A Method for Automated Detection of Chicken Coccidia in Vaccine Environments
Simple Summary
Abstract
1. Introduction
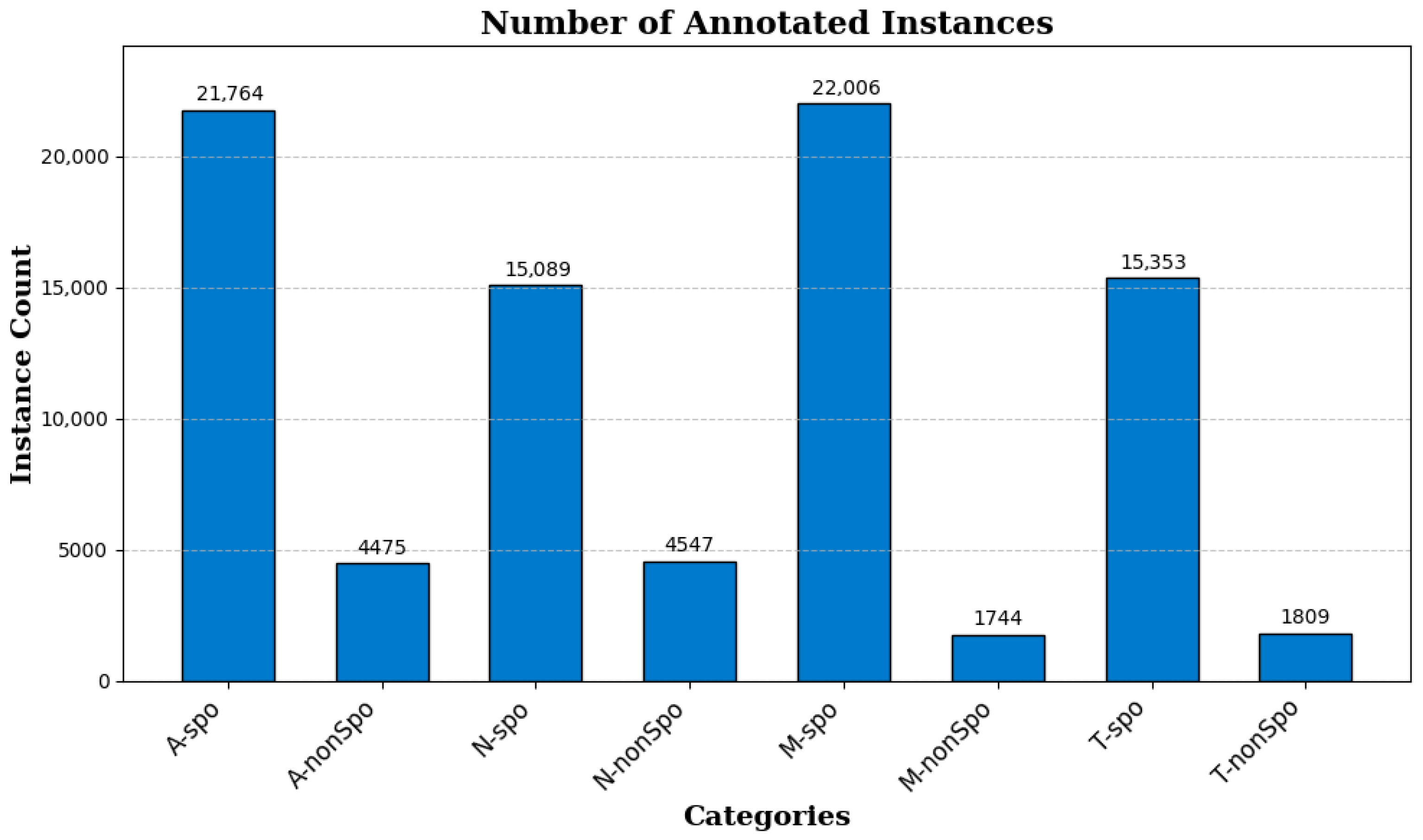
- We constructed a chicken coccidia dataset suitable for vaccine environments. The dataset includes four Eimeria species, and contains both sporulated and non-sporulated morphologies of each species, providing rich and diverse samples for the chicken coccidia detection task.
- The YOLO-Cocci model significantly improves the detection accuracy of chicken coccidia through three key improvements. First, an efficient multi-scale attention (EMA) module is integrated into the backbone to enhance the feature extraction of chicken coccidia oocysts. Second, the original neck is replaced with an inception-style multi-scale fusion pyramid network (IMFPN), which utilizes multi-scale feature fusion and parallel deep convolution to better retain critical features and enhance feature representation ability. Finally, a lightweight feature-reconstructed and partially decoupled detection head (LFPD-Head) is employed to further improve accuracy and optimize performance.
- The results of comparative experiments show that the YOLO-Cocci model outperforms other object detection models on the chicken coccidia dataset. Ablation studies further verifies its advantages in detecting morphologically similar oocysts. To improve user experience, a user-friendly client was developed for automatic detection and visualization of the YOLO-Cocci results. This study provides essential technical support for detecting chicken coccidia in vaccine environments.
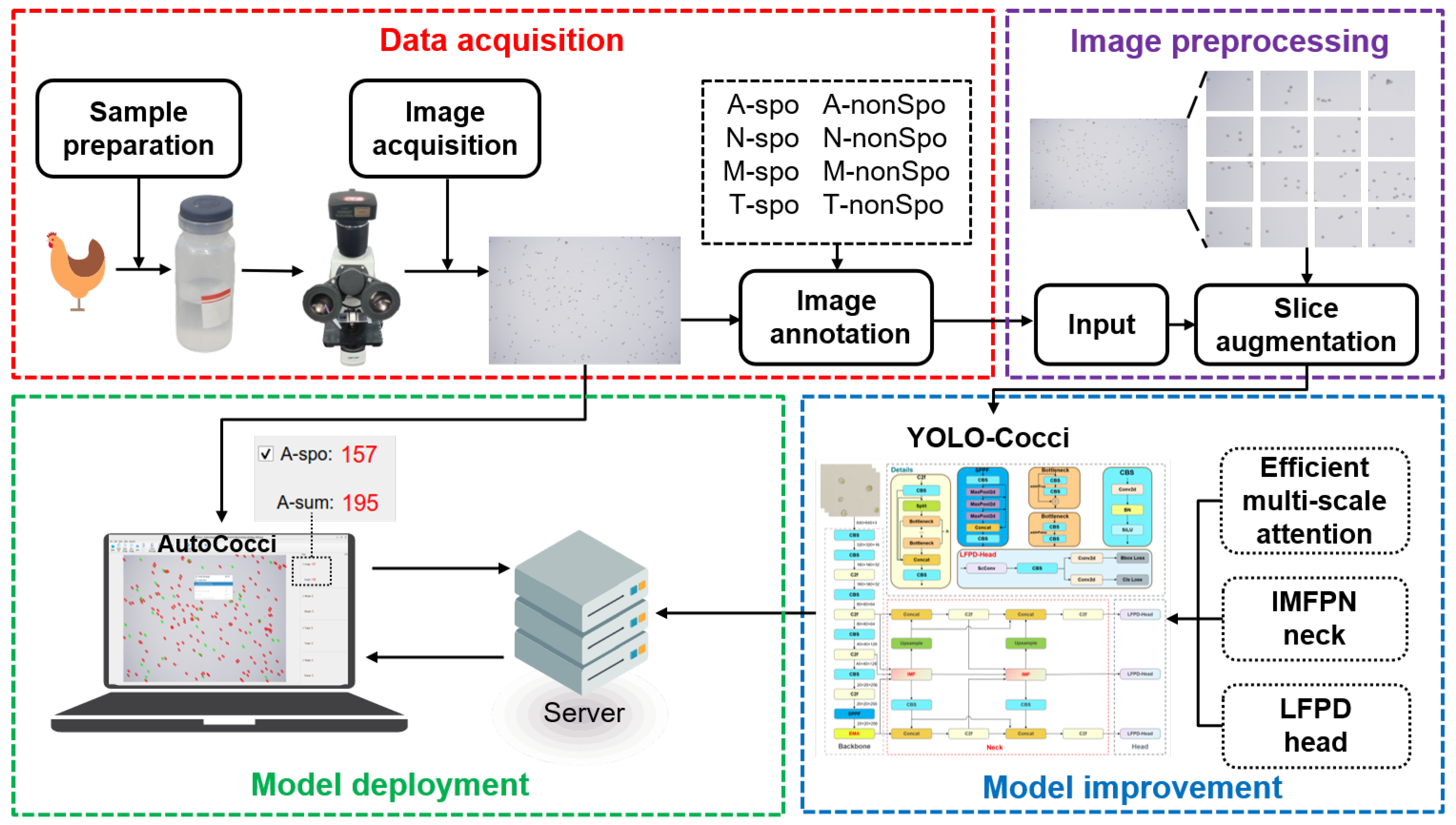
2. Materials and Methods
2.1. Data Acquisition
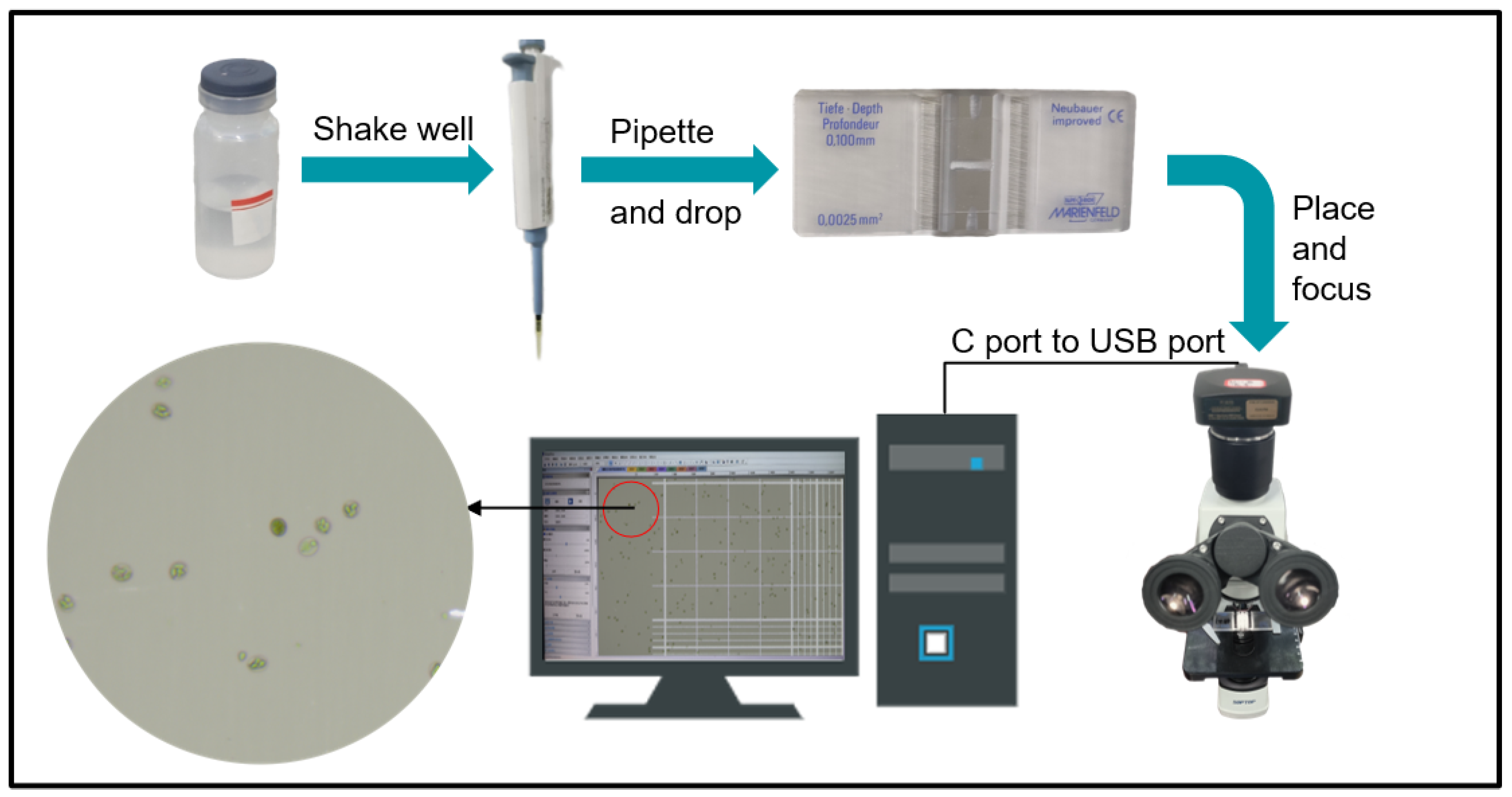
2.1.1. Chicken Eimeria Preparation
2.1.2. Image Acquisition
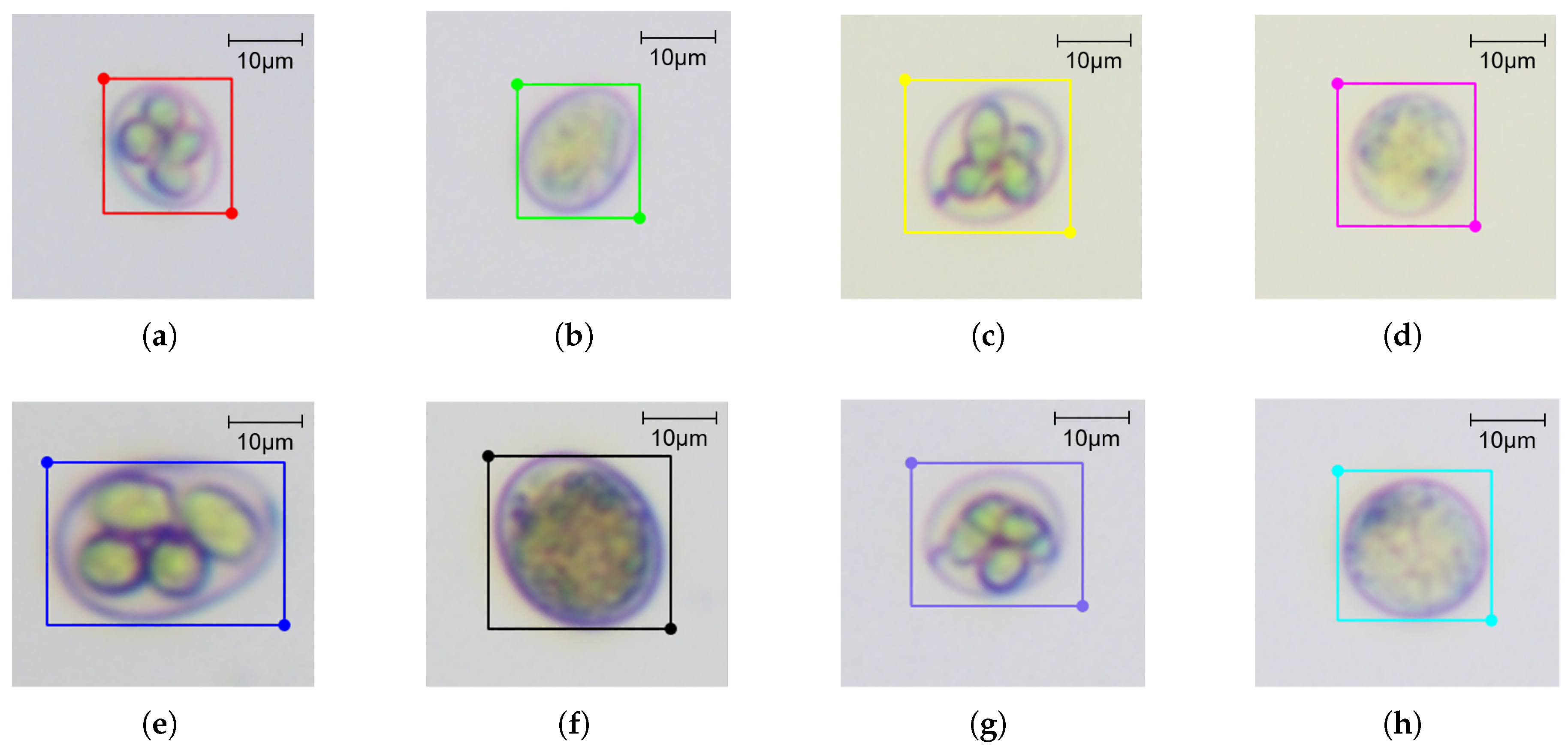
2.1.3. Image Annotation
2.2. Image Preprocessing
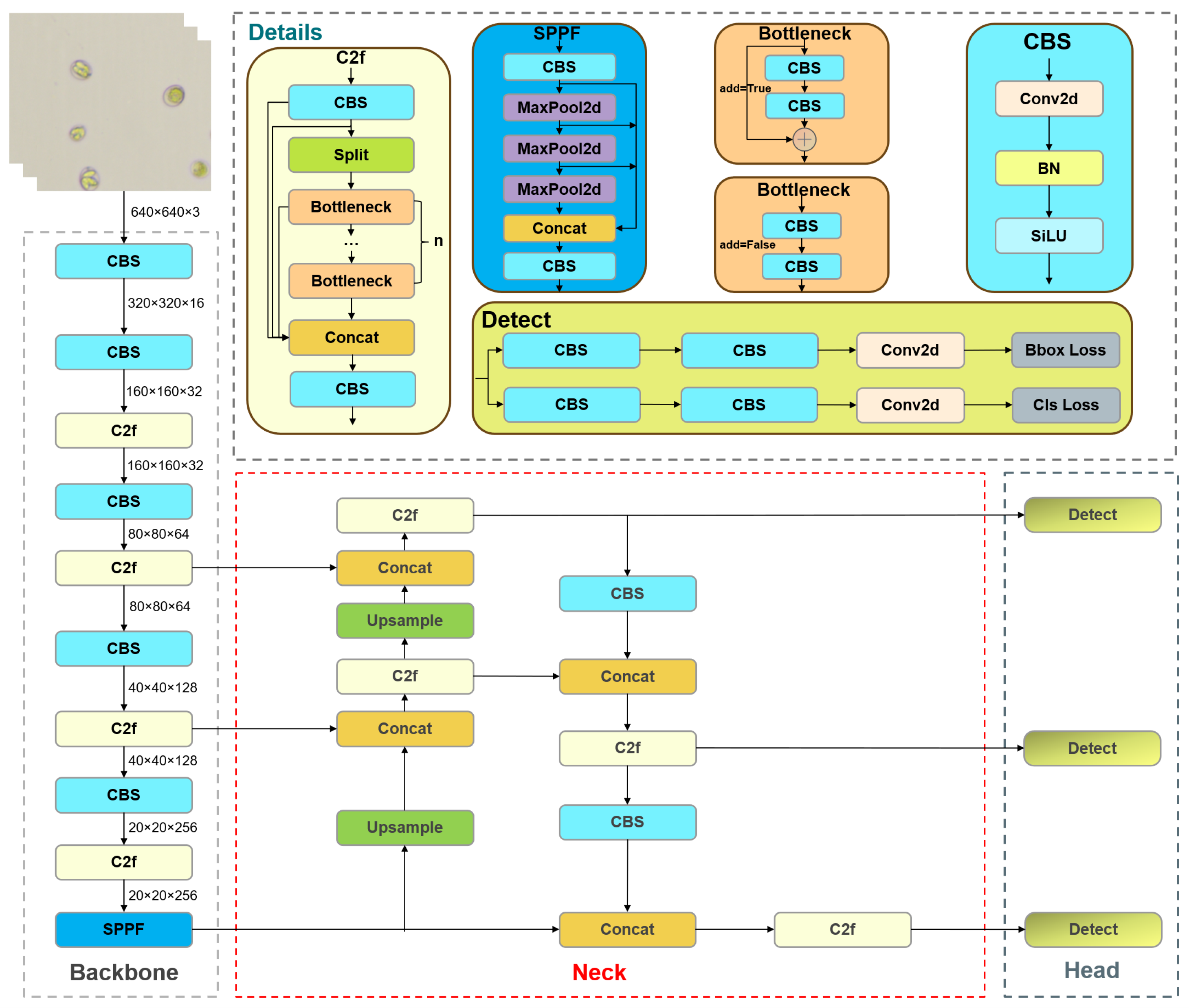
2.3. Baseline Model
2.4. YOLO-Cocci Model
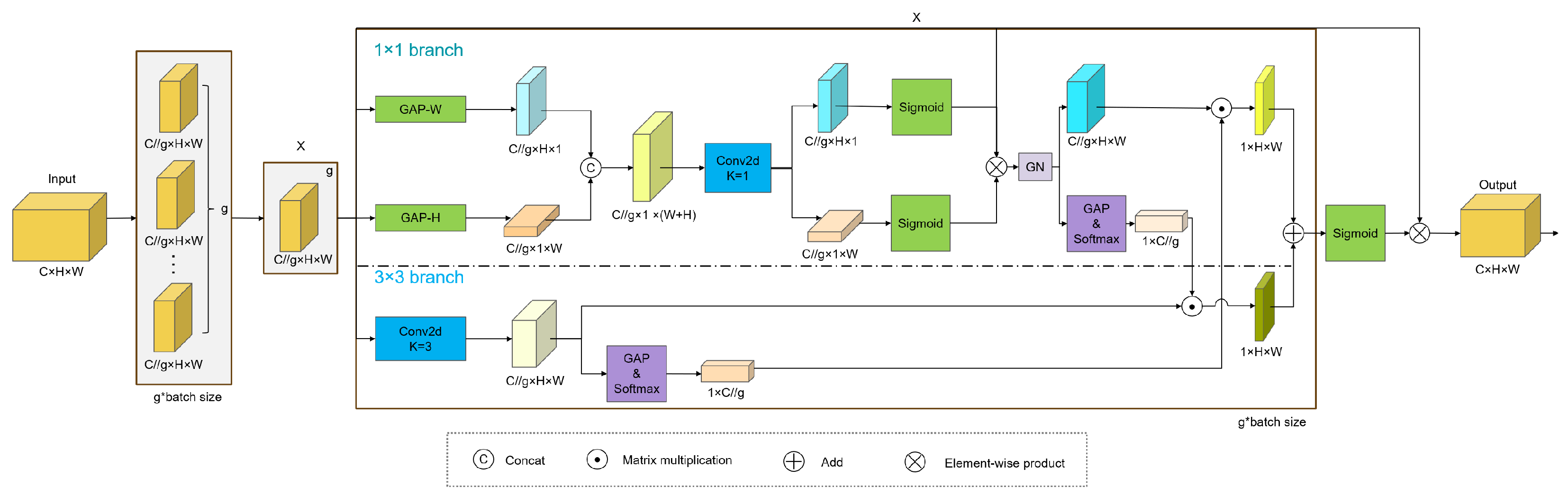
2.4.1. The EMA Module
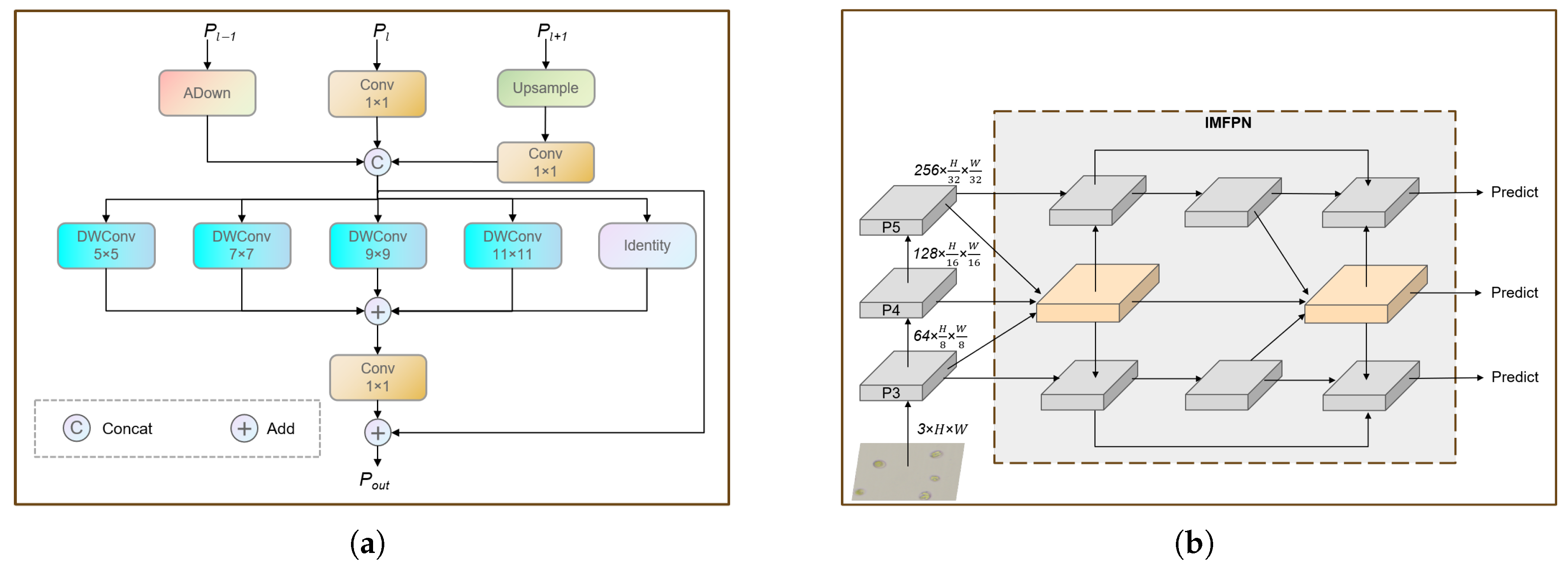
2.4.2. The IMFPN
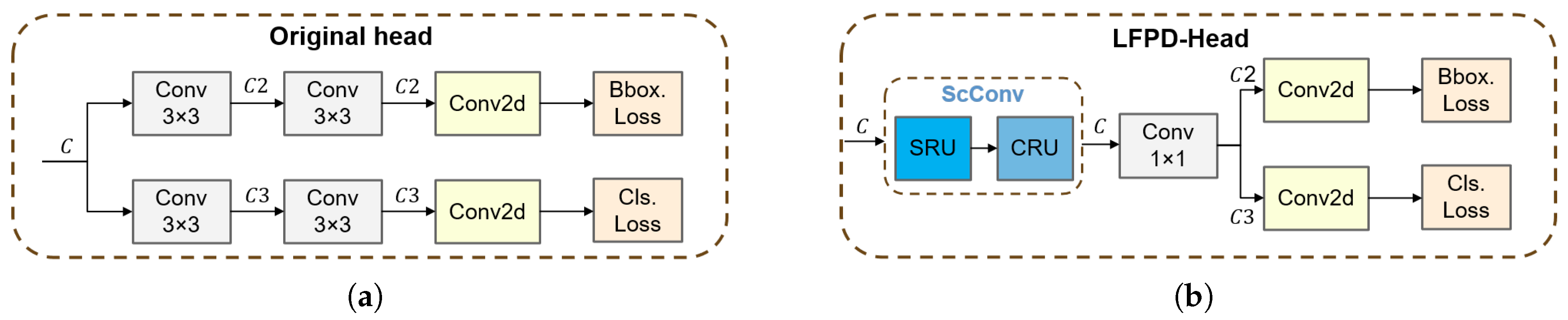
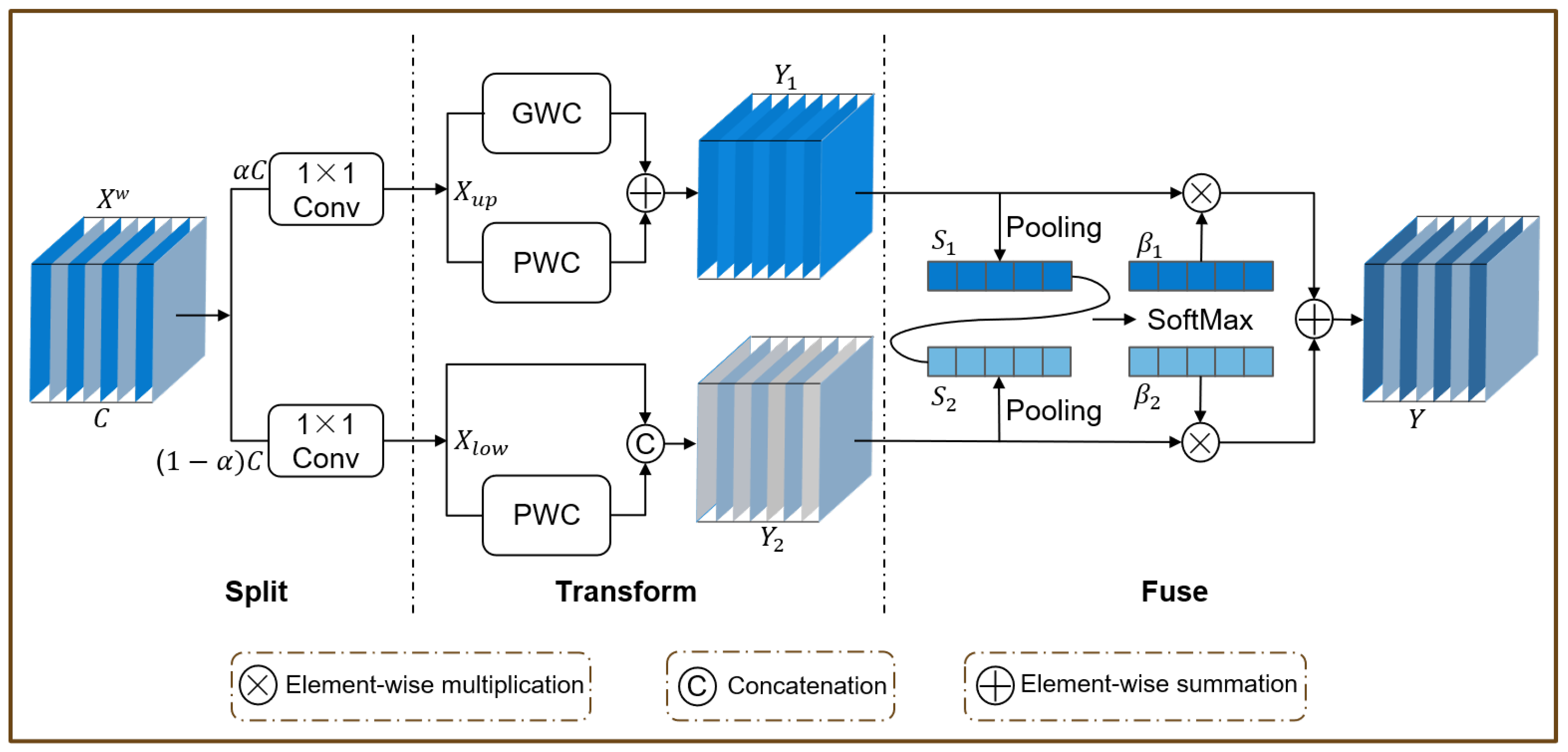
2.4.3. The LFPD-Head Module
3. Results
3.1. Experimental Setup
3.2. Evaluation Metrics
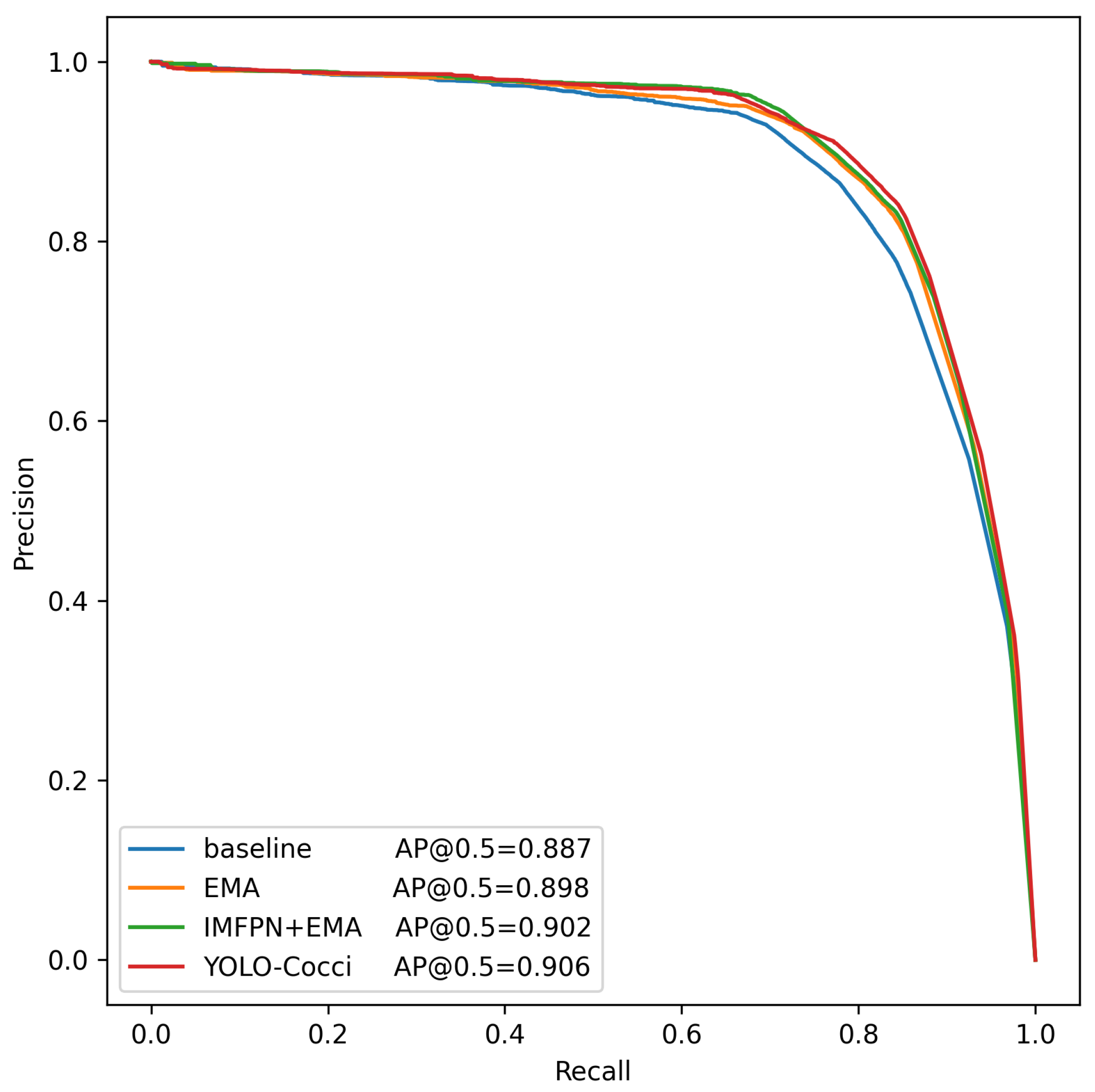
3.3. Comparison Experiment
3.4. Ablation Study
3.4.1. Ablation Experiments for Multi-Scale Kernel Design of IMF Module
3.4.2. Overall Ablation Experiments of the Improved YOLOv8 Model
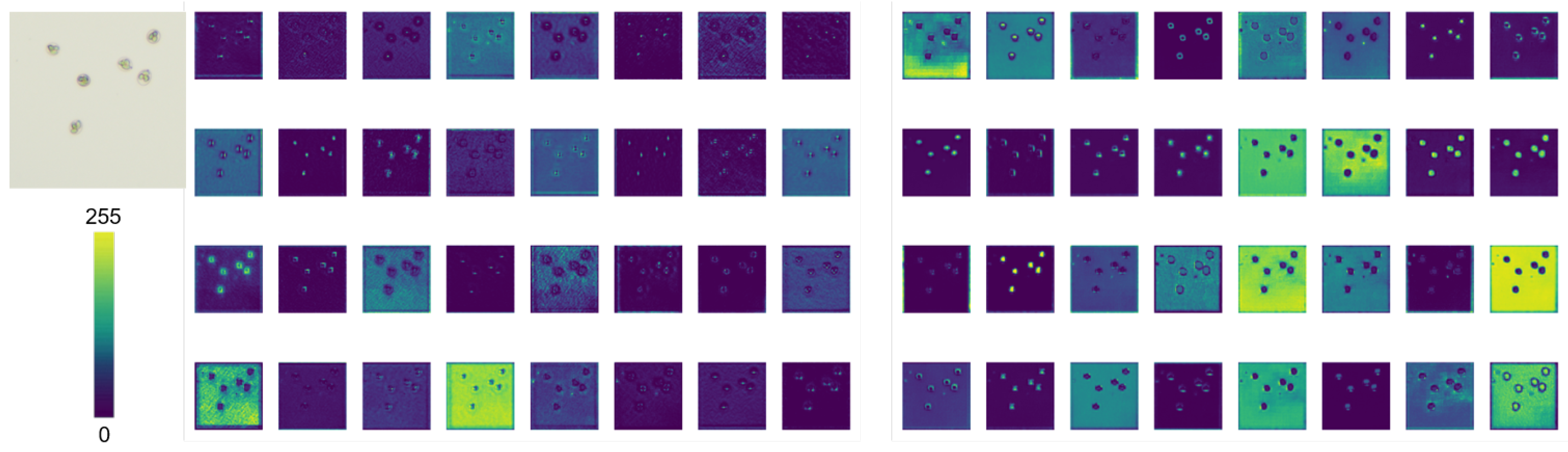
3.5. EMA Visualization
3.6. LFPD-Head Visualization
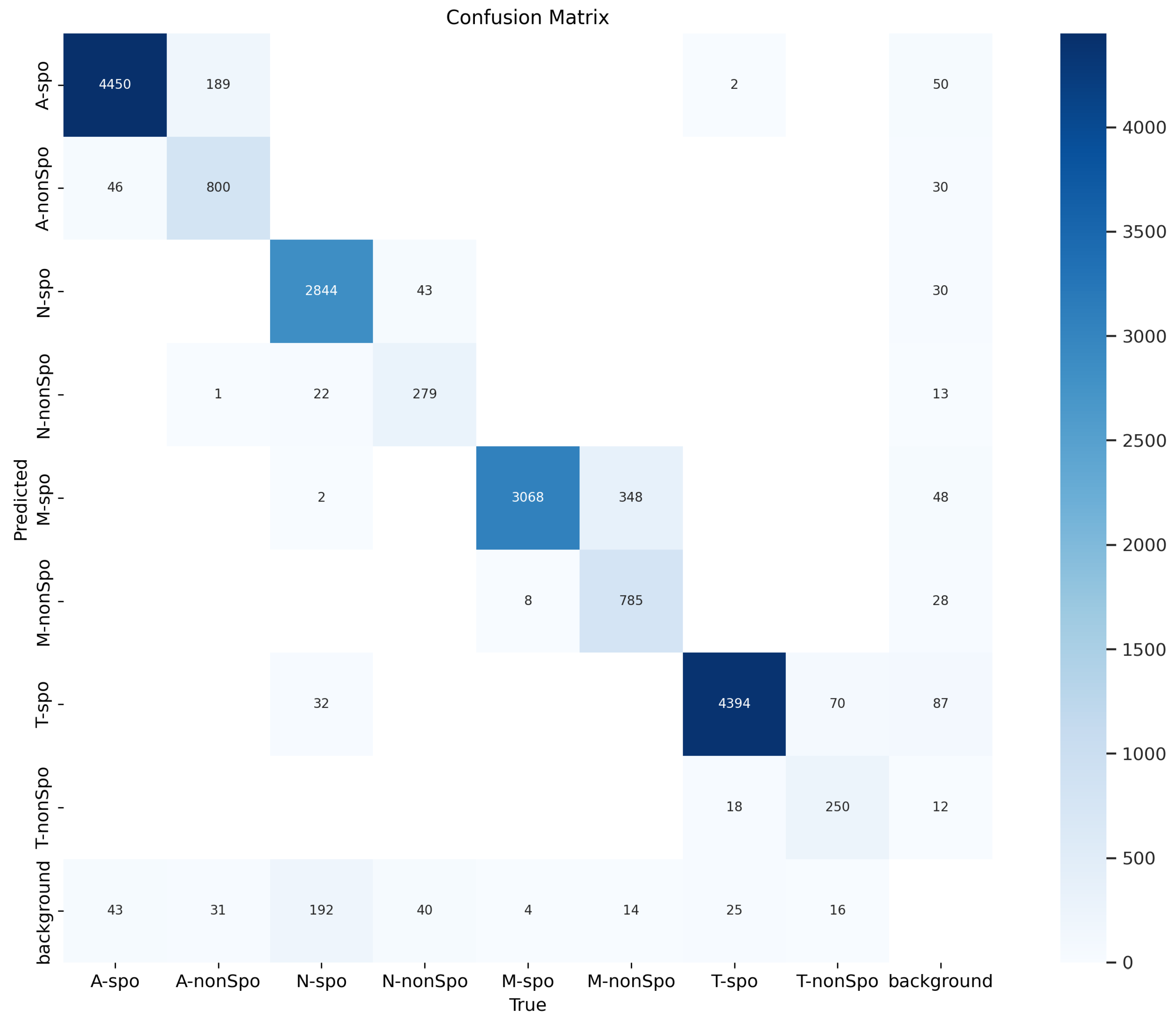
3.7. Confusion Matrix Analysis
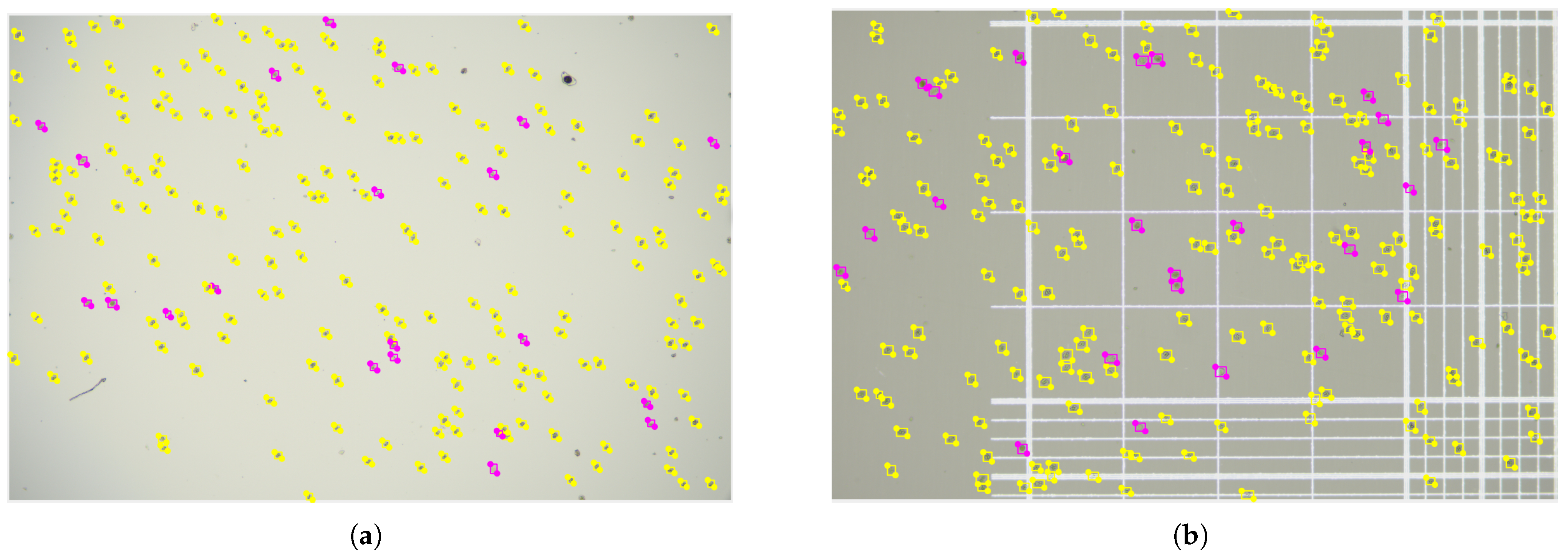
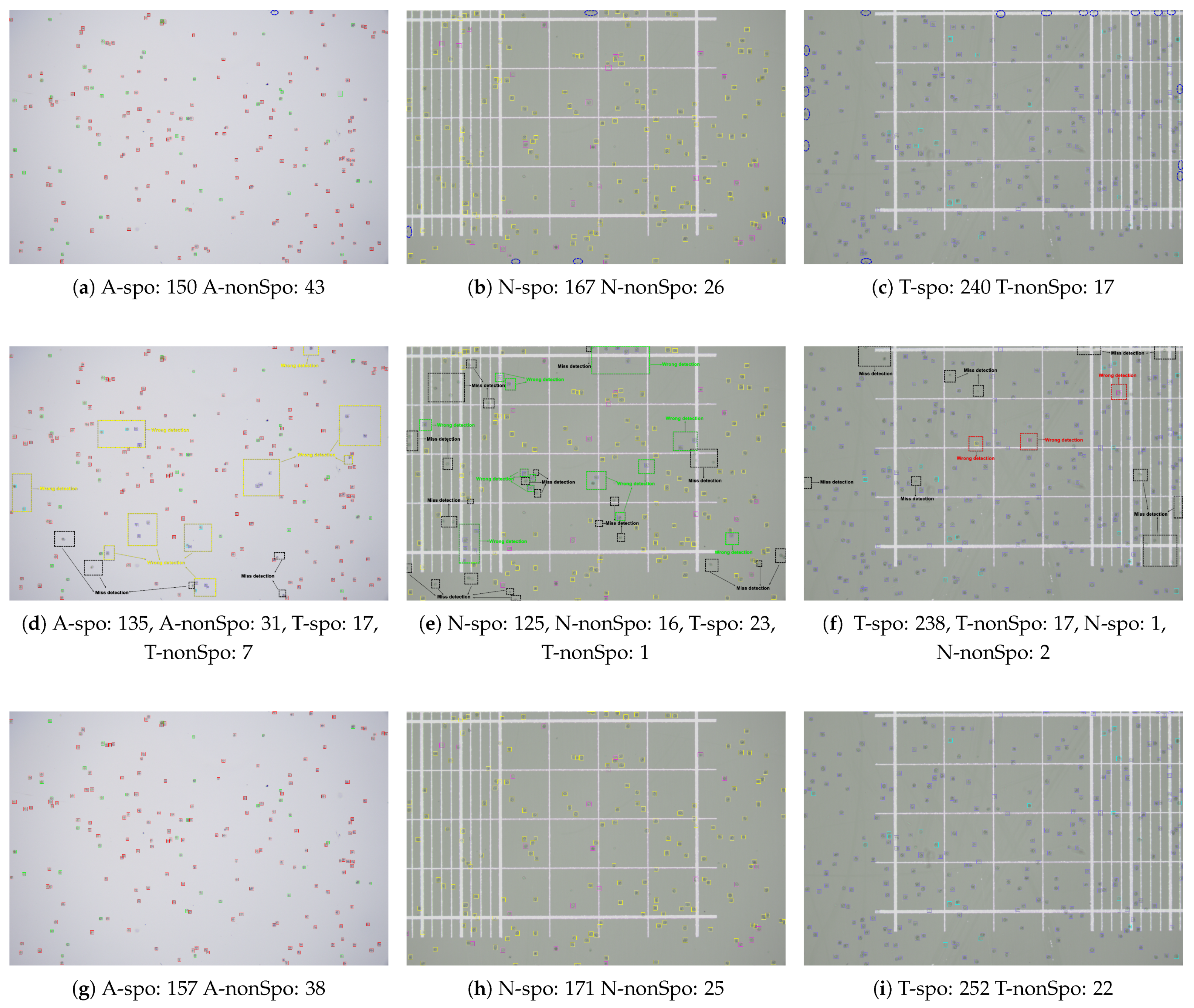
3.8. Visual Analysis of Detection Results
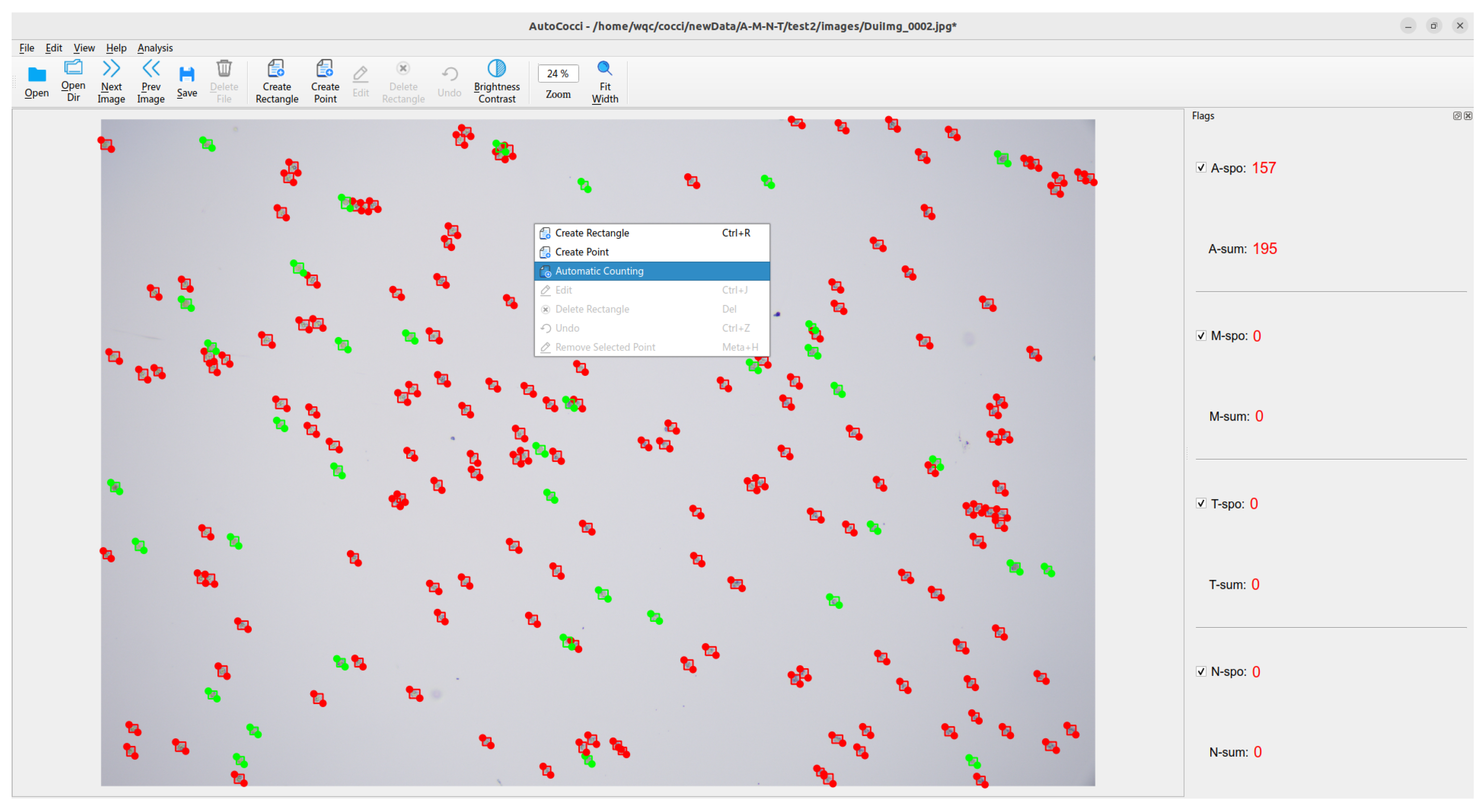
3.9. Deployment and Application of YOLO-Cocci Model
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| YOLO | You Only Look Once |
| EMA | Efficient multi-scale attention |
| IMFPN | Inception-style multi-scale fusion pyramid network |
| LFPD | Lightweight feature-reconstructed and partially decoupled |
References
- Mesa-Pineda, C.; Navarro-Ruíz, J.L.; López-Osorio, S.; Chaparro-Gutiérrez, J.J.; Gómez-Osorio, L.M. Chicken coccidiosis: From the parasite lifecycle to control of the disease. Front. Vet. Sci. 2021, 8, 787653. [Google Scholar] [CrossRef]
- Blake, D.P.; Tomley, F.M. Securing poultry production from the ever-present Eimeria challenge. Trends Parasitol. 2014, 30, 12–19. [Google Scholar] [CrossRef] [PubMed]
- Fatoba, A.J.; Adeleke, M.A. Diagnosis and control of chicken coccidiosis: A recent update. J. Parasit. Dis. 2018, 42, 483–493. [Google Scholar] [CrossRef] [PubMed]
- Abebe, E.; Gugsa, G. A review on poultry coccidiosis. Abyssinia J. Sci. Technol. 2018, 3, 1–12. [Google Scholar]
- Hamid, P.H.; Kristianingrum, Y.P.; Wardhana, A.H.; Prastowo, S.; Silva, L.M.R.d. Chicken coccidiosis in Central Java, Indonesia: A recent update. Vet. Med. Int. 2018, 2018, 8515812. [Google Scholar] [CrossRef]
- Blake, D.P.; Knox, J.; Dehaeck, B.; Huntington, B.; Rathinam, T.; Ravipati, V.; Ayoade, S.; Gilbert, W.; Adebambo, A.O.; Jatau, I.D.; et al. Re-calculating the cost of coccidiosis in chickens. Vet. Res. 2020, 51, 1–14. [Google Scholar] [CrossRef]
- Peek, H.; Landman, W. Coccidiosis in poultry: Anticoccidial products, vaccines and other prevention strategies. Vet. Q. 2011, 31, 143–161. [Google Scholar] [CrossRef]
- Haug, A.; Williams, R.; Larsen, S. Counting coccidial oocysts in chicken faeces: A comparative study of a standard McMaster technique and a new rapid method. Vet. Parasitol. 2006, 136, 233–242. [Google Scholar] [CrossRef]
- Jarquín-Díaz, V.H.; Balard, A.; Ferreira, S.C.M.; Mittné, V.; Murata, J.M.; Heitlinger, E. DNA-based quantification and counting of transmission stages provides different but complementary parasite load estimates: An example from rodent coccidia (Eimeria). Parasites Vectors 2022, 15, 45. [Google Scholar] [CrossRef]
- Ahmed-Laloui, H.; Zaak, H.; Rahmani, A.; Dems, M.A.; Cherb, N. A Simple Spectrophotometric Method for Coccidian Oocysts Counting in Broiler Feces. Acta Parasitol. 2022, 67, 1393–1400. [Google Scholar] [CrossRef]
- Adams, D.S.; Kulkarni, R.R.; Mohammed, J.P.; Crespo, R. A flow cytometric method for enumeration and speciation of coccidia affecting broiler chickens. Vet. Parasitol. 2022, 301, 109634. [Google Scholar] [CrossRef]
- Boyett, T.; Crespo, R.; Vinueza, V.C.; Gaghan, C.; Mohammed, J.P.; Kulkarni, R.R. Enumeration and speciation of coccidia affecting turkeys using flow cytometry method. J. Appl. Poult. Res. 2022, 31, 100270. [Google Scholar] [CrossRef]
- Adams, D.S.; Ruiz-Jimenez, F.; Fletcher, O.J.; Gall, S.; Crespo, R. Image analysis for Eimeria oocyst counts and classification. J. Appl. Poult. Res. 2022, 31, 100260. [Google Scholar] [CrossRef]
- Viet, N.Q.; ThanhTuyen, D.T.; Hoang, T.H. Parasite worm egg automatic detection in microscopy stool image based on Faster R-CNN. In Proceedings of the 3rd International Conference on Machine Learning and Soft Computing, Da Lat, Viet Nam, 25–28 January 2019; pp. 197–202. [Google Scholar]
- Tahir, M.W.; Zaidi, N.A.; Rao, A.A.; Blank, R.; Vellekoop, M.J.; Lang, W. A fungus spores dataset and a convolutional neural network based approach for fungus detection. IEEE Trans. Nanobiosci. 2018, 17, 281–290. [Google Scholar] [CrossRef] [PubMed]
- Panicker, R.O.; Kalmady, K.S.; Rajan, J.; Sabu, M. Automatic detection of tuberculosis bacilli from microscopic sputum smear images using deep learning methods. Biocybern. Biomed. Eng. 2018, 38, 691–699. [Google Scholar] [CrossRef]
- Devi, P.; Subburamu, K.; Giridhari, V.A.; Dananjeyan, B.; Maruthamuthu, T. Integration of AI based tools in dairy quality control: Enhancing pathogen detection efficiency. J. Food Meas. Charact. 2025, 19, 4427–4438. [Google Scholar] [CrossRef]
- Jafar, A.; Bibi, N.; Naqvi, R.A.; Sadeghi-Niaraki, A.; Jeong, D. Revolutionizing agriculture with artificial intelligence: Plant disease detection methods, applications, and their limitations. Front. Plant Sci. 2024, 15, 1356260. [Google Scholar] [CrossRef] [PubMed]
- Oon, Y.L.; Oon, Y.S.; Ayaz, M.; Deng, M.; Li, L.; Song, K. Waterborne pathogens detection technologies: Advances, challenges, and future perspectives. Front. Microbiol. 2023, 14, 1286923. [Google Scholar] [CrossRef]
- Zhou, C.; He, H.; Zhou, H.; Ge, F.; Yu, P. MSRT-DETR: A novel RT-DETR model with multi-scale feature sequence for cell detection. Biomed. Signal Process. Control 2025, 103, 107378. [Google Scholar] [CrossRef]
- Chen, T.; Chefd’Hotel, C. Deep learning based automatic immune cell detection for immunohistochemistry images. In International Workshop on Machine Learning in Medical Imaging; Springer: Berlin/Heidelberg, Germany, 2014; pp. 17–24. [Google Scholar]
- Moen, E.; Bannon, D.; Kudo, T.; Graf, W.; Covert, M.; Van Valen, D. Deep learning for cellular image analysis. Nat. Methods 2019, 16, 1233–1246. [Google Scholar] [CrossRef]
- Abdurahman, F.; Fante, K.A.; Aliy, M. Malaria parasite detection in thick blood smear microscopic images using modified YOLOV3 and YOLOV4 models. BMC Bioinform. 2021, 22, 1–17. [Google Scholar] [CrossRef]
- Ren, S.; He, K.; Girshick, R.; Sun, J. Faster R-CNN: Towards real-time object detection with region proposal networks. IEEE Trans. Pattern Anal. Mach. Intell. 2016, 39, 1137–1149. [Google Scholar] [CrossRef]
- Liu, W.; Anguelov, D.; Erhan, D.; Szegedy, C.; Reed, S.; Fu, C.Y.; Berg, A.C. Ssd: Single shot multibox detector. In Computer Vision–ECCV 2016: 14th European Conference, Amsterdam, The Netherlands, 11–14 October 2016, Proceedings, Part I 14; Springer: Berlin/Heidelberg, Germany, 2016; pp. 21–37. [Google Scholar]
- Kumar, S.; Arif, T.; Ahamad, G.; Chaudhary, A.A.; Khan, S.; Ali, M.A. An efficient and effective framework for intestinal parasite egg detection using YOLOv5. Diagnostics 2023, 13, 2978. [Google Scholar] [CrossRef] [PubMed]
- Smith, M.K.; Buhr, D.L.; Dhlakama, T.A.; Dupraw, D.; Fitz-Coy, S.; Francisco, A.; Ganesan, A.; Hubbard, S.A.; Nederlof, A.; Newman, L.J.; et al. Automated enumeration of Eimeria oocysts in feces for rapid coccidiosis monitoring. Poult. Sci. 2023, 102, 102252. [Google Scholar] [CrossRef] [PubMed]
- Kellogg, I.; Roberts, D.L.; Crespo, R. Automated image analysis for detection of coccidia in poultry. Animals 2024, 14, 212. [Google Scholar] [CrossRef] [PubMed]
- He, K.; Gkioxari, G.; Dollár, P.; Girshick, R. Mask r-cnn. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 2961–2969. [Google Scholar]
- Bochkovskiy, A.; Wang, C.Y.; Liao, H.Y.M. Yolov4: Optimal speed and accuracy of object detection. arXiv 2020, arXiv:2004.10934. [Google Scholar] [CrossRef]
- Zhang, H. mixup: Beyond empirical risk minimization. arXiv 2017, arXiv:1710.09412. [Google Scholar]
- Ouyang, D.; He, S.; Zhang, G.; Luo, M.; Guo, H.; Zhan, J.; Huang, Z. Efficient multi-scale attention module with cross-spatial learning. In Proceedings of the ICASSP 2023–2023 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Rhodes Island, Greece, 4–10 June 2023; pp. 1–5. [Google Scholar]
- Hou, Q.; Zhou, D.; Feng, J. Coordinate attention for efficient mobile network design. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Nashville, TN, USA, 20–25 June 2021; pp. 13713–13722. [Google Scholar]
- Woo, S.; Park, J.; Lee, J.Y.; Kweon, I.S. Cbam: Convolutional block attention module. In Proceedings of the European Conference on Computer Vision (ECCV), Munich, Germany, 8–14 September 2018; pp. 3–19. [Google Scholar]
- Wu, Y.; He, K. Group normalization. In Proceedings of the European conference on computer vision (ECCV), Munich, Germany, 8–14 September 2018; pp. 3–19. [Google Scholar]
- Liu, S.; Qi, L.; Qin, H.; Shi, J.; Jia, J. Path aggregation network for instance segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 8759–8768. [Google Scholar]
- Lin, T.Y.; Dollár, P.; Girshick, R.; He, K.; Hariharan, B.; Belongie, S. Feature pyramid networks for object detection. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 2117–2125. [Google Scholar]
- Li, J.; Wen, Y.; He, L. Scconv: Spatial and channel reconstruction convolution for feature redundancy. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Vancouver, BC, Canada, 17–24 June 2023; pp. 6153–6162. [Google Scholar]
- Li, X.; Wang, W.; Hu, X.; Yang, J. Selective kernel networks. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Long Beach, CA, USA, 15–20 June 2019; pp. 510–519. [Google Scholar]
- Wang, C.Y.; Yeh, I.H.; Mark Liao, H.Y. Yolov9: Learning what you want to learn using programmable gradient information. In European Conference on Computer Vision; Springer: Berlin/Heidelberg, Germany, 2024; pp. 1–21. [Google Scholar]
- Wang, A.; Chen, H.; Liu, L.; Chen, K.; Lin, Z.; Han, J. Yolov10: Real-time end-to-end object detection. Adv. Neural Inf. Process. Syst. 2024, 37, 107984–108011. [Google Scholar]
- Jocher, G. Yolov11. 2024. Available online: https://github.com/ultralytics/ultralytics (accessed on 24 August 2025).
- Lin, T.Y.; Goyal, P.; Girshick, R.; He, K.; Dollár, P. Focal loss for dense object detection. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 2980–2988. [Google Scholar]




| Carrier | Acervulina | Necatrix | Maxima | Tenella |
|---|---|---|---|---|
| Glass slide | 50 | 51 | 59 | 50 |
| Counting chamber | 50 | 56 | 52 | 52 |
| Model | mAP@0.5 | mAP@0.5:0.95 | P | R | Params | FLOPs | FPS |
|---|---|---|---|---|---|---|---|
| YOLOv5n | 87.6 | 65.8 | 89.1 | 83.6 | 1.77 M | 4.2 G | 41 |
| YOLOv9t | 88.6 | 66.6 | 92.3 | 83.9 | 2.60 M | 10.7 G | 19 |
| YOLOv10n | 89.5 | 66.1 | 91.1 | 86.4 | 2.70 M | 8.2 G | 15 |
| YOLOv11n | 88.3 | 66.6 | 92.8 | 84.0 | 2.58 M | 6.3 G | 16 |
| RetinaNet | 60.0 | 40.7 | 92.7 | 60.0 | 56.86 M | 295 G | 3 |
| Faster R-CNN | 73.5 | 48.8 | 85.4 | 76.5 | 60.64 M | 265 G | 4 |
| Mask R-CNN | 74.7 | 49.2 | 86.1 | 75.8 | 62.28 M | 265 G | 2 |
| YOLO-Cocci (ours) | 89.6 | 67.3 | 92.9 | 86.0 | 2.59 M | 7.1 G | 17 |
| Kernel Design | Params | FLOPs | mAP@0.5 |
|---|---|---|---|
| (3, 3, 3, 3) | 2.48 M | 6.7 G | 87.0 |
| (3, 5, 7, 9) | 2.54 M | 6.9 G | 88.6 |
| (5, 7, 9, 11) | 2.59 M | 7.1 G | 89.6 |
| (7, 9, 11, 13) | 2.66 M | 7.3 G | 89.0 |
| (3, 7, 11, 15) | 2.65 M | 7.3 G | 88.0 |
| (3, 5, 7, 9, 11) | 2.60 M | 7.1 G | 87.8 |
| Model | mAP@0.5 | mAP@0.5:0.95 | A-spo | A-nonSpo | N-spo | N-nonSpo | M-spo | M-nonSpo | T-spo | T-nonSpo | Params | FLOPs |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Baseline | 83.1 | 62.3 | 88.6 | 80.9 | 80.9 | 71.4 | 90.3 | 83.0 | 94.1 | 75.7 | 3.01 M | 8.1 G |
| +IMFPN | 86.8 | 65.1 | 93.6 | 85.6 | 87.6 | 78.9 | 91.2 | 82.8 | 95.2 | 79.4 | 3.04 M | 9.4 G |
| +EMA | 88.4 | 66.3 | 94.4 | 85.3 | 91.9 | 80.5 | 90.7 | 83.4 | 97.9 | 83.3 | 3.02 M | 8.2 G |
| +LFPD-Head | 87.6 | 65.3 | 93.5 | 85.3 | 89.6 | 79.0 | 90.1 | 83.1 | 97.4 | 82.5 | 2.53 M | 5.7 G |
| +IMFPN+EMA | 89.1 | 67.1 | 94.7 | 86.2 | 93.9 | 82.1 | 91.4 | 83.2 | 97.5 | 83.9 | 3.05 M | 9.5 G |
| +IMFPN+LFPD-Head | 88.7 | 66.7 | 95.3 | 86.0 | 93.8 | 80.2 | 90.7 | 82.7 | 97.9 | 82.9 | 2.58 M | 7.0 G |
| +EMA+LFPD-Head | 89.0 | 66.3 | 95.0 | 86.1 | 95.7 | 82.8 | 89.8 | 82.5 | 97.3 | 82.4 | 2.54 M | 5.7 G |
| YOLO-Cocci (ours) | 89.6 | 67.3 | 95.6 | 86.6 | 94.9 | 83.9 | 90.6 | 83.2 | 97.7 | 84.0 | 2.59 M | 7.1 G |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, X.; Wang, Q.; Chen, L.; Wang, X.; Zhou, M.; Lin, R.; Guo, Y. A Method for Automated Detection of Chicken Coccidia in Vaccine Environments. Vet. Sci. 2025, 12, 812. https://doi.org/10.3390/vetsci12090812
Li X, Wang Q, Chen L, Wang X, Zhou M, Lin R, Guo Y. A Method for Automated Detection of Chicken Coccidia in Vaccine Environments. Veterinary Sciences. 2025; 12(9):812. https://doi.org/10.3390/vetsci12090812
Chicago/Turabian StyleLi, Ximing, Qianchao Wang, Lanqi Chen, Xinqiu Wang, Mengting Zhou, Ruiqing Lin, and Yubin Guo. 2025. "A Method for Automated Detection of Chicken Coccidia in Vaccine Environments" Veterinary Sciences 12, no. 9: 812. https://doi.org/10.3390/vetsci12090812
APA StyleLi, X., Wang, Q., Chen, L., Wang, X., Zhou, M., Lin, R., & Guo, Y. (2025). A Method for Automated Detection of Chicken Coccidia in Vaccine Environments. Veterinary Sciences, 12(9), 812. https://doi.org/10.3390/vetsci12090812






