Isolation of Lytic Bacteriophages of Escherichia coli and Their Combined Use with Antibiotics Against the Causative Agents of Colibacillosis in Calves
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains
2.2. Isolation of Bacteriophages
2.3. Determination of Bacteriophage Titer
2.4. Plaque Purification and Amplification of Bacteriophages
2.5. Concentration of Bacteriophages by Ultracentrifugation
2.6. Electron Microscopy
2.7. Multiplicity of Infection (MOI)
2.8. Adsorption Assay and One-Step Growth Curve
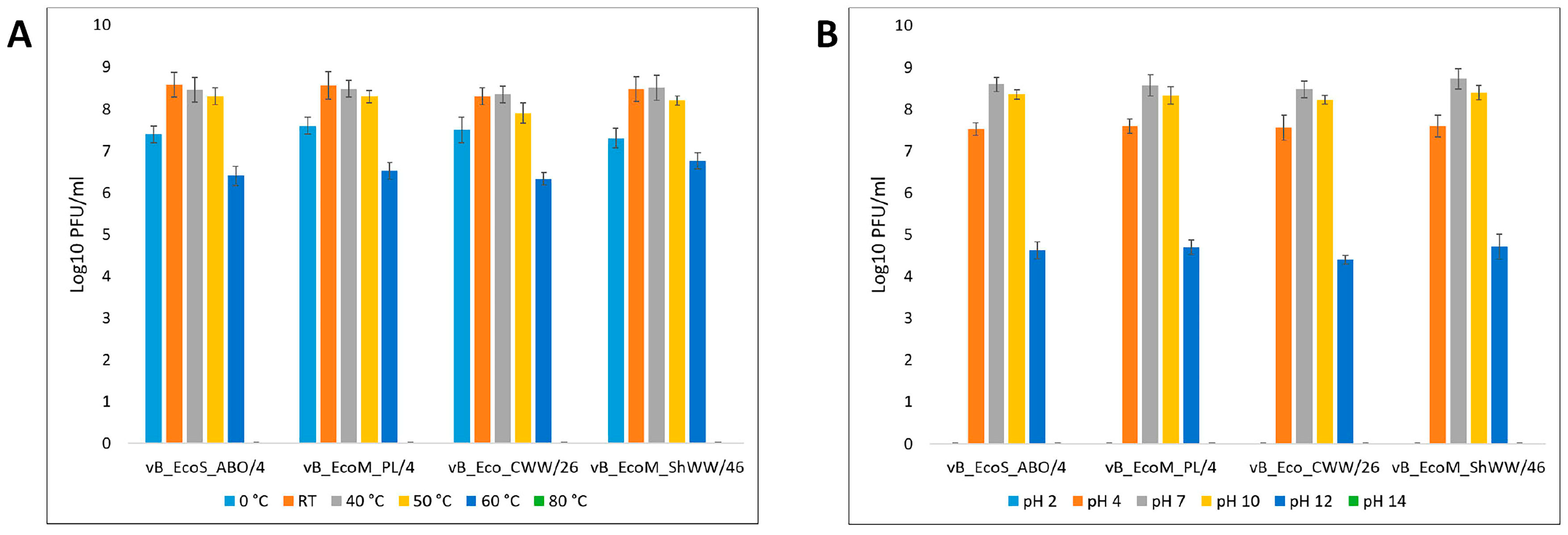
2.9. Thermal and pH Stability of the Isolated Bacteriophages
2.10. Assessment of the Lytic Activity Level of Bacteriophages
2.11. Host Range Determination
2.12. Extraction and Quality Assessment of Bacteriophage DNA
2.13. Whole-Genome Sequencing of Bacteriophages
2.14. Genome Assembly and Analysis
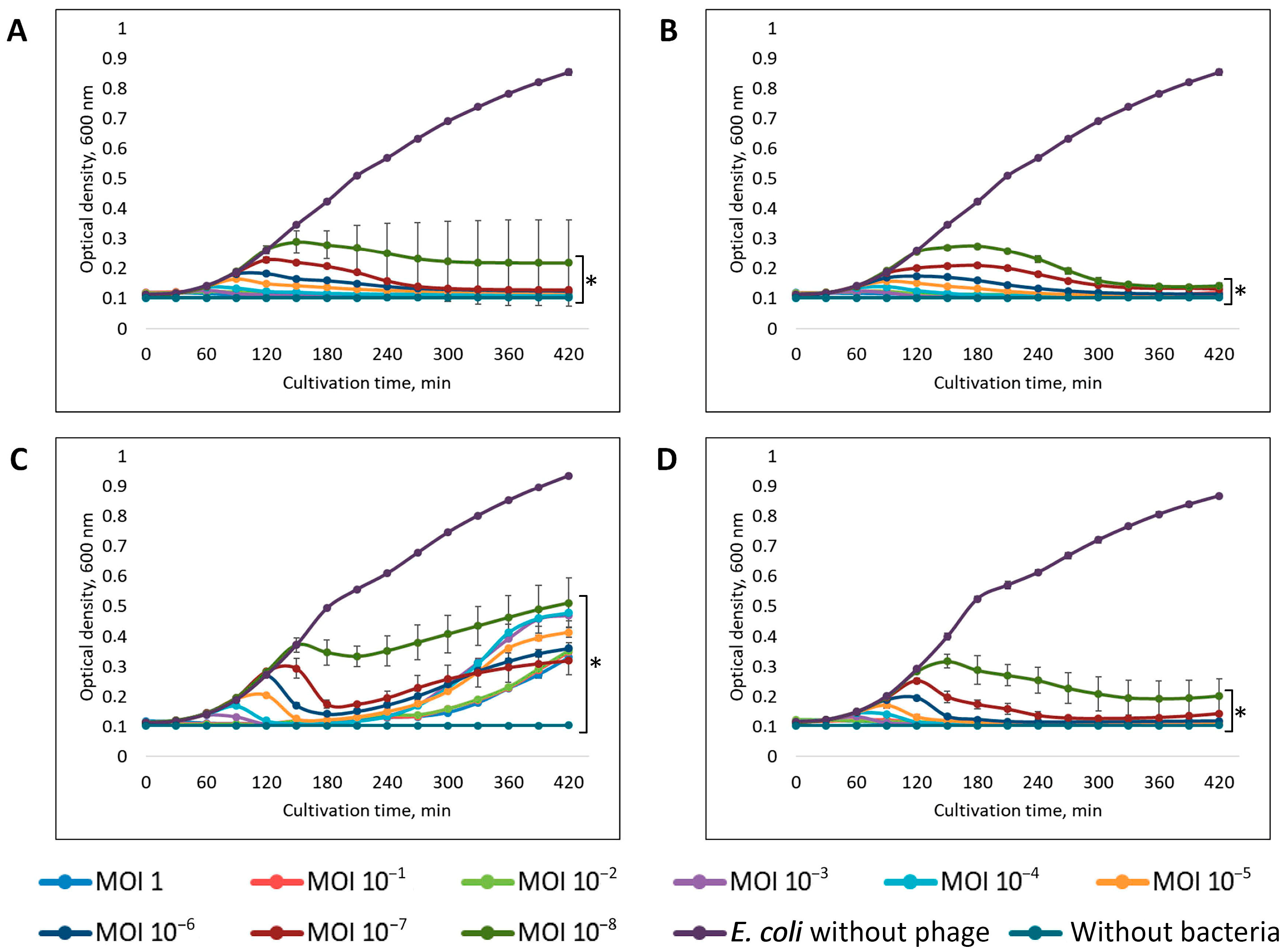
2.15. Assessment of the Synergistic Effect
2.16. Statistical Analysis and Graphical Representation of Results
3. Results
3.1. Isolation of Bacteriophages, Morphological Characterization of Plaques and Viral Particles
3.2. Phage Dynamics and Stability of Isolated Bacteriophages
3.3. Level of Lytic Activity and Host Range of Isolated Bacteriophages
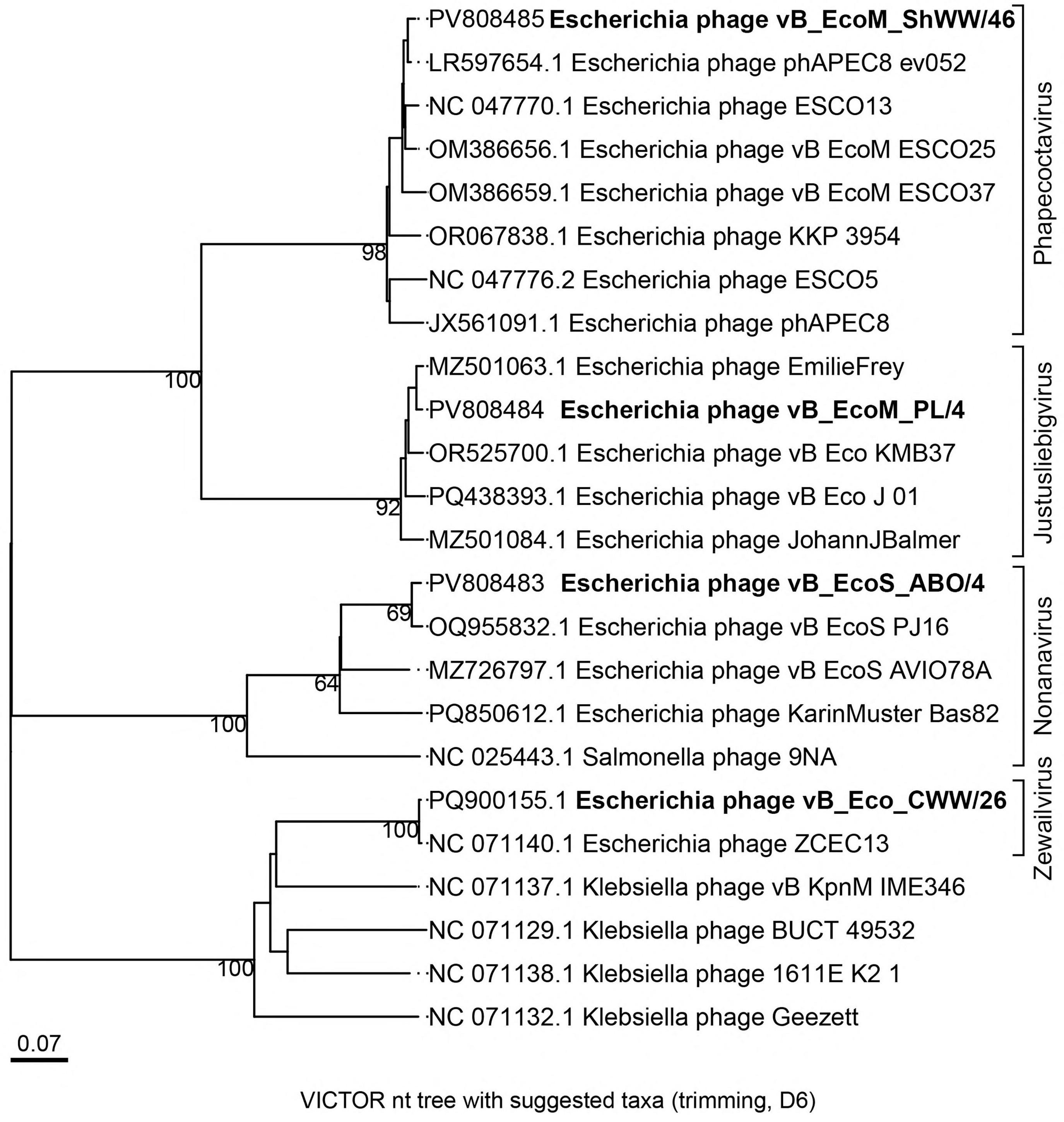
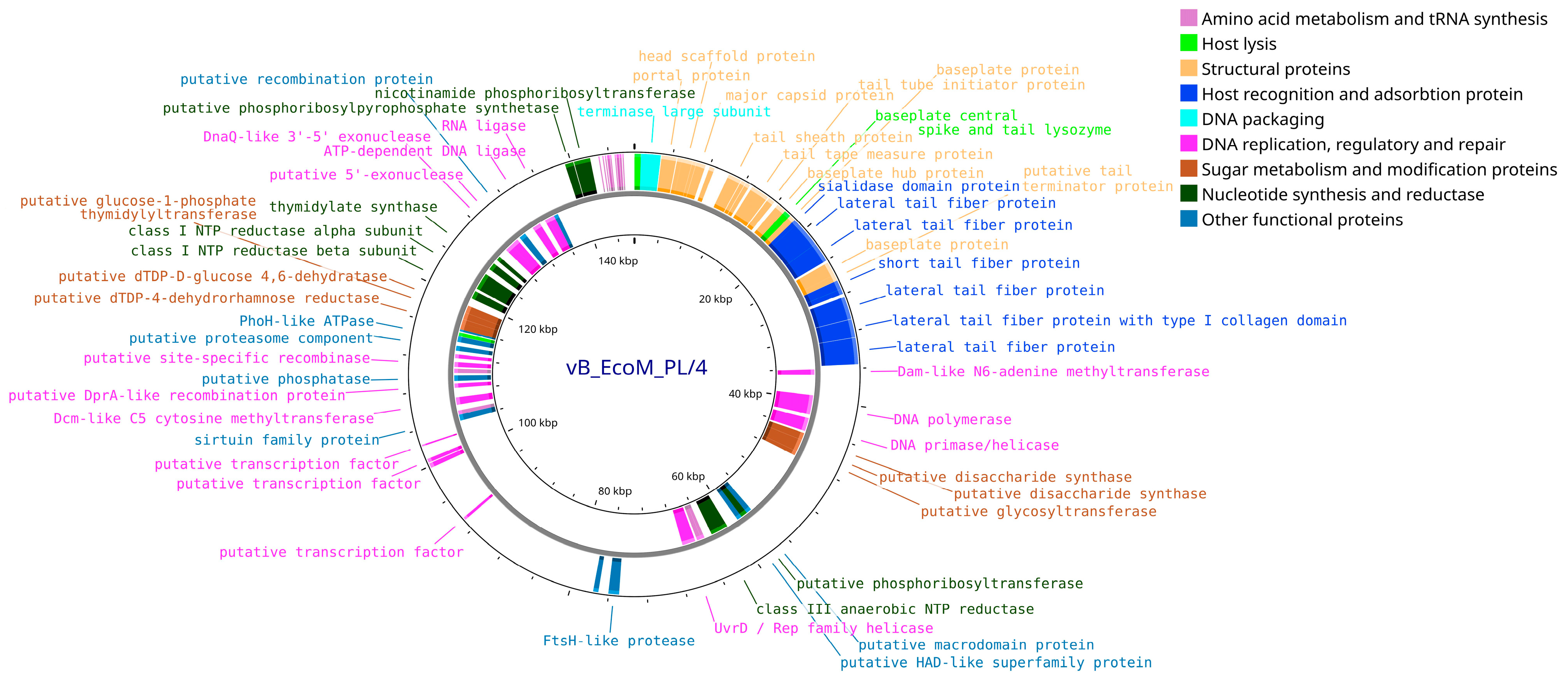
3.4. Bioinformatic Analysis and Genomic Characterization of Isolated Bacteriophages
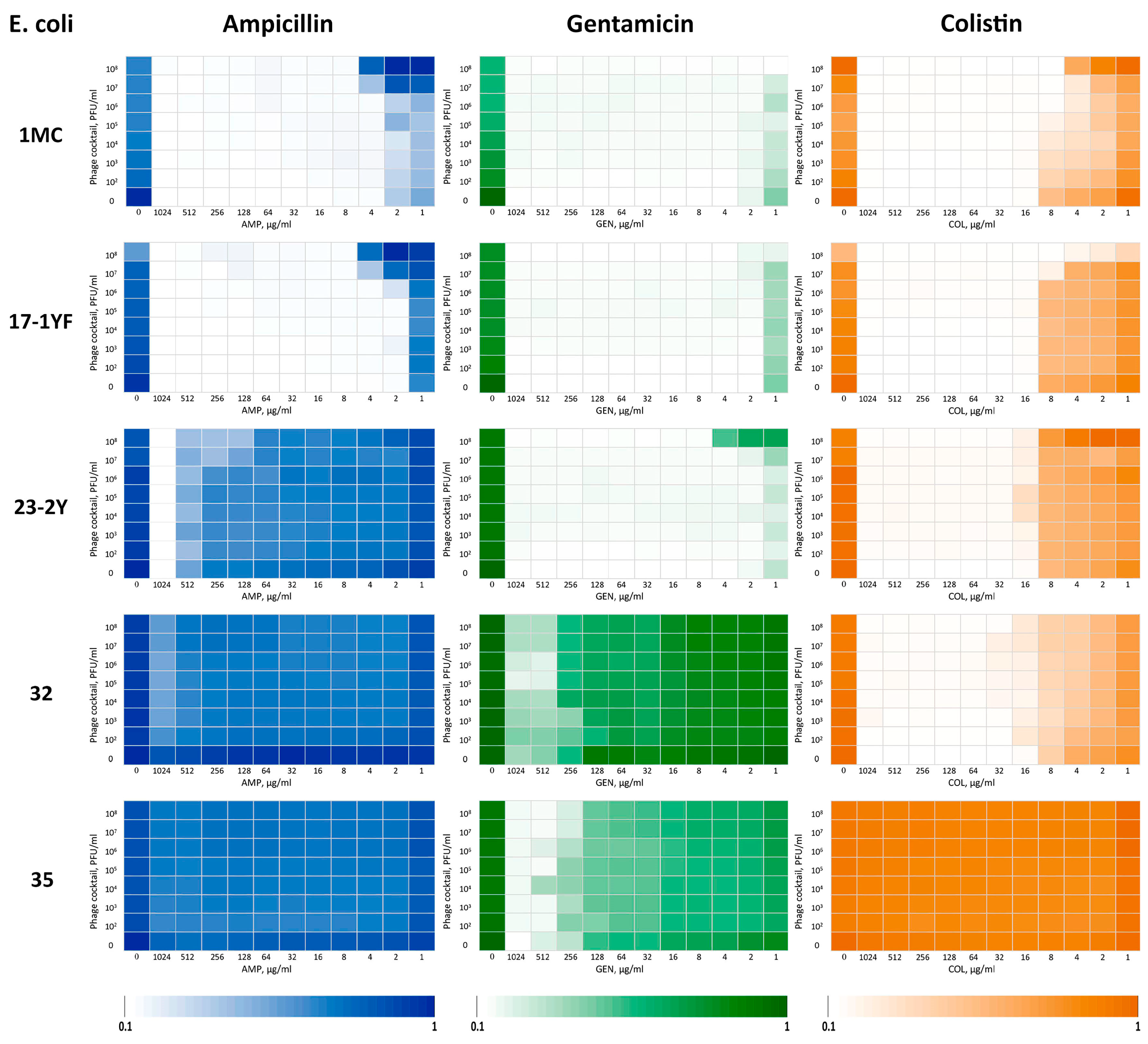
3.5. Assessment of the Synergistic Interaction Between Isolated Bacteriophages and Antibiotics
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| NB | Nutrient Broth |
| NA | Nutrient agar |
| PFU | Plaque forming unit |
| PBS | Phosphate-buffered saline |
| MOI | Multiplicity of infection |
| CFU | Colony forming unit |
| ICTV | International Committee on Taxonomy of Viruses |
| GBDP | Genome-BLAST Distance Phylogeny |
| CDS | Coding sequences |
| AMP | Ampicillin |
| GEN | Gentamicin |
| COL | Colistin |
| FICI | Fractional inhibitory concentration index |
References
- McLeod, A.; Food and Agriculture Organization of the United Nations (Eds.) World Livestock 2011: Livestock in Food Security; Food and Agriculture Organization of the United Nations: Rome, Italy, 2011; ISBN 978-92-5-107013-0. [Google Scholar]
- Alexandratos, N.; Bruinsma, J. World Agriculture Towards 2030/2050: The 2012 Revision; Food and Agriculture Organization of the United Nations: Rome, Italy, 2012. [Google Scholar]
- Tazhibaev, S.; Musabekov, K.; Yesbolova, A.; Ibraimova, S.; Mergenbayeva, A.; Sabdenova, Z.; Seidahmetov, M. Issues in the Development of the Livestock Sector in Kazakhstan. Procedia Soc. Behav. Sci. 2014, 143, 610–614. [Google Scholar] [CrossRef]
- Kazambayeva, A.M.; Aiesheva, G.A.; Yesengaliyeva, S.M. Sustainable Development of Agricultural Production Based on the Use of the Resource Potential of the Region. J. Environ. Manag. Tour. 2020, 10, 1478–1485. [Google Scholar] [CrossRef]
- Yessengaliyeva, S.M.; Mansurova, M.A.; Makhmudov, A.D.; Fedorchenko, L.V. Current state and development trends of livestock in the Republic of Kazakhstan. Econ. Strategy Pract. 2021, 16, 134–144. [Google Scholar] [CrossRef]
- Acres, S.D. Enterotoxigenic Escherichia coli Infections in Newborn Calves: A Review. J. Dairy Sci. 1985, 68, 229–256. [Google Scholar] [CrossRef] [PubMed]
- Kolenda, R.; Burdukiewicz, M.; Schierack, P. A Systematic Review and Meta-Analysis of the Epidemiology of Pathogenic Escherichia coli of Calves and the Role of Calves as Reservoirs for Human Pathogenic E. coli. Front. Cell. Infect. Microbiol. 2015, 5, 23. [Google Scholar] [CrossRef]
- World Health Organization; WHO Advisory Group on Integrated Surveillance of Antimicrobial Resistance (AGISAR). Critically Important Antimicrobials for Human Medicine: Ranking of Antimicrobial Agents for Risk Management of Antimicrobial Resistance due to Non-Human Use, 5th ed.; World Health Organization: Geneva, Switzerland, 2017; ISBN 978-92-4-151222-0. [Google Scholar]
- Alexyuk, P.G.; Bogoyavlenskiy, A.P.; Moldakhanov, Y.S.; Akanova, K.S.; Manakbayeva, A.N.; Kerimov, T.; Berezin, V.E.; Alexyuk, M.S. Genomic and Drug Resistance Profile of Bovine Multidrug-Resistant Escherichia coli Isolated in Kazakhstan. Pathogens 2025, 14, 90. [Google Scholar] [CrossRef]
- Pokharel, S.; Shrestha, P.; Adhikari, B. Antimicrobial Use in Food Animals and Human Health: Time to Implement ‘One Health’ Approach. Antimicrob. Resist. Infect. Control 2020, 9, 181. [Google Scholar] [CrossRef]
- Pandey, S.; Doo, H.; Keum, G.B.; Kim, E.S.; Kwak, J.; Ryu, S.; Choi, Y.; Kang, J.; Kim, S.; Lee, N.R.; et al. Antibiotic Resistance in Livestock, Environment and Humans: One Health Perspective. J. Anim. Sci. Technol 2024, 66, 266–278. [Google Scholar] [CrossRef] [PubMed]
- Czaplewski, L.; Bax, R.; Clokie, M.; Dawson, M.; Fairhead, H.; Fischetti, V.A.; Foster, S.; Gilmore, B.F.; Hancock, R.E.W.; Harper, D.; et al. Alternatives to Antibiotics-a Pipeline Portfolio Review. Lancet Infect. Dis. 2016, 16, 239–251. [Google Scholar] [CrossRef]
- Machowska, A.; Stålsby Lundborg, C. Drivers of Irrational Use of Antibiotics in Europe. Int. J. Environ. Res. Public Health 2019, 16, 27. [Google Scholar] [CrossRef]
- Smith, H.W.; Huggins, M.B. Successful Treatment of Experimental Escherichia coli Infections in Mice Using Phage: Its General Superiority over Antibiotics. J. Gen. Microbiol. 1982, 128, 307–318. [Google Scholar] [CrossRef]
- Smith, H.W.; Huggins, M.B. Effectiveness of Phages in Treating Experimental Escherichia coli Diarrhoea in Calves, Piglets and Lambs. J. Gen. Microbiol. 1983, 129, 2659–2675. [Google Scholar] [CrossRef] [PubMed]
- Dec, M.; Wernicki, A.; Urban-Chmiel, R. Efficacy of Experimental Phage Therapies in Livestock. Anim. Health Res. Rev. 2020, 21, 69–83. [Google Scholar] [CrossRef] [PubMed]
- Jamalludeen, N.; Johnson, R.P.; Shewen, P.E.; Gyles, C.L. Evaluation of Bacteriophages for Prevention and Treatment of Diarrhea Due to Experimental Enterotoxigenic Escherichia coli O149 Infection of Pigs. Vet. Microbiol. 2009, 136, 135–141. [Google Scholar] [CrossRef]
- Zeng, Y.; Wang, Z.; Zou, T.; Chen, J.; Li, G.; Zheng, L.; Li, S.; You, J. Bacteriophage as an Alternative to Antibiotics Promotes Growth Performance by Regulating Intestinal Inflammation, Intestinal Barrier Function and Gut Microbiota in Weaned Piglets. Front. Vet. Sci. 2021, 8, 623899. [Google Scholar] [CrossRef]
- Bicalho, M.L.S.; Machado, V.S.; Nydam, D.V.; Santos, T.M.A.; Bicalho, R.C. Evaluation of Oral Administration of Bacteriophages to Neonatal Calves: Phage Survival and Impact on Fecal Escherichia coli. Livest. Sci. 2012, 144, 294–299. [Google Scholar] [CrossRef]
- Alexyuk, M.; Alexyuk, P.; Moldakhanov, Y.; Akanova, K.; Bogoyavlenskiy, A. Evaluation of Phage Cocktail Efficacy for Controlling Infections Caused by Pathogenic Escherichia coli in in Vivo Experiments. BIO Web Conf. 2024, 100, 2004. [Google Scholar] [CrossRef]
- Skurnik, M.; Pajunen, M.; Kiljunen, S. Biotechnological Challenges of Phage Therapy. Biotechnol. Lett. 2007, 29, 995–1003. [Google Scholar] [CrossRef] [PubMed]
- Abedon, S.T.; Thomas-Abedon, C. Phage Therapy Pharmacology. Curr. Pharm. Biotechnol. 2010, 11, 28–47. [Google Scholar] [CrossRef]
- Gholizadeh, O.; Ghaleh, H.E.G.; Tat, M.; Ranjbar, R.; Dorostkar, R. The Potential Use of Bacteriophages as Antibacterial Agents against Klebsiella pneumoniae. Virol. J. 2024, 21, 191. [Google Scholar] [CrossRef]
- Olawade, D.B.; Fapohunda, O.; Egbon, E.; Ebiesuwa, O.A.; Usman, S.O.; Faronbi, A.O.; Fidelis, S.C. Phage Therapy: A Targeted Approach to Overcoming Antibiotic Resistance. Microb. Pathog. 2024, 197, 107088. [Google Scholar] [CrossRef]
- Alexyuk, P.G.; Bogoyavlenskiy, A.P.; Akanova, K.S.; Kerimov, T.T.; Manakbayeva, A.N.; Moldakhanov, Y.S.; Yershov, I.D.; Alexyuk, M.S. Complete Genome Sequence of the Lytic vB_EcoP_ShWW44 Bacteriophage, Effective against Bovine Pathogenic Escherichia coli. Microbiol. Resour. Announc. 2025, 14, e0019525. [Google Scholar] [CrossRef] [PubMed]
- Merabishvili, M.; Pirnay, J.-P.; Verbeken, G.; Chanishvili, N.; Tediashvili, M.; Lashkhi, N.; Glonti, T.; Krylov, V.; Mast, J.; Van Parys, L.; et al. Quality-Controlled Small-Scale Production of a Well-Defined Bacteriophage Cocktail for Use in Human Clinical Trials. PLoS ONE 2009, 4, e4944. [Google Scholar] [CrossRef] [PubMed]
- Alexyuk, P.; Bogoyavlenskiy, A.; Alexyuk, M.; Akanova, K.; Moldakhanov, Y.; Berezin, V. Isolation and Characterization of Jumbo Coliphage vB_EcoM_Lh1B as a Promising Therapeutic Agent against Chicken Colibacillosis. Microorganisms 2023, 11, 1524. [Google Scholar] [CrossRef]
- Gratia, A. The Numerical Relation between Lysogenic Bacteria and the Phage Particles which they carry. Ann. Inst. Pasteur. 1936, 57, 652–676. [Google Scholar]
- Kropinski, A.M.; Mazzocco, A.; Waddell, T.E.; Lingohr, E.; Johnson, R.P. Enumeration of Bacteriophages by Double Agar Overlay Plaque Assay. In Bacteriophages: Methods and Protocols, Volume 1: Isolation, Characterization, and Interactions; Clokie, M.R.J., Kropinski, A.M., Eds.; Methods in Molecular BiologyTM; Humana Press: Totowa, NJ, USA, 2009; pp. 69–76. ISBN 978-1-60327-164-6. [Google Scholar]
- Green, M.R.; Sambrook, J.; Sambrook, J. Molecular Cloning: A Laboratory Manual, 4th ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2012; ISBN 978-1-936113-41-5. [Google Scholar]
- Ackermann, H.-W. Basic Phage Electron Microscopy. Methods Mol. Biol. 2009, 501, 113–126. [Google Scholar] [CrossRef]
- Abedon, S.T. Phage Therapy Dosing: The Problem(s) with Multiplicity of Infection (MOI). Bacteriophage 2016, 6, e1220348. [Google Scholar] [CrossRef]
- Kim, S.G.; Jun, J.W.; Giri, S.S.; Yun, S.; Kim, H.J.; Kim, S.W.; Kang, J.W.; Han, S.J.; Jeong, D.; Park, S.C. Isolation and Characterisation of pVa-21, a Giant Bacteriophage with Anti-Biofilm Potential against Vibrio Alginolyticus. Sci. Rep. 2019, 9, 6284. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.G.; Lee, S.B.; Giri, S.S.; Kim, H.J.; Kim, S.W.; Kwon, J.; Park, J.; Roh, E.; Park, S.C. Characterization of Novel Erwinia amylovora Jumbo Bacteriophages from Eneladusvirus Genus. Viruses 2020, 12, 1373. [Google Scholar] [CrossRef]
- Wan, S.; Li, N.; Habib, S.; Zheng, P.; Li, Y.; Liang, Y.; Qu, Y. Biological Characteristics and Whole-Genome Analysis of a Porcine E. coli Phage. Vet. Sci. 2025, 12, 57. [Google Scholar] [CrossRef]
- Alves, D.R.; Perez-Esteban, P.; Kot, W.; Bean, J.E.; Arnot, T.; Hansen, L.H.; Enright, M.C.; Jenkins, A.T.A. A Novel Bacteriophage Cocktail Reduces and Disperses Pseudomonas aeruginosa Biofilms under Static and Flow Conditions. Microb. Biotechnol. 2015, 9, 61–74. [Google Scholar] [CrossRef]
- Jakočiūnė, D.; Moodley, A. A Rapid Bacteriophage DNA Extraction Method. Methods Protoc. 2018, 1, 27. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid Prokaryotic Genome Annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Chan, P.P.; Lin, B.Y.; Mak, A.J.; Lowe, T.M. tRNAscan-SE 2.0: Improved Detection and Functional Classification of Transfer RNA Genes. Nucleic Acids Res. 2021, 49, 9077–9096. [Google Scholar] [CrossRef]
- Yukgehnaish, K.; Rajandas, H.; Parimannan, S.; Manickam, R.; Marimuthu, K.; Petersen, B.; Clokie, M.R.J.; Millard, A.; Sicheritz-Pontén, T. PhageLeads: Rapid Assessment of Phage Therapeutic Suitability Using an Ensemble Machine Learning Approach. Viruses 2022, 14, 342. [Google Scholar] [CrossRef]
- Hockenberry, A.J.; Wilke, C.O. BACPHLIP: Predicting Bacteriophage Lifestyle from Conserved Protein Domains. PeerJ 2021, 9, e11396. [Google Scholar] [CrossRef]
- Feldgarden, M.; Brover, V.; Gonzalez-Escalona, N.; Frye, J.G.; Haendiges, J.; Haft, D.H.; Hoffmann, M.; Pettengill, J.B.; Prasad, A.B.; Tillman, G.E.; et al. AMRFinderPlus and the Reference Gene Catalog Facilitate Examination of the Genomic Links among Antimicrobial Resistance, Stress Response, and Virulence. Sci. Rep. 2021, 11, 12728. [Google Scholar] [CrossRef] [PubMed]
- Joensen, K.G.; Scheutz, F.; Lund, O.; Hasman, H.; Kaas, R.S.; Nielsen, E.M.; Aarestrup, F.M. Real-Time Whole-Genome Sequencing for Routine Typing, Surveillance, and Outbreak Detection of Verotoxigenic Escherichia coli. J. Clin. Microbiol. 2014, 52, 1501–1510. [Google Scholar] [CrossRef]
- Seemann, T. ABRicate: Mass Screening of Contigs for Antibiotic Resistance Genes. 2016. Available online: https://github.com/tseemann/abricate (accessed on 1 July 2025).
- Meier-Kolthoff, J.P.; Göker, M. VICTOR: Genome-Based Phylogeny and Classification of Prokaryotic Viruses. Bioinformatics 2017, 33, 3396–3404. [Google Scholar] [CrossRef] [PubMed]
- Meier-Kolthoff, J.P.; Auch, A.F.; Klenk, H.-P.; Göker, M. Genome Sequence-Based Species Delimitation with Confidence Intervals and Improved Distance Functions. BMC Bioinform. 2013, 14, 60. [Google Scholar] [CrossRef] [PubMed]
- Lefort, V.; Desper, R.; Gascuel, O. FastME 2.0: A Comprehensive, Accurate, and Fast Distance-Based Phylogeny Inference Program. Mol. Biol. Evol. 2015, 32, 2798–2800. [Google Scholar] [CrossRef]
- Farris, J.S. Estimating Phylogenetic Trees from Distance Matrices. Am. Nat. 1972, 106, 645–668. [Google Scholar] [CrossRef]
- Yu, G. Using Ggtree to Visualize Data on Tree-Like Structures. Curr. Protoc. Bioinform. 2020, 69, e96. [Google Scholar] [CrossRef] [PubMed]
- Göker, M.; García-Blázquez, G.; Voglmayr, H.; Tellería, M.T.; Martín, M.P. Molecular Taxonomy of Phytopathogenic Fungi: A Case Study in Peronospora. PLoS ONE 2009, 4, e6319. [Google Scholar] [CrossRef] [PubMed]
- Meier-Kolthoff, J.P.; Hahnke, R.L.; Petersen, J.; Scheuner, C.; Michael, V.; Fiebig, A.; Rohde, C.; Rohde, M.; Fartmann, B.; Goodwin, L.A.; et al. Complete Genome Sequence of DSM 30083T, the Type Strain (U5/41T) of Escherichia coli, and a Proposal for Delineating Subspecies in Microbial Taxonomy. Stand. Genom. Sci. 2014, 9, 2. [Google Scholar] [CrossRef]
- Grant, J.R.; Enns, E.; Marinier, E.; Mandal, A.; Herman, E.K.; Chen, C.; Graham, M.; Van Domselaar, G.; Stothard, P. Proksee: In-Depth Characterization and Visualization of Bacterial Genomes. Nucleic Acids Res. 2023, 51, W484–W492. [Google Scholar] [CrossRef]
- Wang, Q.; Chen, R.; Liu, H.; Liu, Y.; Li, J.; Wang, Y.; Jin, Y.; Bai, Y.; Song, Z.; Lu, X.; et al. Isolation and Characterization of Lytic Bacteriophage vB_KpnP_23: A Promising Antimicrobial Candidate against Carbapenem-Resistant Klebsiella pneumoniae. Virus Res. 2024, 350, 199473. [Google Scholar] [CrossRef]
- Butler, M.S.; Buss, A.D. Natural Products--the Future Scaffolds for Novel Antibiotics? Biochem. Pharmacol. 2006, 71, 919–929. [Google Scholar] [CrossRef]
- Lewis, K. Platforms for Antibiotic Discovery. Nat. Rev. Drug Discov. 2013, 12, 371–387. [Google Scholar] [CrossRef]
- Projan, S.J.; Shlaes, D.M. Antibacterial Drug Discovery: Is It All Downhill from Here? Clin. Microbiol. Infect. 2004, 10, 18–22. [Google Scholar] [CrossRef]
- Mellbye, B.; Schuster, M. The Sociomicrobiology of Antivirulence Drug Resistance: A Proof of Concept. mBio 2011, 2, e00131-11. [Google Scholar] [CrossRef]
- Dedrick, R.M.; Guerrero-Bustamante, C.A.; Garlena, R.A.; Russell, D.A.; Ford, K.; Harris, K.; Gilmour, K.C.; Soothill, J.; Jacobs-Sera, D.; Schooley, R.T.; et al. Engineered Bacteriophages for Treatment of a Patient with a Disseminated Drug Resistant Mycobacterium abscessus. Nat. Med. 2019, 25, 730–733. [Google Scholar] [CrossRef]
- Gordillo Altamirano, F.L.; Barr, J.J. Phage Therapy in the Postantibiotic Era. Clin. Microbiol. Rev. 2019, 32, e00066-18. [Google Scholar] [CrossRef]
- Chang, R.Y.K.; Wallin, M.; Lin, Y.; Leung, S.S.Y.; Wang, H.; Morales, S.; Chan, H.-K. Phage Therapy for Respiratory Infections. Adv. Drug Deliv. Rev. 2018, 133, 76–86. [Google Scholar] [CrossRef] [PubMed]
- Zhvania, P.; Hoyle, N.S.; Nadareishvili, L.; Nizharadze, D.; Kutateladze, M. Phage Therapy in a 16-Year-Old Boy with Netherton Syndrome. Front. Med. 2017, 4, 94. [Google Scholar] [CrossRef] [PubMed]
- Necel, A.; Bloch, S.; Nejman-Faleńczyk, B.; Grabski, M.; Topka, G.; Dydecka, A.; Kosznik-Kwaśnicka, K.; Grabowski, Ł.; Jurczak-Kurek, A.; Wołkowicz, T.; et al. Characterization of a Bacteriophage, vB_Eco4M-7, That Effectively Infects Many Escherichia coli O157 Strains. Sci. Rep. 2020, 10, 3743. [Google Scholar] [CrossRef] [PubMed]
- Wójcicki, M.; Średnicka, P.; Błażejak, S.; Gientka, I.; Kowalczyk, M.; Emanowicz, P.; Świder, O.; Sokołowska, B.; Juszczuk-Kubiak, E. Characterization and Genome Study of Novel Lytic Bacteriophages against Prevailing Saprophytic Bacterial Microflora of Minimally Processed Plant-Based Food Products. Int. J. Mol. Sci. 2021, 22, 12460. [Google Scholar] [CrossRef]
- Pires, D.P.; Oliveira, H.; Melo, L.D.R.; Sillankorva, S.; Azeredo, J. Bacteriophage-Encoded Depolymerases: Their Diversity and Biotechnological Applications. Appl. Microbiol. Biotechnol. 2016, 100, 2141–2151. [Google Scholar] [CrossRef]
- Zurabov, F.; Zhilenkov, E. Characterization of Four Virulent Klebsiella Pneumoniae Bacteriophages, and Evaluation of Their Potential Use in Complex Phage Preparation. Virol. J. 2021, 18, 9. [Google Scholar] [CrossRef]
- Maffei, E.; Shaidullina, A.; Burkolter, M.; Heyer, Y.; Estermann, F.; Druelle, V.; Sauer, P.; Willi, L.; Michaelis, S.; Hilbi, H.; et al. Systematic Exploration of Escherichia coli Phage–Host Interactions with the BASEL Phage Collection. PLoS Biol. 2021, 19, e3001424. [Google Scholar] [CrossRef]
- Pereira, C.; Duarte, J.; Costa, P.; Braz, M.; Almeida, A. Bacteriophages in the Control of Aeromonas sp. in Aquaculture Systems: An Integrative View. Antibiotics 2022, 11, 163. [Google Scholar] [CrossRef]
- Khan Mirzaei, M.; Nilsson, A.S. Isolation of Phages for Phage Therapy: A Comparison of Spot Tests and Efficiency of Plating Analyses for Determination of Host Range and Efficacy. PLoS ONE 2015, 10, e0118557. [Google Scholar] [CrossRef]
- Amankwah, S.; Adisu, M.; Gorems, K.; Abdella, K.; Kassa, T. Assessment of Phage-Mediated Inhibition and Removal of Multidrug-Resistant Pseudomonas aeruginosa Biofilm on Medical Implants. Infect. Drug Resist. 2022, 15, 2797–2811. [Google Scholar] [CrossRef]
- Richards, K.; Malik, D.J. Microencapsulation of Bacteriophages Using Membrane Emulsification in Different pH-Triggered Controlled Release Formulations for Oral Administration. Pharmaceuticals 2021, 14, 424. [Google Scholar] [CrossRef]
- Malik, D.J.; Sokolov, I.J.; Vinner, G.K.; Mancuso, F.; Cinquerrui, S.; Vladisavljevic, G.T.; Clokie, M.R.J.; Garton, N.J.; Stapley, A.G.F.; Kirpichnikova, A. Formulation, Stabilisation and Encapsulation of Bacteriophage for Phage Therapy. Adv. Colloid Interface Sci. 2017, 249, 100–133. [Google Scholar] [CrossRef]
- Vila, M.M.D.C.; Balcão, L.M.N.; Balcão, V.M. Phage Delivery Strategies for Biocontrolling Human, Animal, and Plant Bacterial Infections: State of the Art. Pharmaceutics 2024, 16, 374. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Ryu, S. Characterization of a T5-Like Coliphage, SPC35, and Differential Development of Resistance to SPC35 in Salmonella enterica Serovar Typhimurium and Escherichia coli. Appl. Environ. Microbiol. 2011, 77, 2042–2050. [Google Scholar] [CrossRef] [PubMed]
- Kapoor, A.; Mudaliar, S.B.; Bhat, V.G.; Chakraborty, I.; Prasad, A.S.B.; Mazumder, N. Phage Therapy: A Novel Approach against Multidrug-Resistant Pathogens. 3 Biotech 2024, 14, 256. [Google Scholar] [CrossRef]
- Costa, P.; Pereira, C.; Romalde, J.L.; Almeida, A. A Game of Resistance: War between Bacteria and Phages and How Phage Cocktails Can Be the Solution. Virology 2024, 599, 110209. [Google Scholar] [CrossRef] [PubMed]
- Kropinski, A.M. Bacteriophage Research—What We Have Learnt and What Still Needs to Be Addressed. Res. Microbiol. 2018, 169, 481–487. [Google Scholar] [CrossRef]
- Krupovic, M.; Prangishvili, D.; Hendrix, R.W.; Bamford, D.H. Genomics of Bacterial and Archaeal Viruses: Dynamics within the Prokaryotic Virosphere. Microbiol. Mol. Biol. Rev. 2011, 75, 610–635. [Google Scholar] [CrossRef] [PubMed]
- Magar, S.; Barath, S.; Sen, D.; Singari, R.K.; Nagarajan, T.; Parmar, A.; Govindarajan, S. Characterization and Genomic Analysis of Sharanji: A Jumbo Bacteriophage of Escherichia coli. Virol. J. 2025, 22, 67. [Google Scholar] [CrossRef]
- Hatfull, G.F.; Hendrix, R.W. Bacteriophages and Their Genomes. Curr. Opin. Virol. 2011, 1, 298–303. [Google Scholar] [CrossRef]
- Dwivedi, B.; Xue, B.; Lundin, D.; Edwards, R.A.; Breitbart, M. A Bioinformatic Analysis of Ribonucleotide Reductase Genes in Phage Genomes and Metagenomes. BMC Evol. Biol. 2013, 13, 33. [Google Scholar] [CrossRef]
- Bailly-Bechet, M.; Vergassola, M.; Rocha, E. Causes for the Intriguing Presence of tRNAs in Phages. Genome Res. 2007, 17, 1486–1495. [Google Scholar] [CrossRef]
- van den Berg, D.F.; Brouns, S.J.J. Phage tRNAs: Decoding the Enigma. Trends Microbiol. 2025; Epub ahead of print. [Google Scholar] [CrossRef]
- Delesalle, V.A.; Tanke, N.T.; Vill, A.C.; Krukonis, G.P. Testing Hypotheses for the Presence of tRNA Genes in Mycobacteriophage Genomes. Bacteriophage 2016, 6, e1219441. [Google Scholar] [CrossRef]
- Clark, J.R.; March, J.B. Bacteriophages and Biotechnology: Vaccines, Gene Therapy and Antibacterials. Trends Biotechnol. 2006, 24, 212–218. [Google Scholar] [CrossRef]
- Dabrowska, K.; Switała-Jelen, K.; Opolski, A.; Weber-Dabrowska, B.; Gorski, A. Bacteriophage Penetration in Vertebrates. J. Appl. Microbiol. 2005, 98, 7–13. [Google Scholar] [CrossRef]
- Verma, V.; Harjai, K.; Chhibber, S. Restricting Ciprofloxacin-Induced Resistant Variant Formation in Biofilm of Klebsiella pneumoniae B5055 by Complementary Bacteriophage Treatment. J. Antimicrob. Chemother. 2009, 64, 1212–1218. [Google Scholar] [CrossRef] [PubMed]
- Chhibber, S.; Kaur, T. Sandeep Kaur Co-Therapy Using Lytic Bacteriophage and Linezolid: Effective Treatment in Eliminating Methicillin Resistant Staphylococcus aureus (MRSA) from Diabetic Foot Infections. PLoS ONE 2013, 8, e56022. [Google Scholar] [CrossRef] [PubMed]
- Huff, W.E.; Huff, G.R.; Rath, N.C.; Balog, J.M.; Donoghue, A.M. Therapeutic Efficacy of Bacteriophage and Baytril (Enrofloxacin) Individually and in Combination to Treat Colibacillosis in Broilers. Poult. Sci. 2004, 83, 1944–1947. [Google Scholar] [CrossRef]
- Oechslin, F.; Piccardi, P.; Mancini, S.; Gabard, J.; Moreillon, P.; Entenza, J.M.; Resch, G.; Que, Y.-A. Synergistic Interaction Between Phage Therapy and Antibiotics Clears Pseudomonas aeruginosa Infection in Endocarditis and Reduces Virulence. J. Infect. Dis. 2017, 215, 703–712. [Google Scholar] [CrossRef] [PubMed]
- Kamal, F.; Dennis, J.J. Burkholderia cepacia Complex Phage-Antibiotic Synergy (PAS): Antibiotics Stimulate Lytic Phage Activity. Appl. Environ. Microbiol. 2015, 81, 1132–1138. [Google Scholar] [CrossRef]
- De Angelis, G.; Del Giacomo, P.; Posteraro, B.; Sanguinetti, M.; Tumbarello, M. Molecular Mechanisms, Epidemiology, and Clinical Importance of β-Lactam Resistance in Enterobacteriaceae. Int. J. Mol. Sci. 2020, 21, 5090. [Google Scholar] [CrossRef]
- Moryl, M.; Szychowska, P.; Dziąg, J.; Różalski, A.; Torzewska, A. The Combination of Phage Therapy and β-Lactam Antibiotics for the Effective Treatment of Enterococcus faecalis Infections. Int. J. Mol. Sci. 2024, 26, 11. [Google Scholar] [CrossRef]
- Łusiak-Szelachowska, M.; Międzybrodzki, R.; Drulis-Kawa, Z.; Cater, K.; Knežević, P.; Winogradow, C.; Amaro, K.; Jończyk-Matysiak, E.; Weber-Dąbrowska, B.; Rękas, J.; et al. Bacteriophages and Antibiotic Interactions in Clinical Practice: What We Have Learned so Far. J. Biomed. Sci. 2022, 29, 23. [Google Scholar] [CrossRef]
- Rostøl, J.T.; Marraffini, L. (Ph)Ighting Phages—How Bacteria Resist Their Parasites. Cell Host Microbe 2019, 25, 184–194. [Google Scholar] [CrossRef]
- Goldfarb, T.; Sberro, H.; Weinstock, E.; Cohen, O.; Doron, S.; Charpak-Amikam, Y.; Afik, S.; Ofir, G.; Sorek, R. BREX Is a Novel Phage Resistance System Widespread in Microbial Genomes. EMBO J. 2015, 34, 169–183. [Google Scholar] [CrossRef]
- Ofir, G.; Melamed, S.; Sberro, H.; Mukamel, Z.; Silverman, S.; Yaakov, G.; Doron, S.; Sorek, R. DISARM Is a Widespread Bacterial Defence System with Broad Anti-Phage Activities. Nat. Microbiol. 2018, 3, 90–98. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Gu, P. Overview of Phage Defense Systems in Bacteria and Their Applications. Int. J. Mol. Sci. 2024, 25, 13316. [Google Scholar] [CrossRef] [PubMed]
- Supina, B.S.I.; Dennis, J.J. The Current Landscape of Phage–Antibiotic Synergistic (PAS) Interactions. Antibiotics 2025, 14, 545. [Google Scholar] [CrossRef] [PubMed]
- Klimenko, A.I.; Matushkin, Y.G.; Kolchanov, N.A.; Lashin, S.A. Bacteriophages Affect Evolution of Bacterial Communities in Spatially Distributed Habitats: A Simulation Study. BMC Microbiol. 2016, 16, S10. [Google Scholar] [CrossRef] [PubMed]
- Koskella, B.; Hernandez, C.A.; Wheatley, R.M. Understanding the Impacts of Bacteriophage Viruses: From Laboratory Evolution to Natural Ecosystems. Annu. Rev. Virol. 2022, 9, 57–78. [Google Scholar] [CrossRef] [PubMed]
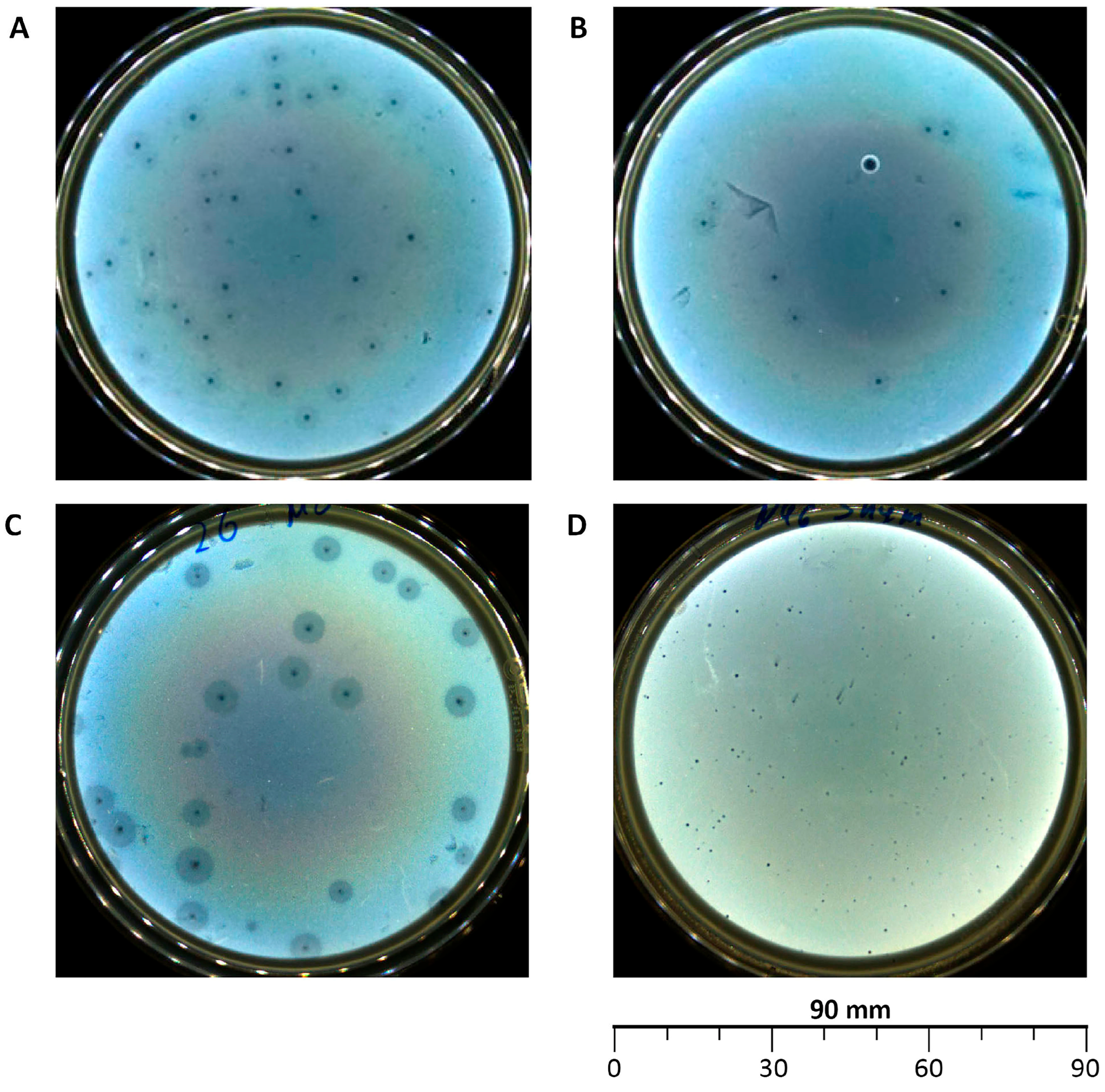
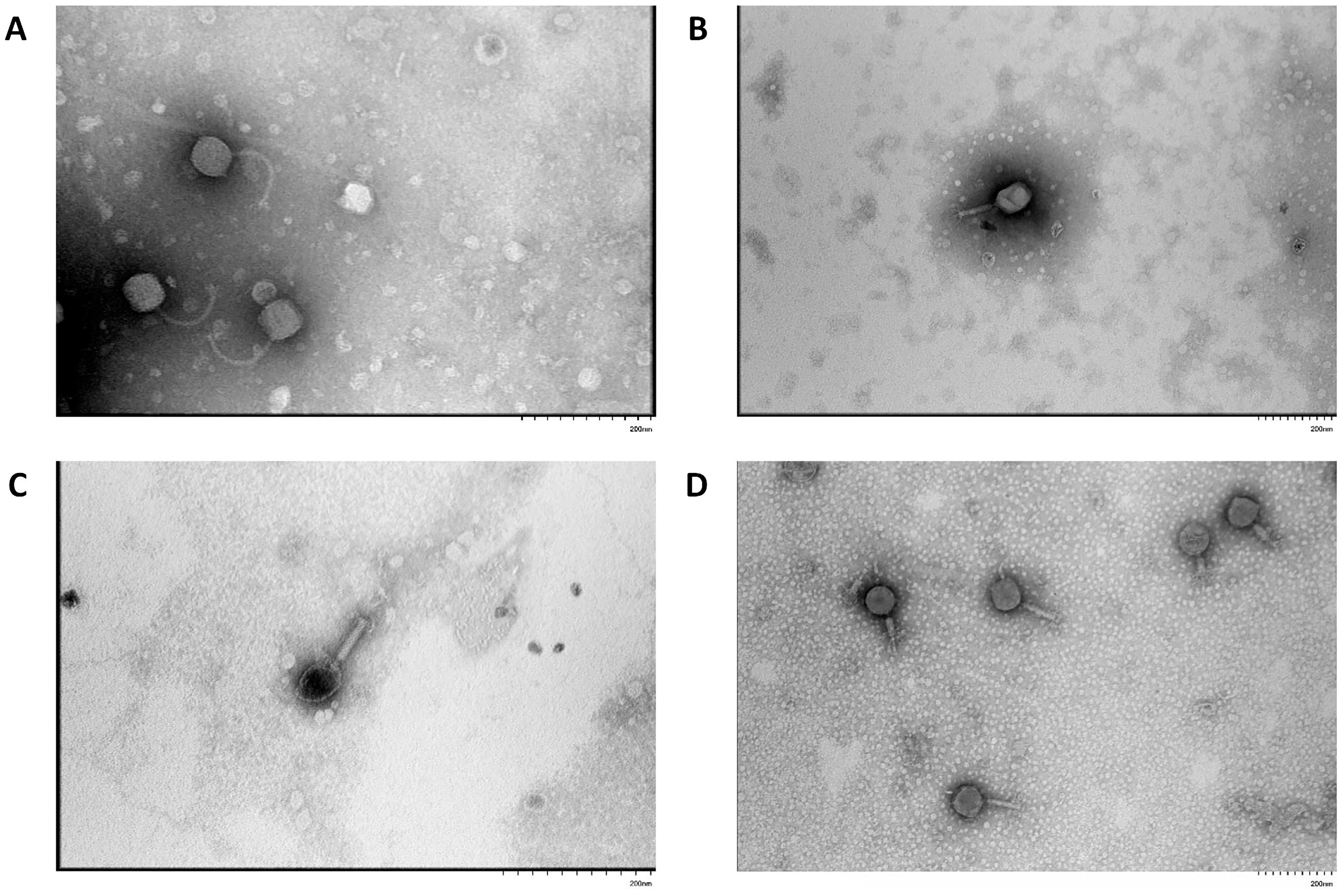
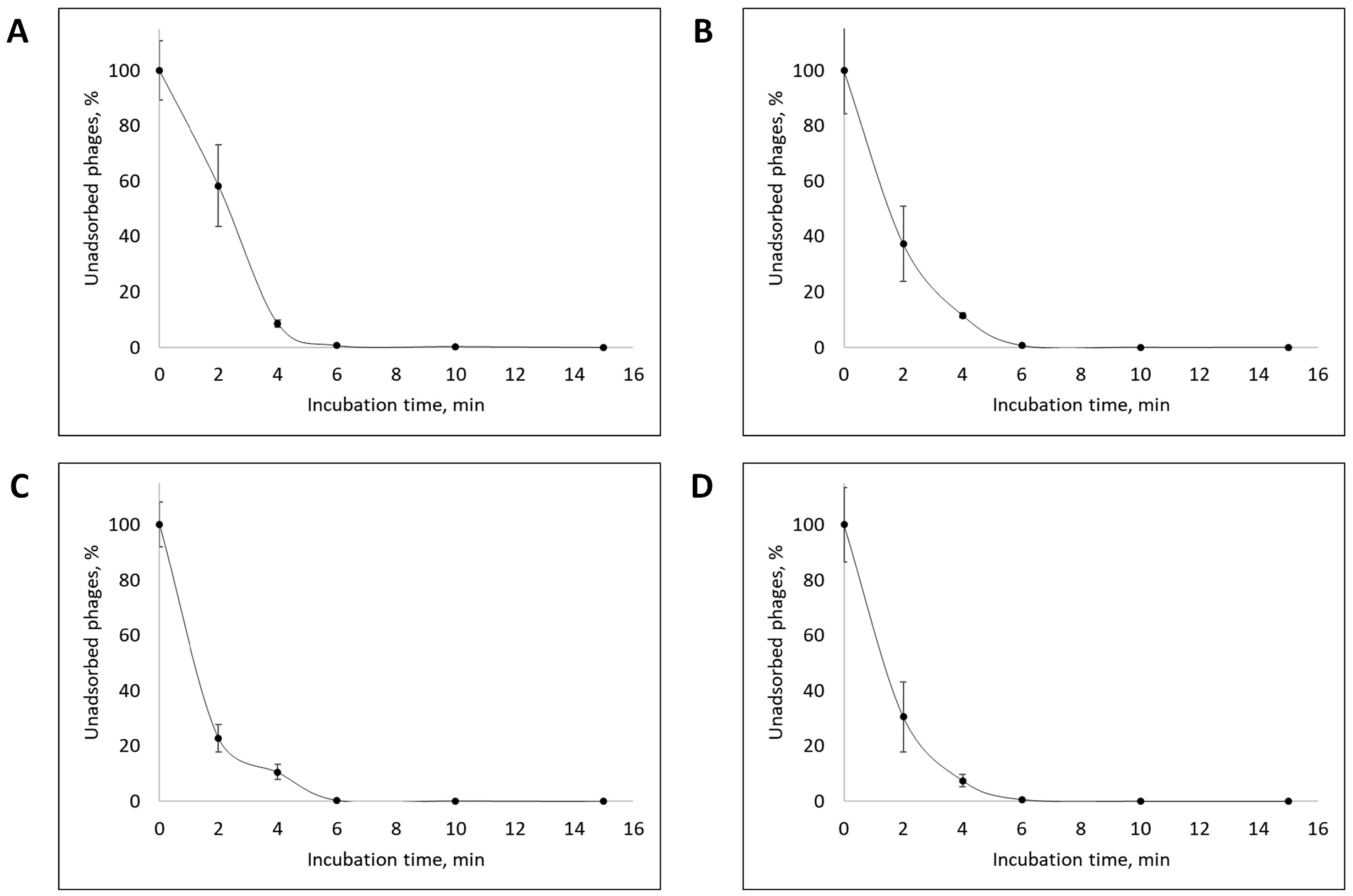
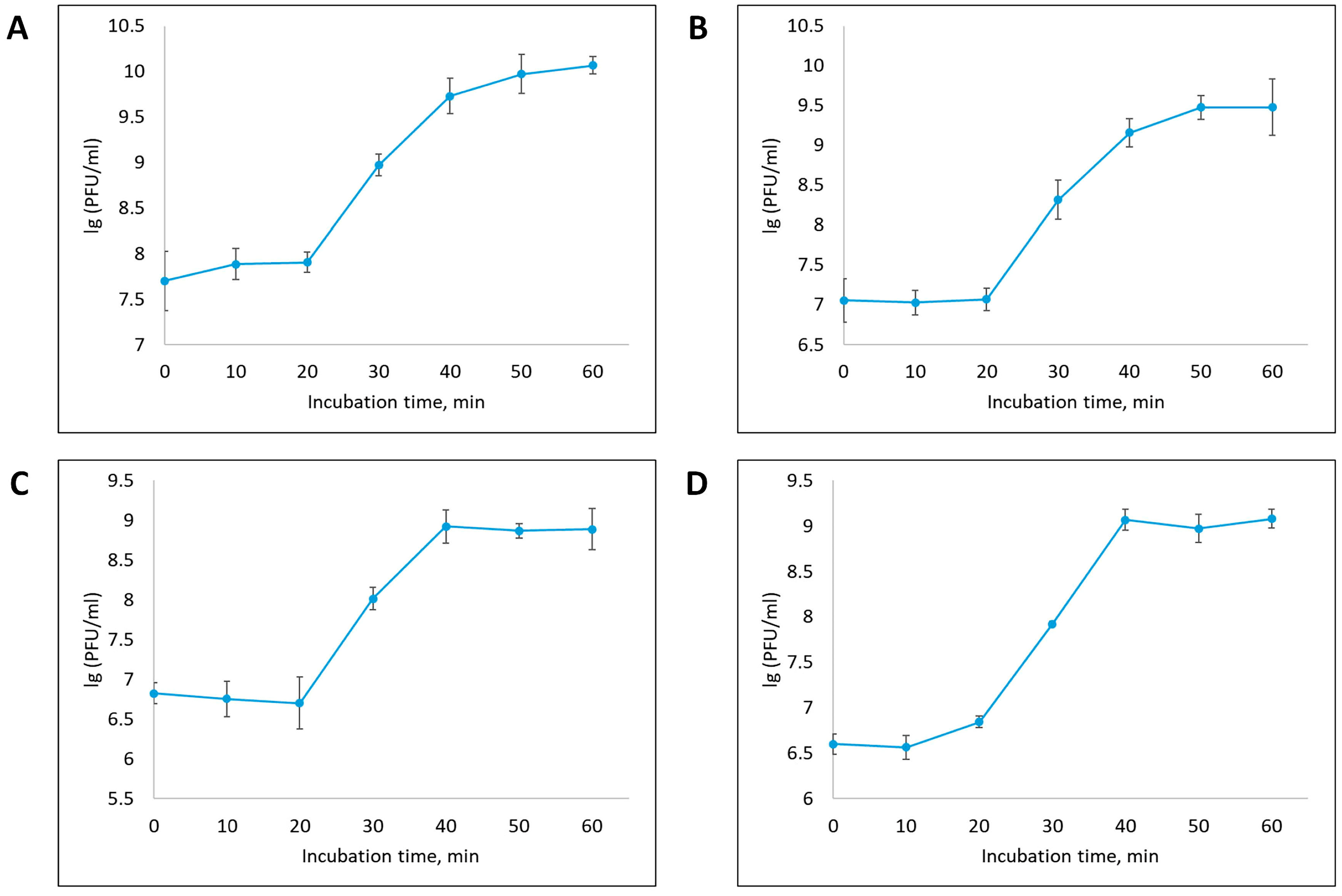

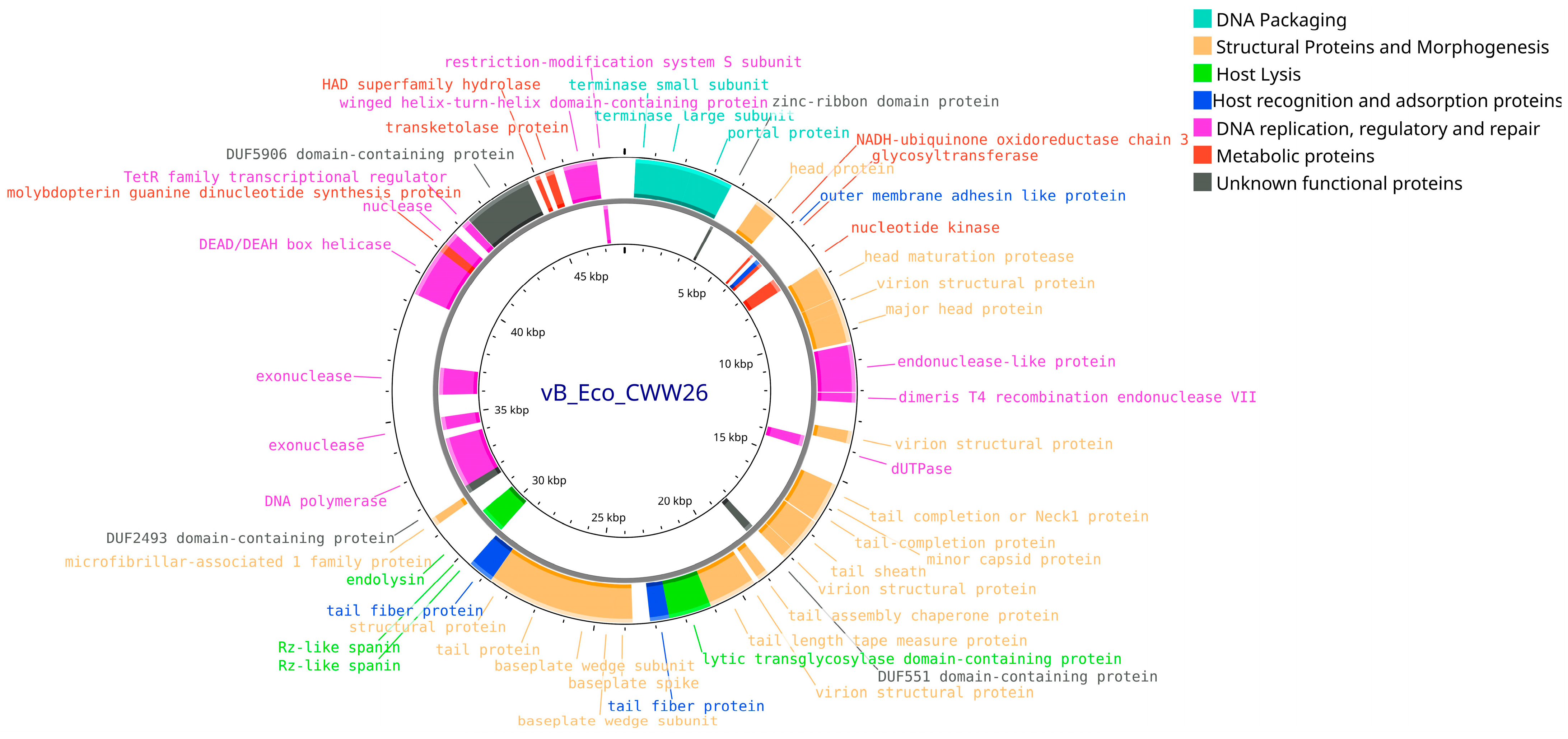
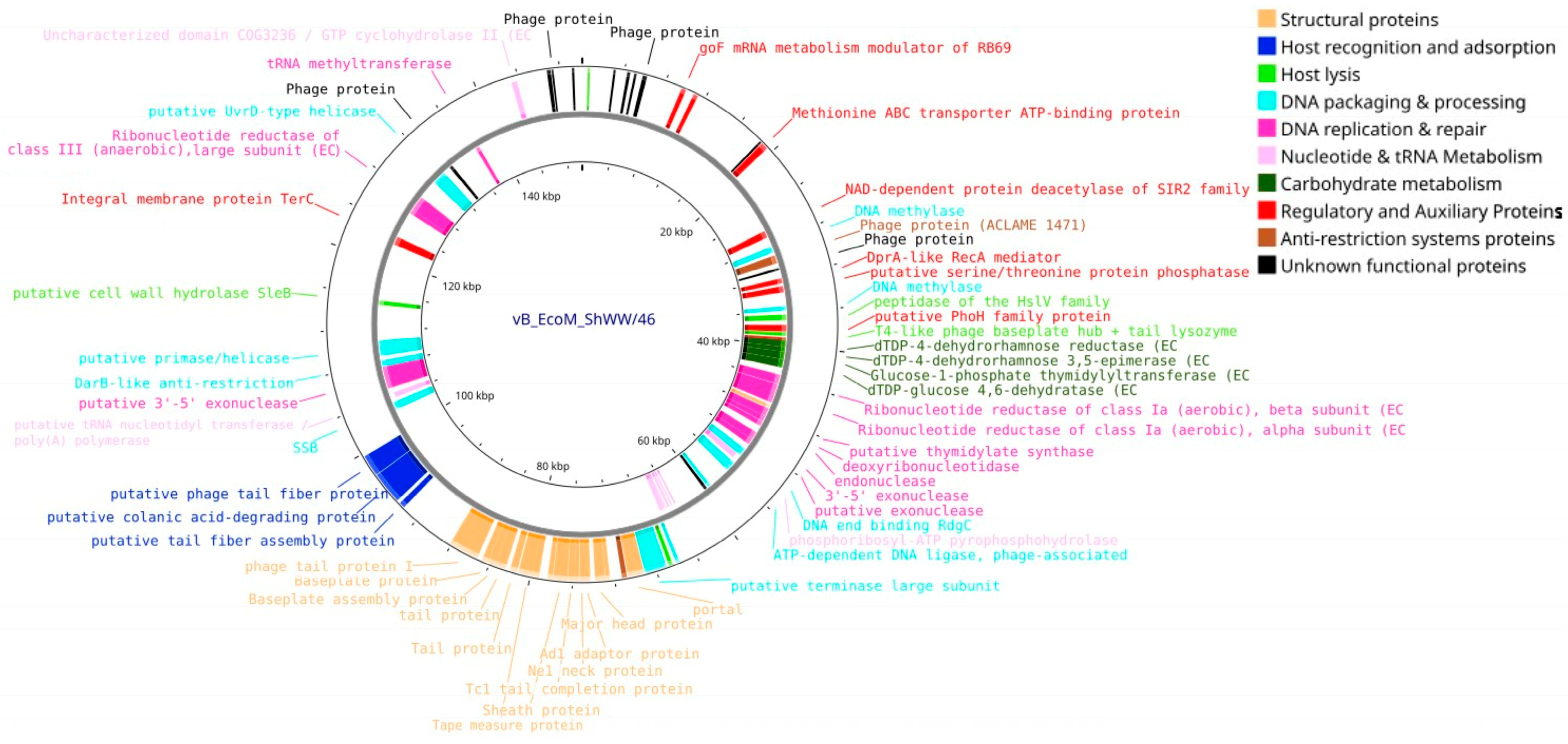
| Designation of Isolated Bacteriophage | Bacterial Host Strain | City (Source of Phage-Containing Wastewater) |
|---|---|---|
| vB_EcoS_ABO/4 | E. coli 4 | Almaty |
| vB_EcoM_PL/4 | Talgar | |
| vB_Eco_CWW/26 | E. coli 26 | Konayev |
| vB_EcoM_ShWW/46 | E. coli 46 | Shymkent |
| Bacteriophage | MOI | Number PFU | Bacteriophage | MOI | Number PFU |
|---|---|---|---|---|---|
| vB_EcoS_ABO/4 | 10−6 | 98 ± 15 * | vB_EcoM_PL/4 | 10−6 | 38 ± 6 |
| 10−5 | 8.2 ± 0.4 × 102 | 10−5 | 3.6 ± 0.4 × 102 | ||
| 10−4 | 4.7 ± 0.3 × 104 | 10−4 | 2.7 ± 0.1 × 104 | ||
| 10−3 | 5.1 ± 0.3 × 105 | 10−3 | 4.1 ± 0.2 × 105 | ||
| 10−2 | 2.5 ± 0.1 × 107 | 10−2 | 3.8 ± 0.1 × 107 | ||
| 10−1 | 4.1 ± 0.2 × 108 | 10−1 | 7.5 ± 0.2 × 107 | ||
| 1 | 8.2 ± 0.3 × 108 | 1 | 4.9 ± 0.4 × 108 | ||
| 10 | 5.5 ± 0.3 × 108 | 10 | 2.1 ± 0.1 × 108 | ||
| vB_Eco_CWW/26 | 10−6 | 13 ± 4 | vB_EcoM_ShWW/46 | 10−6 | 54 ± 9 |
| 10−5 | 51 ± 11 | 10−5 | 5.6 ± 0.4 × 102 | ||
| 10−4 | 2.7 ± 0.2 × 104 | 10−4 | 2.7 ± 0.1 × 104 | ||
| 10−3 | 4.8 ± 0.3 × 105 | 10−3 | 4.5 ± 0.3 × 105 | ||
| 10−2 | 3.2 ± 0.2 × 107 | 10−2 | 3.2 ± 0.1 × 107 | ||
| 10−1 | 6.4 ± 0.2 × 107 | 10−1 | 8.6 ± 0.4 × 107 | ||
| 1 | 2.9 ± 0.1 × 108 | 1 | 6.7 ± 0.3 × 108 | ||
| 10 | 1.6 ± 0.1 × 108 | 10 | 3.3 ± 0.1 × 108 |
| № | E. coli Strains | Bacteriophages | |||
|---|---|---|---|---|---|
| vB_EcoS_ABO/4 | vB_EcoM_PL/4 | vB_Eco_CWW/26 | vB_EcoM_ShWW/46 | ||
| 1 | 1MC | +++* | +++ | ++ | +++ |
| 2 | 4MY | +++ | +++ | +++ | — |
| 3 | 8MY | ++ | — | — | + |
| 4 | 9MY | — | — | + | — |
| 5 | 12MC | ++ | — | — | + |
| 6 | 16-1ML | — | — | — | — |
| 7 | 16-1P | ++ | ++ | ++ | — |
| 8 | 16-2CF | — | — | ++ | — |
| 9 | 17-1YF | + | + | — | ++ |
| 10 | 18 YML | +++ | +++ | +++ | — |
| 11 | 19-AY | — | — | ++ | — |
| 12 | 19-BYF | — | — | — | + |
| 13 | 20-1 YF | — | — | — | — |
| 14 | 20-2 Y | — | — | — | — |
| 15 | 21-1C | — | — | — | — |
| 16 | 21-2YF | +++ | + | — | + |
| 17 | 22-1YF | ++ | + | — | ++ |
| 18 | 23-1YF | — | — | — | ++ |
| 19 | 23-2Y | ++ | + | — | — |
| 20 | 24CF | — | — | ++ | + |
| 21 | 25-1CF | — | — | ++ | — |
| 22 | 25-2 P | +++ | + | — | — |
| 23 | 25-2W | +++ | — | — | + |
| 24 | 26 | — | — | +++ | + |
| 25 | 27 | — | — | + | — |
| 26 | 32 | — | — | — | — |
| 27 | 33 | — | — | — | — |
| 28 | 35 | — | — | — | — |
| 29 | 37 | — | — | — | — |
| 30 | 38 | — | — | — | ++ |
| 31 | 39 | +++ | +++ | — | + |
| 32 | 44 | +++ | +++ | — | +++ |
| 33 | 45 | +++ | +++ | — | — |
| 34 | 46 | +++ | +++ | — | +++ |
| 35 | 48 | +++ | +++ | — | ++ |
| Spectrum Index | 17 | 14 | 11 | 16 | |
| Feature | vB_Eco_CWW/26 | vB_EcoS_ABO/4 | vB_EcoM_PL/4 | vB_EcoM_ShWW/46 |
|---|---|---|---|---|
| Genome size, bp | 48,021 | 57,756 | 146,666 | 150,152 |
| Number of contig N50 | 1 | 1 | 1 | 1 |
| N50 length (bp) | 48,021 | 57,756 | 146,666 | 150,152 |
| CheckV completeness (%) | 100 | 100 | 100 | 100 |
| CheckV quality | High-quality | High-quality | High-quality | High-quality |
| ORF number | 88 | 98 | 264 | 276 |
| GC content (%) | 48.9 | 43.5 | 37.5 | 39 |
| tRNAs | - | - | 14 | 10 |
| Bacphlip life cycle (%) | Virulent (85) | Virulent (91.4) | Virulent (91.2) | Virulent (93.7) |
| E. coli | FICI | ||
|---|---|---|---|
| AMP | GEN | COL | |
| 1MC | 1 * | 0.5 | 0.13 |
| 17-1YF | 1 | 0.5 | 0.12 |
| 23-2Y | 0.25 | 1 | 1 |
| 32 | 2 | 0.5 | 0.5 |
| 35 | 2 | 0.5 | 2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alexyuk, P.G.; Bogoyavlenskiy, A.P.; Akanova, K.S.; Moldakhanov, Y.S.; Kerimov, T.T.; Sokolova, N.S.; Berezin, V.E.; Alexyuk, M.S. Isolation of Lytic Bacteriophages of Escherichia coli and Their Combined Use with Antibiotics Against the Causative Agents of Colibacillosis in Calves. Vet. Sci. 2025, 12, 817. https://doi.org/10.3390/vetsci12090817
Alexyuk PG, Bogoyavlenskiy AP, Akanova KS, Moldakhanov YS, Kerimov TT, Sokolova NS, Berezin VE, Alexyuk MS. Isolation of Lytic Bacteriophages of Escherichia coli and Their Combined Use with Antibiotics Against the Causative Agents of Colibacillosis in Calves. Veterinary Sciences. 2025; 12(9):817. https://doi.org/10.3390/vetsci12090817
Chicago/Turabian StyleAlexyuk, Pavel G., Andrey P. Bogoyavlenskiy, Kuralay S. Akanova, Yergali S. Moldakhanov, Timur T. Kerimov, Nadezhda S. Sokolova, Vladimir E. Berezin, and Madina S. Alexyuk. 2025. "Isolation of Lytic Bacteriophages of Escherichia coli and Their Combined Use with Antibiotics Against the Causative Agents of Colibacillosis in Calves" Veterinary Sciences 12, no. 9: 817. https://doi.org/10.3390/vetsci12090817
APA StyleAlexyuk, P. G., Bogoyavlenskiy, A. P., Akanova, K. S., Moldakhanov, Y. S., Kerimov, T. T., Sokolova, N. S., Berezin, V. E., & Alexyuk, M. S. (2025). Isolation of Lytic Bacteriophages of Escherichia coli and Their Combined Use with Antibiotics Against the Causative Agents of Colibacillosis in Calves. Veterinary Sciences, 12(9), 817. https://doi.org/10.3390/vetsci12090817







