MicroRNA Expression Profiling in Canine Myxomatous Mitral Valve Disease Highlights Potential Diagnostic Tool and Molecular Pathways
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Selection and Histological Classification
2.2. Computational Prediction of miRNAs
2.3. RNA Extraction and Purification
2.4. cDNA Synthesis and qPCR Amplification
2.5. Reference Gene Selection
2.6. Statistical Analysis
3. Results
3.1. Morphological Assessment at the Gross and Histological Level
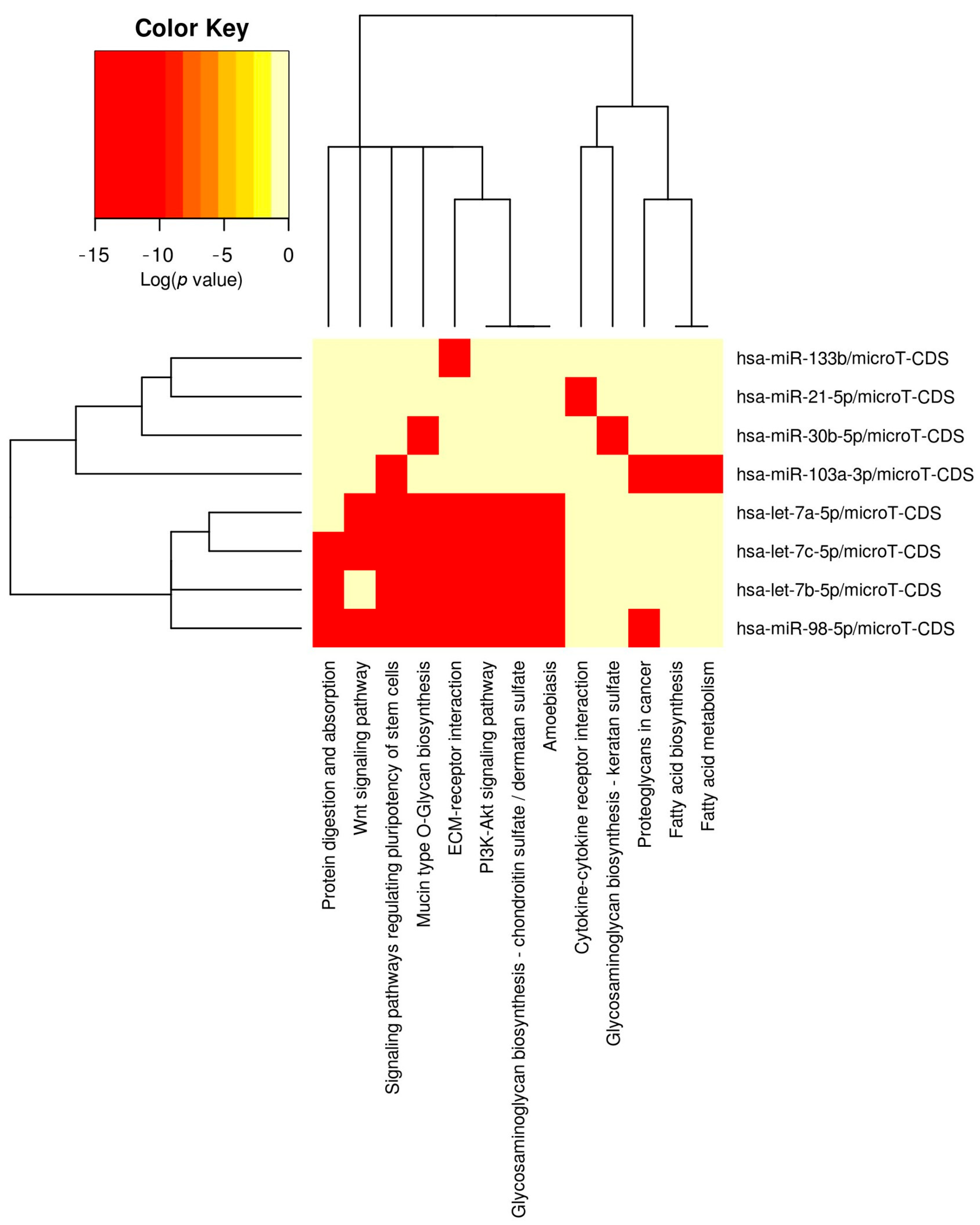
3.2. Selection and Bioinformatic Enrichment of Candidate miRNAs
3.3. RNA Integrity Analysis and Reference Gene Validation
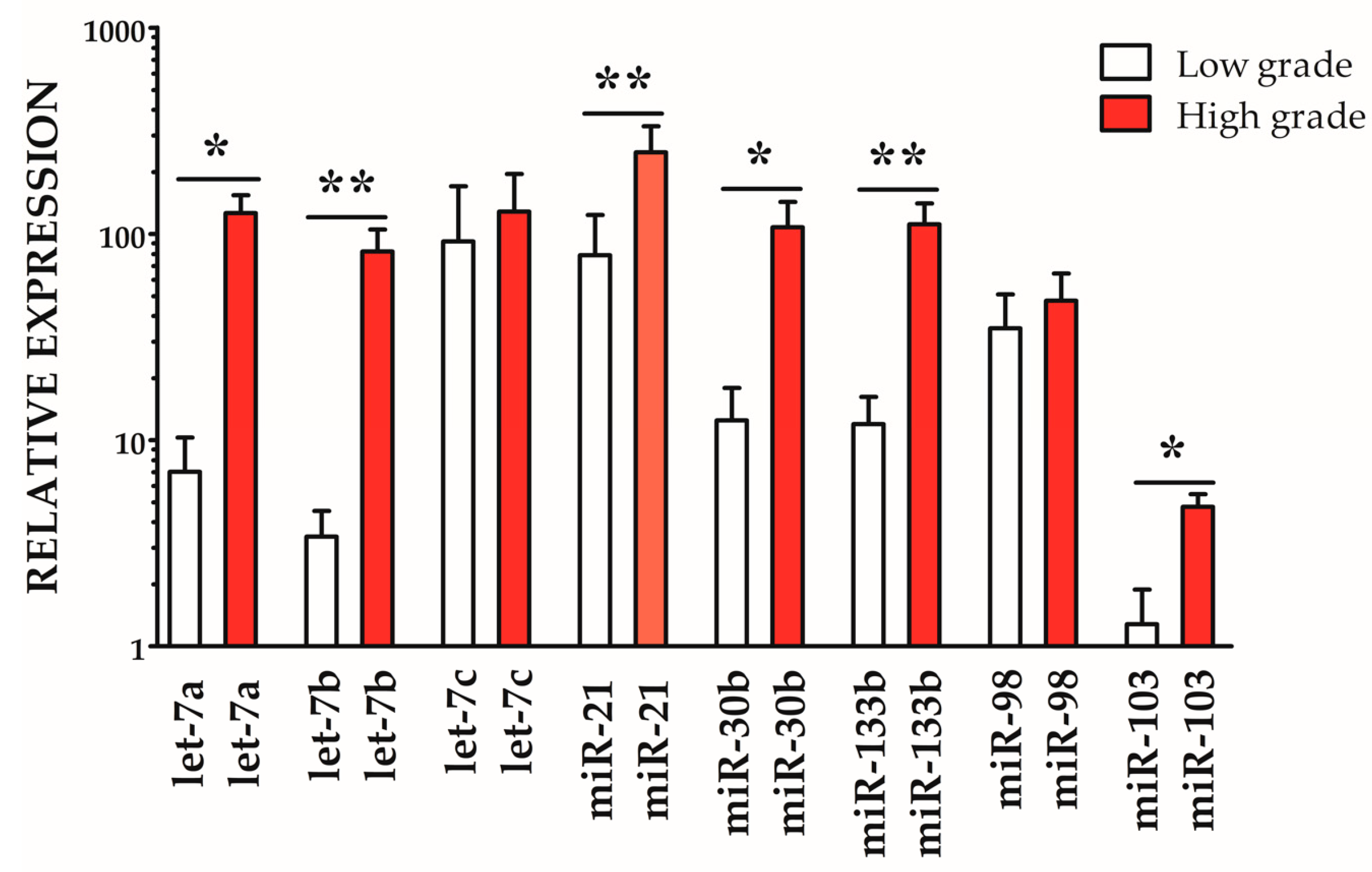
3.4. MiRNA Expression Signatures
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lee, S.-Y.; Lee, S.; Kim, S.-H.; Chang, H.; Cho, W.-Y.; Ryu, M.-O.; Choi, J.; Yoon, H.-Y.; Seo, K.-W. Deep Learning-Based Evaluation of the Severity of Mitral Regurgitation in Canine Myxomatous Mitral Valve Disease Patients Using Digital Stethoscope Recordings. BMC Vet. Res. 2025, 21, 326. [Google Scholar] [CrossRef] [PubMed]
- Oyama, M.A.; Elliott, C.; Loughran, K.A.; Kossar, A.P.; Castillero, E.; Levy, R.J.; Ferrari, G. Comparative Pathology of Human and Canine Myxomatous Mitral Valve Degeneration: 5HT and TGF-β Mechanisms. Cardiovasc. Pathol. 2020, 46, 107196. [Google Scholar] [CrossRef]
- Fox, P.R. Pathology of Myxomatous Mitral Valve Disease in the Dog. J. Vet. Cardiol. 2012, 14, 103–126. [Google Scholar] [CrossRef] [PubMed]
- Levine, R.A.; Hagége, A.A.; Judge, D.P.; Padala, M.; Dal-Bianco, J.P.; Aikawa, E.; Beaudoin, J.; Bischoff, J.; Bouatia-Naji, N.; Bruneval, P.; et al. Mitral Valve Disease—Morphology and Mechanisms. Nat. Rev. Cardiol. 2015, 12, 689–710. [Google Scholar] [CrossRef]
- Pedersen, H. Mitral Valve Prolapse in the Dog: A Model of Mitral Valve Prolapse in Man. Cardiovasc. Res. 2000, 47, 234–243. [Google Scholar] [CrossRef]
- Markby, G.; Summers, K.; MacRae, V.; Corcoran, B. Comparative Transcriptomic Profiling and Gene Expression for Myxomatous Mitral Valve Disease in the Dog and Human. Vet. Sci. 2017, 4, 34. [Google Scholar] [CrossRef]
- Kumiega, E.; Kobak, K.A.; Noszczyk-Nowak, A.; Kasztura, M. Iron Parameters Analysis in Dogs with Myxomatous Mitral Valve Disease. BMC Vet. Res. 2024, 20, 210. [Google Scholar] [CrossRef] [PubMed]
- Sayed, D.; Abdellatif, M. MicroRNAs in Development and Disease. Physiol. Rev. 2011, 91, 827–887. [Google Scholar] [CrossRef]
- Mitchell, P.S.; Parkin, R.K.; Kroh, E.M.; Fritz, B.R.; Wyman, S.K.; Pogosova-Agadjanyan, E.L.; Peterson, A.; Noteboom, J.; O’Briant, K.C.; Allen, A.; et al. Circulating microRNAs as Stable Blood-Based Markers for Cancer Detection. Proc. Natl. Acad. Sci. USA 2008, 105, 10513–10518. [Google Scholar] [CrossRef]
- Chen, X.; Ba, Y.; Ma, L.; Cai, X.; Yin, Y.; Wang, K.; Guo, J.; Zhang, Y.; Chen, J.; Guo, X.; et al. Characterization of microRNAs in Serum: A Novel Class of Biomarkers for Diagnosis of Cancer and Other Diseases. Cell Res. 2008, 18, 997–1006. [Google Scholar] [CrossRef]
- Tijsen, A.J.; Creemers, E.E.; Moerland, P.D.; De Windt, L.J.; Van Der Wal, A.C.; Kok, W.E.; Pinto, Y.M. MiR423-5p As a Circulating Biomarker for Heart Failure. Circ. Res. 2010, 106, 1035–1039. [Google Scholar] [CrossRef]
- Guelfi, G.; Venanzi, N.; Capaccia, C.; Stefanetti, V.; Brachelente, C.; Sforna, M.; Porciello, F.; Lepri, E. Feline Hypertrophic Cardiomyopathy: Does the microRNA–mRNA Regulatory Network Contribute to Heart Sarcomeric Protein Remodelling? Int. J. Exp. Path 2024, 105, 170–183. [Google Scholar] [CrossRef] [PubMed]
- Segev, G.; Daminet, S.; Meyer, E.; De Loor, J.; Cohen, A.; Aroch, I.; Bruchim, Y. Characterization of Kidney Damage Using Several Renal Biomarkers in Dogs with Naturally Occurring Heatstroke. Vet. J. 2015, 206, 231–235. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Freeman, L.; Rush, J.; Laflamme, D. Expression Profiling of Circulating MicroRNAs in Canine Myxomatous Mitral Valve Disease. Int. J. Mol. Sci. 2015, 16, 14098–14108. [Google Scholar] [CrossRef]
- Yang, V.K.; Loughran, K.A.; Meola, D.M.; Juhr, C.M.; Thane, K.E.; Davis, A.M.; Hoffman, A.M. Circulating Exosome microRNA Associated with Heart Failure Secondary to Myxomatous Mitral Valve Disease in a Naturally Occurring Canine Model. J. Extracell. Vesicles 2017, 6, 1350088. [Google Scholar] [CrossRef] [PubMed]
- Bagardi, M.; Ghilardi, S.; Zamarian, V.; Ceciliani, F.; Brambilla, P.G.; Lecchi, C. Circulating MiR-30b-5p Is Upregulated in Cavalier King Charles Spaniels Affected by Early Myxomatous Mitral Valve Disease. PLoS ONE 2022, 17, e0266208. [Google Scholar] [CrossRef]
- Palarea-Albaladejo, J.; Bode, E.F.; Partington, C.; Basili, M.; Mederska, E.; Hodgkiss-Geere, H.; Capewell, P.; Chauché, C.; Coultous, R.M.; Hanks, E.; et al. Assessing the Use of Blood microRNA Expression Patterns for Predictive Diagnosis of Myxomatous Mitral Valve Disease in Dogs. Front. Vet. Sci. 2024, 11, 1443847. [Google Scholar] [CrossRef]
- Kim, T.-S.; Hong, C.-Y.; Oh, S.-J.; Choe, Y.-H.; Hwang, T.-S.; Kim, J.; Lee, S.-L.; Yoon, H.; Bok, E.-Y.; Cho, A.; et al. RNA Sequencing Provides Novel Insights into the Pathogenesis of Naturally Occurring Myxomatous Mitral Valve Disease Stage B1 in Beagle Dogs. PLoS ONE 2024, 19, e0300813. [Google Scholar] [CrossRef]
- Ma, R.; Jiang, T.; Kang, X. Circulating microRNAs in Cancer: Origin, Function and Application. J. Exp. Clin. Cancer Res. 2012, 31, 38. [Google Scholar] [CrossRef]
- Nappi, F.; Avtaar Singh, S.S.; Jitendra, V.; Alzamil, A.; Schoell, T. The Roles of microRNAs in the Cardiovascular System. Int. J. Mol. Sci. 2023, 24, 14277. [Google Scholar] [CrossRef]
- Connell, P.S.; Han, R.I.; Grande-Allen, K.J. Differentiating the Aging of the Mitral Valve from Human and Canine Myxomatous Degeneration. J. Vet. Cardiol. 2012, 14, 31–45. [Google Scholar] [CrossRef]
- Whitney, J.C. Observations on the Effect of Age on the Severity of Heart Valve Lesions in the Dog. J. Small Anim. Pract. 1974, 15, 511–522. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Vlachos, I.S.; Zagganas, K.; Paraskevopoulou, M.D.; Georgakilas, G.; Karagkouni, D.; Vergoulis, T.; Dalamagas, T.; Hatzigeorgiou, A.G. DIANA-miRPath v3.0: Deciphering microRNA Function with Experimental Support. Nucleic Acids Res. 2015, 43, W460–W466. [Google Scholar] [CrossRef]
- Kozomara, A.; Griffiths-Jones, S. miRBase: Annotating High Confidence microRNAs Using Deep Sequencing Data. Nucleic Acids Res. 2014, 42, D68–D73. [Google Scholar] [CrossRef]
- Chew, D.S.; Leung, M.-Y.; Choi, K.P. AT Excursion: A New Approach to Predict Replication Origins in Viral Genomes by Locating AT-Rich Regions. BMC Bioinform. 2007, 8, 163. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple Sequence Alignment with High Accuracy and High Throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Guelfi, G.; Capaccia, C.; Santoro, M.M.; Diverio, S. Identification of Appropriate Endogenous Controls for Circulating miRNA Quantification in Working Dogs under Physiological Stress Conditions. Animals 2023, 13, 576. [Google Scholar] [CrossRef] [PubMed]
- Silver, N.; Best, S.; Jiang, J.; Thein, S.L. Selection of Housekeeping Genes for Gene Expression Studies in Human Reticulocytes Using Real-Time PCR. BMC Mol. Biol. 2006, 7, 33. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W.; Tichopad, A.; Prgomet, C.; Neuvians, T.P. Determination of Stable Housekeeping Genes, Differentially Regulated Target Genes and Sample Integrity: BestKeeper--Excel-Based Tool Using Pair-Wise Correlations. Biotechnol. Lett. 2004, 26, 509–515. [Google Scholar] [CrossRef]
- Andersen, C.L.; Jensen, J.L.; Ørntoft, T.F. Normalization of Real-Time Quantitative Reverse Transcription-PCR Data: A Model-Based Variance Estimation Approach to Identify Genes Suited for Normalization, Applied to Bladder and Colon Cancer Data Sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate Normalization of Real-Time Quantitative RT-PCR Data by Geometric Averaging of Multiple Internal Control Genes. Genome Biol. 2002, 3, research0034.1. [Google Scholar] [CrossRef]
- Xie, F.; Xiao, P.; Chen, D.; Xu, L.; Zhang, B. miRDeepFinder: A miRNA Analysis Tool for Deep Sequencing of Plant Small RNAs. Plant Mol. Biol. 2012, 80, 75–84. [Google Scholar] [CrossRef]
- Reis-Ferreira, A.; Neto-Mendes, J.; Brás-Silva, C.; Lobo, L.; Fontes-Sousa, A.P. Emerging Roles of Micrornas in Veterinary Cardiology. Vet. Sci. 2022, 9, 533. [Google Scholar] [CrossRef]
- Ghilardi, S.; Lecchi, C.; Bagardi, M.; Romito, G.; Colombo, F.M.; Polli, M.; Franco, C.; Brambilla, P.G. Prospective Pilot Study on the Predictive Significance of Plasma miR-30b-5p through the Study of Echocardiographic Modifications in Cavalier King Charles Spaniels Affected by Different Stages of Myxomatous Mitral Valve Disease: The PRIME Study. PLoS ONE 2022, 17, e0274724. [Google Scholar] [CrossRef] [PubMed]
- Yang, V.K.; Tai, A.K.; Huh, T.P.; Meola, D.M.; Juhr, C.M.; Robinson, N.A.; Hoffman, A.M. Dysregulation of Valvular Interstitial Cell Let-7c, miR-17, miR-20a, and miR-30d in Naturally Occurring Canine Myxomatous Mitral Valve Disease. PLoS ONE 2018, 13, e0188617. [Google Scholar] [CrossRef] [PubMed]
- Hulanicka, M.; Garncarz, M.; Parzeniecka-Jaworska, M.; Jank, M. Plasma miRNAs as Potential Biomarkers of Chronic Degenerative Valvular Disease in Dachshunds. BMC Vet. Res. 2014, 10, 205. [Google Scholar] [CrossRef] [PubMed]
- Varvil, M.S.; Dos Santos, A.P. A Review on microRNA Detection and Expression Studies in Dogs. Front. Vet. Sci. 2023, 10, 1261085. [Google Scholar] [CrossRef]

| CASE | SEX | BREED | AGE | MMDV Grade | Type (I–IV) |
|---|---|---|---|---|---|
| C-1 | M | Lagotto | 4–6 | Control | - |
| C-2 | F | NA | 4–6 | Control | - |
| C-3 | F | Mixed-breed | 5 | Control | - |
| C-4 | F | Pitbull | 4–6 | Control | - |
| C-5 | M | English Setter | 13 | Control | - |
| C-6 | F | Golden Retriever | 4–6 | Control | - |
| C-7 | M | Mixed-breed | 18 | Control | - |
| L-1 | M | Mixed-breed | 12 | Low | II |
| L-2 | M | NA | 12 | Low | I |
| L-3 | M | Hound | 8 | Low | I |
| L-4 | M | Bracco | 8 | Low | I |
| L-5 | F | Pekingese | 10 | Low | II |
| L-6 | M | Brittany | 4–6 | Low | II |
| L-7 | M | Bobtail | 10 | Low | II |
| L-8 | F | Golden Retriever | 11 | Low | I |
| H-1 | F | Pointing Dog | 5 | High | III |
| H-2 | M | NA | 8 | High | IV |
| H-3 | M | Mixed-breed | 15 | High | III |
| H-4 | F | NA | 4–6 | High | III |
| H-5 | F | NA | 4–6 | High | IV |
| Grade | Valvular Leaflet Lesions | Chordae Tendineae |
|---|---|---|
| I | Small nodules at the chordae-leaflet junction | Unaffected |
| II | Coalescing nodules in the leaflet edge. | Unaffected |
| III | Thickened and moderately distorted plaques. | Mild segmental thickening |
| IV | Larger plaques severely distorting leaflet profile. | Severe thickening, possible rupture |
| Primer | Recommended for | GeneGlobe ID |
|---|---|---|
| UniSp2 | Spike-in (to assess RNA isolation efficiency) | YP00203950 |
| UniSp4 | Spike-in (to assess RNA isolation efficiency) | YP00203953 |
| UniSp6 | Spike-in (to assess RT and PCR inhibitors) | YP00203954 |
| hsa-miR-29a-5p | Candidate EC microRNA | YP00204676 |
| hsa-miR-16-5p | Candidate EC microRNA | YP00205702 |
| hsa-miR-186-5p | Candidate EC microRNA | YP00206053 |
| hsa-let-7a-5p | Target microRNA | YP00205727 |
| hsa-let-7b-5p | Target microRNA | YP00205728 |
| hsa-let-7c-5p | Target microRNA | YP00205729 |
| hsa-miR-21-5p | Target microRNA | YP00204230 |
| hsa-miR-30b-5p | Target microRNA | YP00205949 |
| hsa-miR-133b | Target microRNA | YP00206011 |
| hsa-miR-98-5p | Target microRNA | YP00206127 |
| hsa-miR-103a-3p | Target microRNA | YP00206450 |
| Procedures | Better | Good | Average |
|---|---|---|---|
| Delta CT | miR-16 | miR-29a | miR-186-5p |
| BestKeeper | miR-29a | miR-16 | miR-186-5p |
| Normfinder | miR-29a | miR-16 | miR-186-5p |
| Geonorm | miR-29a | miR-16 | miR-186-5p |
| RefFinder Ranking | miR-29a | miR-16 | miR-186-5p |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guelfi, G.; Santarelli, N.; Capaccia, C.; Valeri, F.; Caivano, D.; Lepri, E. MicroRNA Expression Profiling in Canine Myxomatous Mitral Valve Disease Highlights Potential Diagnostic Tool and Molecular Pathways. Vet. Sci. 2025, 12, 1029. https://doi.org/10.3390/vetsci12111029
Guelfi G, Santarelli N, Capaccia C, Valeri F, Caivano D, Lepri E. MicroRNA Expression Profiling in Canine Myxomatous Mitral Valve Disease Highlights Potential Diagnostic Tool and Molecular Pathways. Veterinary Sciences. 2025; 12(11):1029. https://doi.org/10.3390/vetsci12111029
Chicago/Turabian StyleGuelfi, Gabriella, Noemi Santarelli, Camilla Capaccia, Federica Valeri, Domenico Caivano, and Elvio Lepri. 2025. "MicroRNA Expression Profiling in Canine Myxomatous Mitral Valve Disease Highlights Potential Diagnostic Tool and Molecular Pathways" Veterinary Sciences 12, no. 11: 1029. https://doi.org/10.3390/vetsci12111029
APA StyleGuelfi, G., Santarelli, N., Capaccia, C., Valeri, F., Caivano, D., & Lepri, E. (2025). MicroRNA Expression Profiling in Canine Myxomatous Mitral Valve Disease Highlights Potential Diagnostic Tool and Molecular Pathways. Veterinary Sciences, 12(11), 1029. https://doi.org/10.3390/vetsci12111029








