Presence of Methicillin-Resistant Staphylococci and Carbapenemase-Positive Acinetobacter Isolates on Surfaces in German Dog Daycare Facilities and Correlation with Cleaning Practices
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Participating Dog Daycare Facilities
2.2. Sample Collection
2.3. Isolation and Identification of Bacterial Isolates
2.4. Genotypic Investigations
2.5. Statistical Methods
3. Results
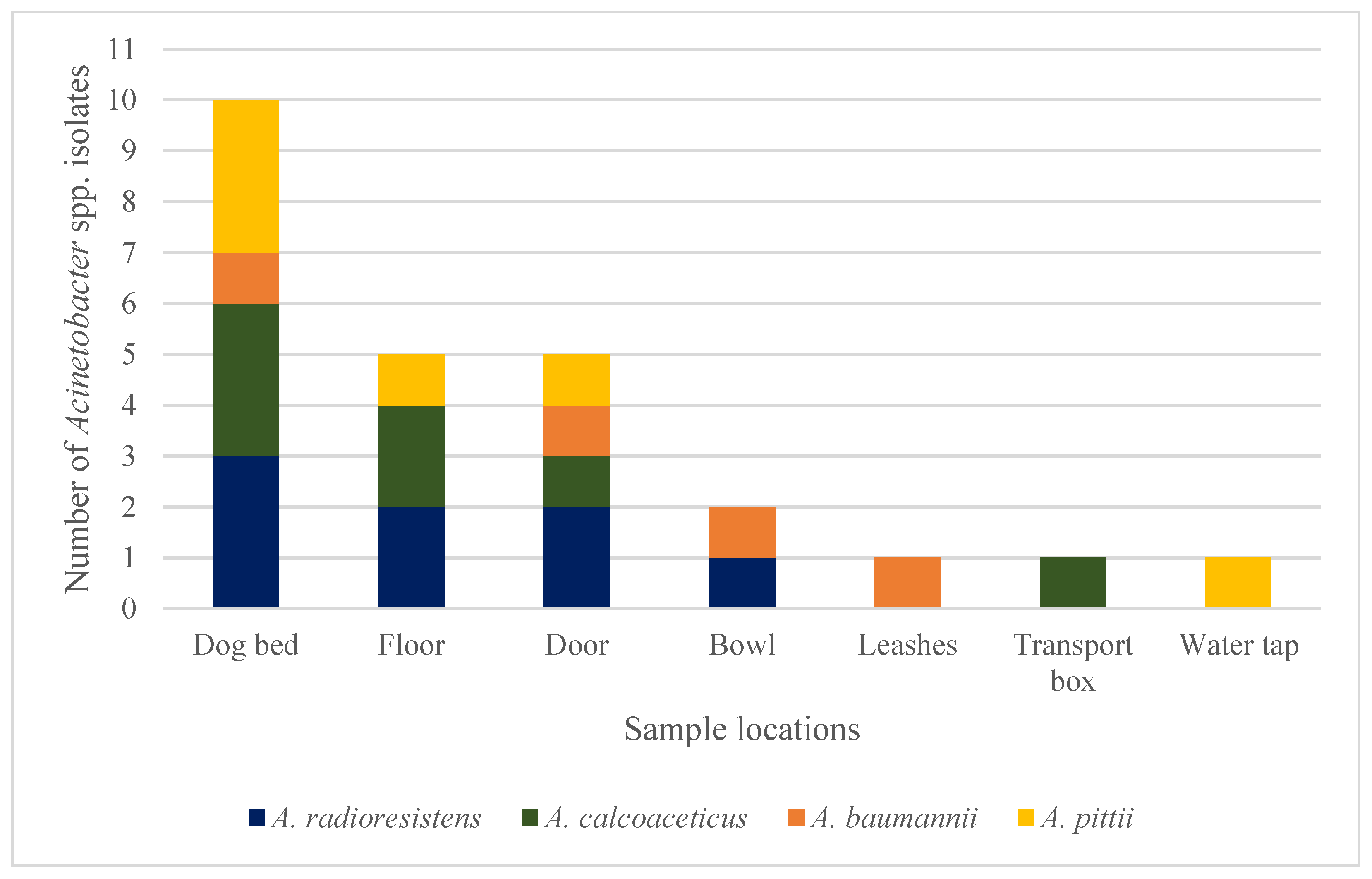
3.1. Detected Staphylococci and Acinetobacter spp. Isolates
3.2. Cleaning Protocols of Dog Daycare Facilities
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Velazquez-Meza, M.E.; Galarde-López, M.; Carrillo-Quiróz, B.; Alpuche-Aranda, C.M. Antimicrobial resistance: One Health approach. Vet. World 2022, 15, 743–749. [Google Scholar] [CrossRef] [PubMed]
- Magiorakos, A.-P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.; Giske, C.; Harbarth, S.; Hindler, J.; Kahlmeter, G.; Olsson-Liljequist, B. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef] [PubMed]
- Morris, D.O.; Loeffler, A.; Davis, M.F.; Guardabassi, L.; Weese, J.S. Recommendations for approaches to meticillin-resistant staphylococcal infections of small animals: Diagnosis, therapeutic considerations and preventative measures. Clinical Consensus Guidelines of the World Association for Veterinary Dermatology. Vet. Dermatol. 2017, 28, 304-e369. [Google Scholar] [CrossRef] [PubMed]
- Uehara, Y. Current status of staphylococcal cassette chromosome mec (SCC mec). Antibiotics 2022, 11, 86. [Google Scholar] [CrossRef]
- Frosini, S.M.; Bond, R.; King, R.; Feudi, C.; Schwarz, S.; Loeffler, A. Effect of topical antimicrobial therapy and household cleaning on meticillin-resistant Staphylococcus pseudintermedius carriage in dogs. Vet. Rec. 2022, 190, e937. [Google Scholar] [CrossRef]
- Windahl, U.; Reimegård, E.; Holst, B.S.; Egenvall, A.; Fernström, L.; Fredriksson, M.; Trowald-Wigh, G.; Andersson, U.G. Carriage of methicillin-resistant Staphylococcus pseudintermedius in dogs—A longitudinal study. BMC Vet. Res. 2012, 8, 34. [Google Scholar] [CrossRef]
- Frosini, S.M.; Bond, R.; King, R.H.; Loeffler, A. The nose is not enough: Multi-site sampling is best for MRSP detection in dogs and households. Vet. Dermatol. 2022, 33, 576–580. [Google Scholar] [CrossRef]
- Gould, A.P.; Coyner, K.S.; Trimmer, A.M.; Weese, J.S.; Budke, C.M. Recovery of meticillin-resistant Staphylococcus species from pet-grooming salons. Vet. Dermatol. 2020, 31, 262-e260. [Google Scholar] [CrossRef]
- Asokan, G.V.; Ramadhan, T.; Ahmed, E.; Sanad, H. WHO global priority pathogens list: A bibliometric analysis of Medline-PubMed for knowledge mobilization to infection prevention and control practices in Bahrain. Oman Med. J. 2019, 34, 184–193. [Google Scholar] [CrossRef]
- Shi, J.; Cheng, J.; Liu, S.; Zhu, Y.; Zhu, M. Acinetobacter baumannii: An evolving and cunning opponent. Front. Microbiol. 2024, 15, 1332108. [Google Scholar] [CrossRef]
- Leelapsawas, C.; Yindee, J.; Nittayasut, N.; Chueahiran, S.; Boonkham, P.; Suanpairintr, N.; Chanchaithong, P. Emergence and multi-lineages of carbapenemase-producing Acinetobacter baumannii-calcoaceticus complex from canine and feline origins. J. Vet. Med. Sci. 2022, 84, 1377–1384. [Google Scholar] [CrossRef] [PubMed]
- Kuzi, S.; Blum, S.; Kahane, N.; Adler, A.; Hussein, O.; Segev, G.; Aroch, I. Multidrug-resistant Acinetobacter calcoaceticus-Acinetobacter baumannii complex infection outbreak in dogs and cats in a veterinary hospital. J. Small Anim. Pract. 2016, 57, 617–625. [Google Scholar] [CrossRef] [PubMed]
- Müller, S.; Janssen, T.; Wieler, L.H. Multidrug resistant Acinetobacter baumannii in veterinary medicine–emergence of an underestimated pathogen. Berl. Muenchener Tieraerztliche Wochenschr. 2014, 127, 435–446. [Google Scholar] [CrossRef]
- Ewers, C.; Klotz, P.; Leidner, U.; Stamm, I.; Prenger-Berninghoff, E.; Göttig, S.; Semmler, T.; Scheufen, S. OXA-23 and ISAba1–OXA-66 class D β-lactamases in Acinetobacter baumannii isolates from companion animals. Int. J. Antimicrob. Agents 2017, 49, 37–44. [Google Scholar] [CrossRef] [PubMed]
- Jacobmeyer, L.; Semmler, T.; Stamm, I.; Ewers, C. Genomic Analysis of Acinetobacter baumannii Isolates Carrying OXA-23 and OXA-58 Genes from Animals Reveals ST1 and ST25 as Major Clonal Lineages. Antibiotics 2022, 11, 1045. [Google Scholar] [CrossRef]
- Lupo, A.; Valot, B.; Saras, E.; Drapeau, A.; Robert, M.; Bour, M.; Haenni, M.; Plésiat, P.; Madec, J.-Y.; Potron, A. Multiple host colonization and differential expansion of multidrug-resistant ST25-Acinetobacter baumannii clades. Sci. Rep. 2023, 13, 21854. [Google Scholar] [CrossRef]
- Ahuatzin-Flores, O.E.; Torres, E.; Chávez-Bravo, E. Acinetobacter baumannii, a Multidrug-Resistant Opportunistic Pathogen in New Habitats: A Systematic Review. Microorganisms 2024, 12, 644. [Google Scholar] [CrossRef]
- Genath, A.; Hackmann, C.; Denkel, L.; Weber, A.; Maechler, F.; Kola, A.; Schwarz, S.; Gastmeier, P.; Leistner, R. The genetic relationship between human and pet isolates: A core genome multilocus sequence analysis of multidrug-resistant bacteria. Antimicrob. Resist. Infect. Control. 2024, 13, 107. [Google Scholar] [CrossRef]
- Soedarmanto, I.; Kanbar, T.; Ülbegi-Mohyla, H.; Hijazin, M.; Alber, J.; Lämmler, C.; Akineden, Ö.; Weiss, R.; Moritz, A.; Zschöck, M. Genetic relatedness of methicillin-resistant Staphylococcus pseudintermedius (MRSP) isolated from a dog and the dog owner. Res. Vet. Sci. 2011, 91, e25–e27. [Google Scholar] [CrossRef]
- Nocera, F.P.; Pizzano, F.; Masullo, A.; Cortese, L.; De Martino, L. Antimicrobial resistant staphylococcus species colonization in dogs, their owners, and veterinary staff of the veterinary teaching hospital of Naples, Italy. Pathogens 2023, 12, 1016. [Google Scholar] [CrossRef]
- Da Silva, J.M.; Menezes, J.; Fernandes, L.; Marques, C.; Costa, S.S.; Timofte, D.; Amaral, A.; Pomba, C. Dynamics of blaOXA-23 gene transmission in Acinetobacter spp. from contaminated veterinary environmental surfaces: An emerging One Health threat? J. Hosp. Infect. 2024, 146, 116–124. [Google Scholar] [CrossRef] [PubMed]
- Lehner, G.; Linek, M.; Bond, R.; Lloyd, D.H.; Prenger-Berninghoff, E.; Thom, N.; Straube, I.; Verheyen, K.; Loeffler, A. Case–control risk factor study of methicillin-resistant Staphylococcus pseudintermedius (MRSP) infection in dogs and cats in Germany. Vet. Microbiol. 2014, 168, 154–160. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, T.; Tsubakishita, S.; Tanaka, Y.; Sakusabe, A.; Ohtsuka, M.; Hirotaki, S.; Kawakami, T.; Fukata, T.; Hiramatsu, K. Multiplex-PCR method for species identification of coagulase-positive staphylococci. J. Clin. Microbiol. 2010, 48, 765–769. [Google Scholar] [CrossRef] [PubMed]
- Cuny, C.; Layer, F.; Strommenger, B.; Witte, W. Rare occurrence of methicillin-resistant Staphylococcus aureus CC130 with a novel mecA homologue in humans in Germany. PLoS ONE 2011, 6, e24360. [Google Scholar] [CrossRef] [PubMed]
- Higgins, P.G.; Lehmann, M.; Seifert, H. Inclusion of OXA-143 primers in a multiplex polymerase chain reaction (PCR) for genes encoding prevalent OXA carbapenemases in Acinetobacter spp. Int. J. Antimicrob. Agents 2010, 35, 305–314. [Google Scholar] [CrossRef] [PubMed]
- Kern, A.; Perreten, V. Clinical and molecular features of methicillin-resistant, coagulase-negative staphylococci of pets and horses. J. Antimicrob. Chemother. 2013, 68, 1256–1266. [Google Scholar] [CrossRef]
- Moodley, A.; Guardabassi, L. Clonal spread of methicillin-resistant coagulase-negative staphylococci among horses, personnel and environmental sites at equine facilities. Vet. Microbiol. 2009, 137, 397–401. [Google Scholar] [CrossRef]
- Nemeghaire, S.; Argudín, M.A.; Fessler, A.T.; Hauschild, T.; Schwarz, S.; Butaye, P. The ecological importance of the Staphylococcus sciuri species group as a reservoir for resistance and virulence genes. Vet. Microbiol. 2014, 171, 342–356. [Google Scholar] [CrossRef]
- Mazal, C.; Sieger, B. Staphylococcus lentus: The troublemaker. Int. J. Infect. Dis. 2010, 14, e397. [Google Scholar] [CrossRef]
- Teixeira, I.M.; de Oliveira Ferreira, E.; de Araújo Penna, B. Dogs as reservoir of methicillin resistant coagulase negative staphylococci strains–A possible neglected risk. Microb. Pathog. 2019, 135, 103616. [Google Scholar] [CrossRef]
- Ehlers, S.; Merrill, S.A. Staphylococcus saprophyticus Infection. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2023. [Google Scholar] [PubMed]
- Ray, M.D.; Boundy, S.; Archer, G.L. Transfer of the methicillin resistance genomic island among staphylococci by conjugation. Mol. Microbiol. 2016, 100, 675–685. [Google Scholar] [CrossRef] [PubMed]
- Rossi, C.C.; Ahmad, F.; Giambiagi-deMarval, M. Staphylococcus haemolyticus: An updated review on nosocomial infections, antimicrobial resistance, virulence, genetic traits, and strategies for combating this emerging opportunistic pathogen. Microbiol. Res. 2024, 282, 127652. [Google Scholar] [CrossRef]
- Hackmann, C.; Genath, A.; Gruhl, D.; Weber, A.; Maechler, F.; Kola, A.; Schwab, F.; Schwarz, S.; Lübke-Becker, A.; Schneider, T. The transmission risk of multidrug-resistant organisms between hospital patients and their pets–a case−control study, Germany, 2019 to 2022. Eurosurveillance 2024, 29, 2300714. [Google Scholar] [CrossRef] [PubMed]
- Schwerkoetting, F. Explorative Untersuchungen zur Nachweishäufigkeit von Methicillin-Resistenten Staphylococcus Aureus (MRSA) bei Kleintieren in Einer Ländlich Gelegenen Kleintierpraxis und in Einem Tierheim; DVG: Hannover, Germany, 2011. [Google Scholar]
- Kaspar, U.; von Lützau, A.; Schlattmann, A.; Roesler, U.; Köck, R.; Becker, K. Zoonotic multidrug-resistant microorganisms among small companion animals in Germany. PLoS ONE 2018, 13, e0208364. [Google Scholar] [CrossRef] [PubMed]
- Poirel, L.; Figueiredo, S.; Cattoir, V.; Carattoli, A.; Nordmann, P. Acinetobacter radioresistens as a silent source of carbapenem resistance for Acinetobacter spp. Antimicrob. Agents Chemother. 2008, 52, 1252–1256. [Google Scholar] [CrossRef] [PubMed]
- Lazarev, A.; Hyun, J.; Sanchez, J.L.; Verda, L. Community-acquired Acinetobacter radioresistens bacteremia in an immunocompetent host. Cureus 2022, 14, e29650. [Google Scholar] [CrossRef]
- Lopes, M.C.; Évora, B.S.; Cidral, T.A.; Botelho, L.B.; Melo, M.C.N. Bloodstream infection by Acinetobacter radioresistens: The first case report in Brazil. J. Bras. Patol. Med. Lab. 2020, 55, 669–674. [Google Scholar] [CrossRef]
- Lane, H.E.; Weese, J.S.; Stull, J.W. Canine oral papillomavirus outbreak at a dog daycare facility. Can. Vet. J. 2017, 58, 747–749. [Google Scholar]
- Iverson, S.A.; Levy, C.; Yaglom, H.D.; Venkat, H.L.; Artus, A.; Galloway, R.; Guagliardo, S.A.J.; Reynolds, L.; Kretschmer, M.J.; Jenni, M.E.L. Clinical, diagnostic, and epidemiological features of a community-wide outbreak of canine leptospirosis in a low-prevalence region (Maricopa County, Arizona). J. Am. Vet. Med. Assoc. 2021, 258, 616–629. [Google Scholar] [CrossRef]
- Traverse, M.; Aceto, H. Environmental cleaning and disinfection. Vet. Clin. Small Anim. 2015, 45, 299–330. [Google Scholar] [CrossRef]
- Fraise, A.P.; Wilkinson, M.; Bradley, C.; Oppenheim, B.; Moiemen, N. The antibacterial activity and stability of acetic acid. J. Hosp. Infect. 2013, 84, 329–331. [Google Scholar] [CrossRef] [PubMed]
- Aboualizadeh, E.; Bumah, V.V.; Masson-Meyers, D.S.; Eells, J.T.; Hirschmugl, C.J.; Enwemeka, C.S. Understanding the antimicrobial activity of selected disinfectants against methicillin-resistant Staphylococcus aureus (MRSA). PLoS ONE 2017, 12, e0186375. [Google Scholar] [CrossRef] [PubMed]
- Valizadeh, M.; Beigomi, M.; Fazeli-Nasab, B. Antibacterial and Anti biofilm effects of ethanol and aceton leaf extract of Momordica charantia and Tecomella undulata against Acinetobacter baumannii. Int. J. Adv. Biol. Biomed. Res. 2020, 8, 403–418. [Google Scholar]
- Nwugo, C.C.; Arivett, B.A.; Zimbler, D.L.; Gaddy, J.A.; Richards, A.M.; Actis, L.A. Effect of Ethanol on Differential Protein Production and Expression of Potential Virulence Functions in the Opportunistic Pathogen Acinetobacter baumannii. PLoS ONE 2012, 7, e51936. [Google Scholar] [CrossRef]
- Gerba, C.P. Quaternary ammonium biocides: Efficacy in application. Appl. Environ. Microbiol. 2015, 81, 464–469. [Google Scholar] [CrossRef] [PubMed]
- Varga, Z.; Nicol, E.; Bouchonnet, S. Photodegradation of benzisothiazolinone: Identification and biological activity of degradation products. Chemosphere 2020, 240, 124862. [Google Scholar] [CrossRef]
- Zhulenkovs, D.; Rudevica, Z.; Jaudzems, K.; Turks, M.; Leonchiks, A. Discovery and structure–activity relationship studies of irreversible benzisothiazolinone-based inhibitors against Staphylococcus aureus sortase A transpeptidase. Bioorg. Med. Chem. 2014, 22, 5988–6003. [Google Scholar] [CrossRef]
- Noel, D. The Antibacterial Efficacy of Disinfectants Used for Infection Control; University of Southampton: Southampton, UK, 2023; p. 273. [Google Scholar]
- Stanojević-Nikolić, S.; Dimić, G.; Mojović, L.; Pejin, J.; Djukić-Vuković, A.; Kocić-Tanackov, S. Antimicrobial Activity of Lactic Acid Against Pathogen and Spoilage Microorganisms. J. Food Process. Preserv. 2016, 40, 990–998. [Google Scholar] [CrossRef]
- Puschmann, J.; Herbig, M.E.; Müller-Goymann, C.C. Correlation of antimicrobial effects of phenoxyethanol with its free concentration in the water phase of o/w-emulsion gels. Eur. J. Pharm. Biopharm. 2018, 131, 152–161. [Google Scholar] [CrossRef]
- Falk, N.A. Surfactants as Antimicrobials: A Brief Overview of Microbial Interfacial Chemistry and Surfactant Antimicrobial Activity. J. Surfactants Deterg. 2019, 22, 1119–1127. [Google Scholar] [CrossRef]
- Xie, J.; Wang, L.; Zhang, X.; Li, Y.; Liao, X.; Yang, C.; Tang, R.-Y. Discovery of New Anti-MRSA Agents Based on Phenoxyethanol and Its Mechanism. ACS Infect. Dis. 2022, 8, 2291–2306. [Google Scholar] [CrossRef] [PubMed]
- Smart, K.; Pieper, J.B.; Viall, A.K.; Noxon, J.O.; Berger, D.J. Comparison of commercial next-generation sequencing assays to conventional culture methods for bacterial identification and antimicrobial susceptibility of samples obtained from clinical cases of canine superficial bacterial folliculitis. Vet. Dermatol. 2024, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Su, M.; Satola, S.W.; Read, T.D. Genome-based prediction of bacterial antibiotic resistance. J. Clin. Microbiol. 2019, 57, e01405-18. [Google Scholar] [CrossRef] [PubMed]
- Van Camp, P.-J.; Haslam, D.B.; Porollo, A. Prediction of antimicrobial resistance in gram-negative bacteria from whole-genome sequencing data. Front. Microbiol. 2020, 11, 1013. [Google Scholar] [CrossRef]
- Bortolaia, V.; Kaas, R.S.; Ruppe, E.; Roberts, M.C.; Schwarz, S.; Cattoir, V.; Philippon, A.; Allesoe, R.L.; Rebelo, A.R.; Florensa, A.F. ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 2020, 75, 3491–3500. [Google Scholar] [CrossRef]


| Product Name | Active Ingredient | Frequency of DDFs with Bacteria Isolated | |
|---|---|---|---|
| MRS | Carbapenemase-Positive Acinetobacter spp. | ||
| Commercial universal cleaning agent | Surfactants | 2 | 0 |
| Sagrotan (B Hygiene Home Deutschland GmbH, Heidelberg, Germany) Prowin (proWIN Winter GmbH, Illingen, Germany) | Surfactants and Lactic acid | 1 | 1 |
| Kärcher (Alfred Kärcher Vertriebs-GmbH, Winnenden, Germany) | Phenoxyethanol and benzisothiazolinone | 1 | 1 |
| Vinegar cleaner | Acetic acid | 0 | 0 |
| Biguanid Surface N (Dr. Schumacher GmbH, Malsfeld, Germany) | Alkyl-dimethyl-benzyl-ammonium-chloride | 0 | 0 |
| Biodor (Biodor GmbH, Bocholt, Germany) | PermaZym® | 0 | 0 |
| Dr. Becher Rapid disinfection (Dr. Becher GmbH, Seelze, Germany) | Ethanol and Ethyl-alcohol | 0 | 0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Forbes, S.; Prenger-Berninghoff, E.; Ewers, C.; Doelle, M.; Roethig, A. Presence of Methicillin-Resistant Staphylococci and Carbapenemase-Positive Acinetobacter Isolates on Surfaces in German Dog Daycare Facilities and Correlation with Cleaning Practices. Vet. Sci. 2024, 11, 568. https://doi.org/10.3390/vetsci11110568
Forbes S, Prenger-Berninghoff E, Ewers C, Doelle M, Roethig A. Presence of Methicillin-Resistant Staphylococci and Carbapenemase-Positive Acinetobacter Isolates on Surfaces in German Dog Daycare Facilities and Correlation with Cleaning Practices. Veterinary Sciences. 2024; 11(11):568. https://doi.org/10.3390/vetsci11110568
Chicago/Turabian StyleForbes, Stephanie, Ellen Prenger-Berninghoff, Christa Ewers, Maren Doelle, and Anja Roethig. 2024. "Presence of Methicillin-Resistant Staphylococci and Carbapenemase-Positive Acinetobacter Isolates on Surfaces in German Dog Daycare Facilities and Correlation with Cleaning Practices" Veterinary Sciences 11, no. 11: 568. https://doi.org/10.3390/vetsci11110568
APA StyleForbes, S., Prenger-Berninghoff, E., Ewers, C., Doelle, M., & Roethig, A. (2024). Presence of Methicillin-Resistant Staphylococci and Carbapenemase-Positive Acinetobacter Isolates on Surfaces in German Dog Daycare Facilities and Correlation with Cleaning Practices. Veterinary Sciences, 11(11), 568. https://doi.org/10.3390/vetsci11110568






