo, T.F.; Mao, S.J.; Chuang, T.F.; Lin, C.S.; Chu, R.M. Immunopathogenic Behaviors of Canine Transmissible Venereal Tumor in Dogs Following an Immunotherapy Using Dendritic/Tumor Cell Hybrid. Vet. Immunol. Immunopathol. 2011, 139, 187–199. [Google Scholar] [CrossRef] [PubMed]
- Pellerin, O.; Déan, C.; Reb, P.; Chaix, C.; Floch, F.; Tierny, D.; Sapoval, M. Prostate Artery Chemoembolization in Prostate Cancer: A Proof of Concept Study in Spontaneous Prostate Cancer in a Canine Model. Diagn. Interv. Imaging 2021, 102, 709–715. [Google Scholar] [CrossRef]
- Rivera-Calderón, L.G.; Fonseca-Alves, C.E.; Kobayashi, P.E.; Carvalho, M.; Drigo, S.A.; de Oliveira Vasconcelos, R.; Laufer-Amorim, R. Alterations in PTEN, MDM2, TP53 and AR protein and gene expression are associated with Canine Prostate Carcinogenesis. Res. Vet. Sci. 2016, 106, 56–61. [Google Scholar] [CrossRef]
- Lin, H.Y.; Palmieri, C. Is STAT3 and PTEN Expression Altered in Canine Prostate Cancer? J. Comp. Pathol. 2016, 155, 185–189. [Google Scholar] [CrossRef] [PubMed]
- Cavalca, A.M.B.; Brandi, A.; Fonseca-Alves, R.H.; Laufer-Amorim, R.; Fonseca-Alves, C.E. P-Glycoprotein and Androgen Receptor Expression Reveals Independence of Canine Prostate Cancer from Androgen Hormone Stimulation. Int. J. Mol. Sci. 2022, 23, 1163. [Google Scholar] [CrossRef]
- Kobayashi, M.; Onozawa, M.; Watanabe, S.; Nagashima, T.; Tamura, K.; Kubo, Y.; Ikeda, A.; Ochiai, K.; Michishita, M.; Bonkobara, M.; et al. Establishment of a BRAF V595E-Mutant Canine Prostate Cancer Cell Line and the Antitumor Effects of MEK Inhibitors Against Canine Prostate Cancer. Vet. Comp. Oncol. 2023, 21, 221–230. [Google Scholar] [CrossRef] [PubMed]
- Kealy, R.D.; Lawler, D.F.; Ballam, J.M.; Mantz, S.L.; Biery, D.N.; Greeley, E.H.; Lust, G.; Segre, M.; Smith, G.K.; Stowe, H.D. Effects of Diet Restriction on Life Span and Age-Related Changes in Dogs. JAVMA Sci. Rep. 2002, 220, 1315–1320. [Google Scholar] [CrossRef] [PubMed]
- Di Francesco, A.; Deighan, A.G.; Litichevskiy, L.; Chen, Z.; Luciano, A.; Robinson, L.; Garland, G.; Donato, H.; Vincent, M.; Schott, W.; et al. Dietary Restriction Impacts Health and Lifespan of Genetically Diverse Mice. Nature 2024, 634, 684–692. [Google Scholar] [CrossRef] [PubMed]
| Common Canine Cancer Genes | |
|---|---|
| Oncogenes | |
| Epidermal Growth Factor Receptor Gene Family | c-erbB-1/EGFR |
| c-erbB-2/HER2 | |
| c-erbB-3 | |
| c-erbB-4 | |
| Tumor Suppressor Genes | |
| INK4A/p16 | |
| P14ARF | |
| p53 |
| Human and Canine Breast/Mammary Cancer Phenotype Definitions | |||
|---|---|---|---|
| Phenotype | ER Estrogen Receptor Alpha | PR Progesterone Receptor | HER2 c-erbB-2 |
| Luminal A | + | + | − |
| − | + | − | |
| + | − | − | |
| Luminal B | + | + | + |
| − | + | + | |
| + | − | + | |
| HER2 | − | − | + |
| Triple Negative | − | − | − |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bird, R.C.; Smith, B.F. Comparative Genetics of Canine and Human Cancers. Vet. Sci. 2025, 12, 875. https://doi.org/10.3390/vetsci12090875
Bird RC, Smith BF. Comparative Genetics of Canine and Human Cancers. Veterinary Sciences. 2025; 12(9):875. https://doi.org/10.3390/vetsci12090875
Chicago/Turabian StyleBird, Richard Curtis, and Bruce F. Smith. 2025. "Comparative Genetics of Canine and Human Cancers" Veterinary Sciences 12, no. 9: 875. https://doi.org/10.3390/vetsci12090875
APA StyleBird, R. C., & Smith, B. F. (2025). Comparative Genetics of Canine and Human Cancers. Veterinary Sciences, 12(9), 875. https://doi.org/10.3390/vetsci12090875






