Polymorphisms of the IL-17A Gene Influence Milk Production Traits and Somatic Cell Score in Chinese Holstein Cows
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animal Housing
2.2. Test Sample
2.3. Data and Sample Collection
2.4. Primer Design
2.5. DNA Extraction and SNP Genotyping
2.6. Amplification, Sequencing, and Genotyping of a Single DNA Sample
2.7. Statistical Analyses
3. Results
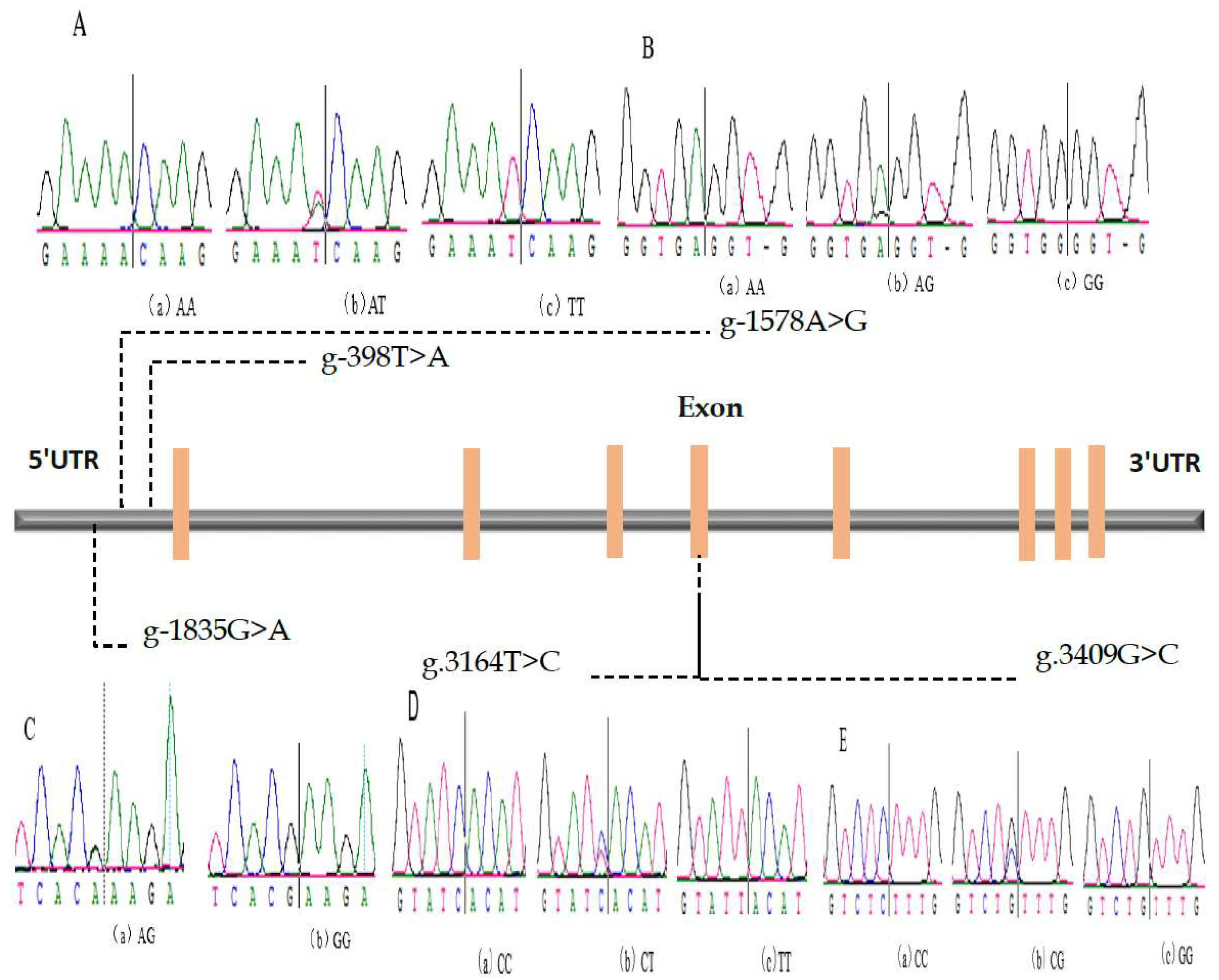
3.1. SNP of Bovine IL-17A Gene (Genotype Frequency, Allele Frequency, and Hardy–Weinberg’s Law)
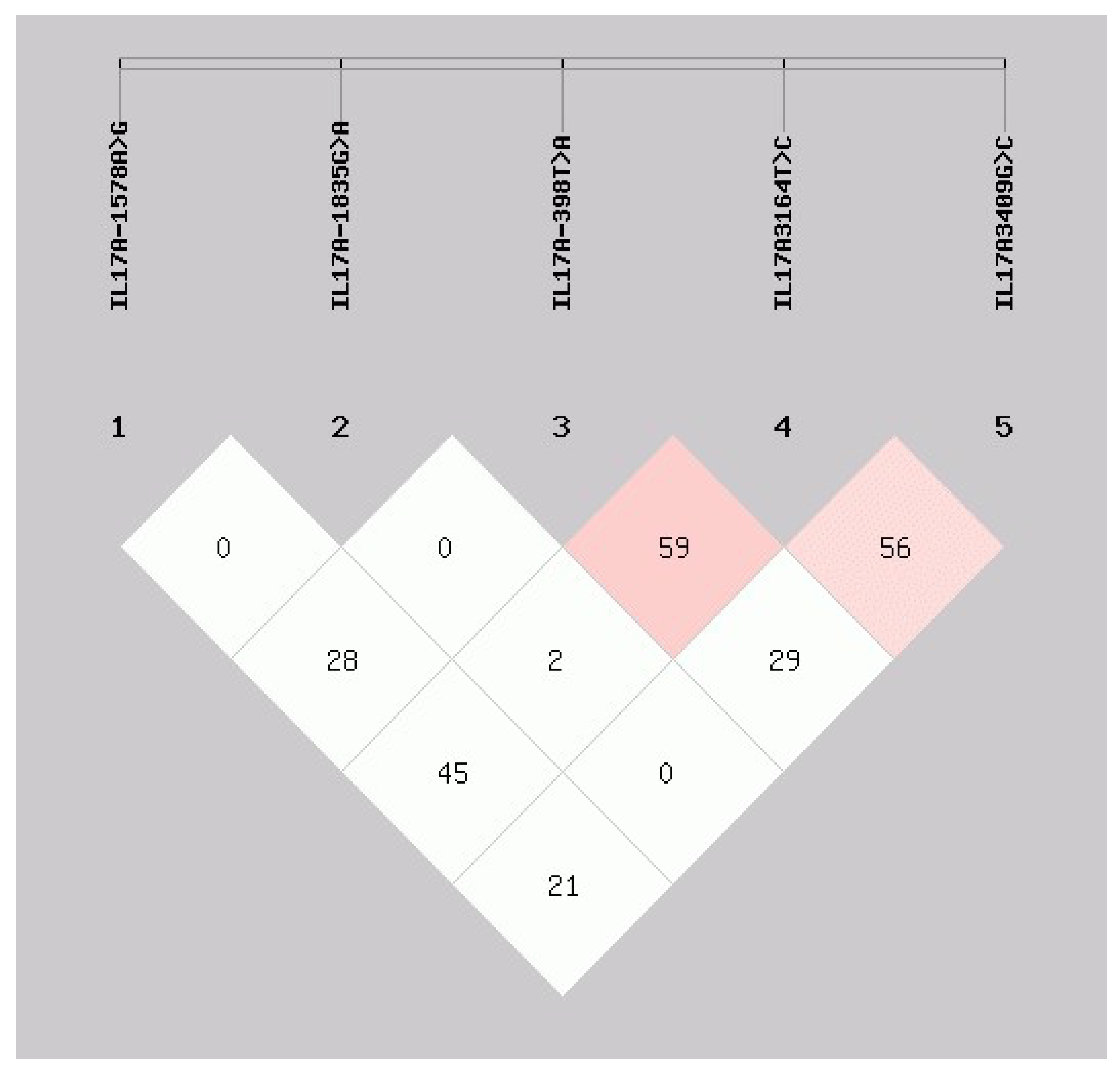
3.2. Haplotype Analysis of Single-SNPS of IL-17A Gene of Chinese Holstein Cows
3.3. Associations’ of SNPs in IL17A Gene with Milking Traits and Somatic Cell Score
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Du, X.X.; Sherein, S.A.; Liu, P.; Haque, M.A.; Khan, A. Bovine Mastitis: Behavioral Changes, Treatment and Control. Cont. Vet J. 2022, 2, 15–23. [Google Scholar]
- Sordillo, L.M.; Streicher, K.L. Mammary Gland Immunity and Mastitis Susceptibility. J. Mammary Gland Biol. Neoplasia 2002, 7, 135–146. [Google Scholar] [CrossRef] [PubMed]
- Mimoune, N.; Saidi, R.; Benadjel, O.; Khelef, D.; Kaidi, R. Alternative Treatment of Bovine Mastitis. Vet. stanica 2021, 52, 639–649. [Google Scholar] [CrossRef]
- Lamari, I.; Mimoune, N.; Khelef, D. Effect of Feed Additive Supplementation on Bovine Subclinical Mastitis. Vet. stanica 2021, 52, 445–460. [Google Scholar] [CrossRef]
- Sulabh, S.; Panigrahi, M.; Varshney, R.; Gupta, J.P.; Kumar, S.; Verma, A.; Kumar, A.; Asaf, V.N.M.; Kumar, P.; Bhushan, B. In-Vitro Analysis of Interleukin-10 Expression in Cell Cultures of Crossbred Cattle, Tharparkar Cattle and Murrah Buffalo in Response to Mastitis Causing Antigens Derived from Staphylococcus Aureus and Escherichia Coli. Biol. Rhythm. Res. 2022, 53, 197–206. [Google Scholar] [CrossRef]
- Gussmann, M.; Steeneveld, W.; Kirkeby, C.; Hogeveen, H.; Farre, M.; Halasa, T. Economic and Epidemiological Impact of Different Intervention Strategies for Subclinical and Clinical Mastitis. Prev. Vet. Med. 2019, 166, 78–85. [Google Scholar] [CrossRef]
- Cvetnić, L.; Samardžija, M.; Duvnjak, S.; Habrun, B.; Cvetnić, M.; Tkalec, V.J.; Đuričić, D.; Benić, M. Multi Locus Sequence Typing and Spa Typing of Staphylococcus Aureus Isolated from the Milk of Cows with Subclinical Mastitis in Croatia. Microorganisms 2021, 9, 725. [Google Scholar] [CrossRef]
- Saidi, R.; Khelef, D.; Solmaz, H.; Ergun, Y.; Mimoune, N.; Cantekin, Z.; Saidi, R. Investigation of the Presence of Slime Production, VanA Gene and Antiseptic Resistance Genes in Staphylococci Isolated from Bovine Mastitis in Algeria. Vet. stanica 2020, 52, 57–63. [Google Scholar] [CrossRef]
- Gernand, E.; König, S. Random Regression Test-Day Model for Clinical Mastitis: Genetic Parameters, Model Comparison, and Correlations with Indicator Traits. J. Dairy Sci. 2014, 97, 3953–3963. [Google Scholar] [CrossRef]
- Oliveira, H.R.; Cant, J.P.; Brito, L.F.; Feitosa, F.L.B.; Chud, T.C.S.; Fonseca, P.A.S.; Jamrozik, J.; Silva, F.F.; Lourenco, D.A.L.; Schenkel, F.S. Genome-Wide Association for Milk Production Traits and Somatic Cell Score in Different Lactation Stages of Ayrshire, Holstein, and Jersey Dairy Cattle. J. Dairy Sci. 2019, 102, 8159–8174. [Google Scholar] [CrossRef]
- Bobbo, T.; Penasa, M.; Cassandro, M. Genetic Parameters of Bovine Milk Fatty Acid Profile, Yield, Composition, Total and Differential Somatic Cell Count. Animals 2020, 10, 2406. [Google Scholar] [CrossRef]
- Jattawa, D.; Koonawootrittriron, S.; Elzo, M.A.; Suwanasopee, T. Somatic Cells Count and Its Genetic Association with Milk Yield in Dairy Cattle Raised under Thai Tropical Environmental Conditions. Asian-Australasian. J. Anim. Sci. 2012, 25, 1216–1222. [Google Scholar] [CrossRef] [PubMed]
- Akers, R.M. A 100-Year Review: Mammary Development and Lactation. J. Dairy Sci. 2017, 100, 10332–10352. [Google Scholar] [CrossRef]
- Brookes, A.J. Single Nucleotide Polymorphism (SNP). In Encyclopedia of Life Sciences; John Wiley & Sons, Ltd.: Chichester, UK, 2007. [Google Scholar]
- Clancey, E.; Kiser, J.N.; Moraes, J.G.N.; Dalton, J.C.; Spencer, T.E.; Neibergs, H.L. Genome-wide Association Analysis and Gene Set Enrichment Analysis with SNP Data Identify Genes Associated with 305-day Milk Yield in Holstein Dairy Cows. Anim. Genet. 2019, 50, 254–258. [Google Scholar] [CrossRef] [PubMed]
- Mohsin, M.; Swar, S.O.; Imran, M.; Ali, W.; Sultan, M.D.; Asrar, R.; Abbas, R.Z.; Aleem, M.T.; Aguilar-Marcelino, L.; Aslam, A.; et al. Chronic mastitis: Leading cause of udder fibrosis and different means of its management. Agrobiol. Records. 2022, 8, 13–21. [Google Scholar] [CrossRef]
- Stocco, G.; Summer, A.; Cipolat-Gotet, C.; Zanini, L.; Vairani, D.; Dadousis, C.; Zecconi, A. Differential somatic cell count as a novel indicator of milk quality in dairy cows. Animals 2020, 10, 753. [Google Scholar] [CrossRef]
- Yuan, Z.; Li, J.; Li, J.; Zhang, L.; Gao, X.; Gao, H.J.; Xu, S. Investigation on BRCA1 SNPs and Its Effects on Mastitis in Chinese Commercial Cattle. Gene 2012, 505, 190–194. [Google Scholar] [CrossRef]
- Chu, M.X.; Ye, S.C.; Qiao, L.; Wang, J.X.; Feng, T.; Huang, D.W.; Cao, G.L.; Di, R.; Fang, L.; Chen, G.H. Polymorphism of Exon 2 of BoLA-DRB3 Gene and Its Relationship with Somatic Cell Score in Beijing Holstein Cows. Mol. Biol. Rep. 2012, 39, 2909–2914. [Google Scholar] [CrossRef]
- Kerami, Z.; Duijvis, N.W.; Vogels, E.W.; van Dooren, F.H.; Moerland, P.D.; te Velde, A.A. Effect of Interleukin-17 on Gene Expression Profile of Fibroblasts from Crohn’s Disease Patients. J. Crohn’s Colitis 2014, 8, 1208–1216. [Google Scholar] [CrossRef]
- Fujino, S.; Andoh, A.; Bamba, S.; Ogawa, A.; Hata, K.; Araki, Y.; Bamba, T.; Fujiyama, Y. Increased Expression of Interleukin 17 in Inflammatory Bowel Disease. Gut 2003, 52, 65–70. [Google Scholar] [CrossRef]
- Jin, E.-H.; Choi, E.-Y.; Yang, J.Y.; Chung, H.-T.; Yang, Y.-S. Significant Association between IL-17F Promoter Region Polymorphism and Susceptibility to Asthma in a Korean Population. Int. Arch. Allergy Immunol. 2011, 155, 106–110. [Google Scholar] [CrossRef] [PubMed]
- Nisar, H.; Pasha, U.; Mirza, M.U.; Abid, R.; Hanif, K.; Kadarmideen, H.N.; Sadaf, S. Impact of IL-17F 7488T/C Functional Polymorphism on Progressive Rheumatoid Arthritis: Novel Insight from the Molecular Dynamic Simulations. Immunol. Invest. 2021, 50, 416–426. [Google Scholar] [CrossRef]
- Guo, N.; Zhang, J. Interleukin-17 Promotes Ovarian Carcinoma SKOV3 Cells via MTA1-Induced Epithelial-to-Mesenchymal Transition. Eur. J. Gynaecol. Oncol. 2020, 41, 70. [Google Scholar] [CrossRef]
- Tong, Z.; Yang, X.O.; Yan, H.; Liu, W.; Niu, X.; Shi, Y.; Fang, W.; Xiong, B.; Wan, Y.; Dong, C. A Protective Role by Interleukin-17F in Colon Tumorigenesis. PLoS One 2012, 7, e34959. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Jiang, Y.; Zhang, Y.; Wang, Y.; Huang, S.; Wang, Z.; Tian, B.; Yang, Y.; Jiang, W.; Pang, D. Association Analysis of IL-17A and IL-17F Polymorphisms in Chinese Han Women with Breast Cancer. PLoS One 2012, 7, e34400. [Google Scholar] [CrossRef]
- Yao, Z.; Painter, S.L.; Fanslow, W.C.; Ulrich, D.; Macduff, B.M.; Spriggs, M.K.; Armitage, R.J. Human IL-17: A Novel Cytokine Derived from T Cells. J. Immunol. 1995, 155, 5483–5486. [Google Scholar]
- Hayashi, R.; Tahara, T.; Shiroeda, H.; Matsue, Y.; Minato, T.; Nomura, T.; Yamada, H.; Saito, T.; Matsunaga, K.; Fukuyama, T.; et al. Association of Genetic Polymorphisms in IL17A and IL17F with Gastro-Duodenal Diseases. J. Gastrointestin. Liver Dis. 2012, 21, 243–249. [Google Scholar]
- Yu, H.; Sun, S.; Liu, F.; Xu, Q.-H. Meta-Analysis of Associations between Interleukin-17 Gene Polymorphisms and Risk of Gastric Cancer. Asian Pac. J. Cancer Prev. 2014, 15, 8709–8713. [Google Scholar] [CrossRef]
- Schnyder-Candrian, S.; Togbe, D.; Couillin, I.; Mercier, I.; Brombacher, F.; Quesniaux, V.; Fossiez, F.; Ryffel, B.; Schnyder, B. Interleukin-17 Is a Negative Regulator of Established Allergic Asthma. J. Exp. Med. 2006, 203, 2715–2725. [Google Scholar] [CrossRef]
- Usman, T.; Wang, Y.; Liu, C.; He, Y.; Wang, X.; Dong, Y.; Wu, H.; Liu, A.; Yu, Y. Novel SNPs in IL-17F and IL-17A Genes Associated with Somatic Cell Count in Chinese Holstein and Inner-Mongolia Sanhe Cattle. J. Anim. Sci. Biotechnol. 2017, 8, 5. [Google Scholar] [CrossRef]
- Zhou, F.; Liang, Y.; Arbab, A.A.I.; Li, M.; Yang, Z.; Karrow, N.A.; Mao, Y. Analysis of Non-Genetic Factors Affecting Wood’s Model of Daily Milk Fat Percentage of Holstein Cattle. Vet. Sci. 2022, 9, 188. [Google Scholar] [CrossRef]
- Liang, Y.; Gao, Q.; Zhang, Q.; Arbab, A.A.I.; Li, M.; Yang, Z.; Karrow, N.A.; Mao, Y. Polymorphisms of the ACSL1 Gene Influence Milk Production Traits and Somatic Cell Score in Chinese Holstein Cows. Animals 2020, 10, 2282. [Google Scholar] [CrossRef]
- Rai, P.; Arya, H. Molecular Cloning. In The Design and Development of Novel Drugs and Vaccines; Elsevier Inc.: Amsterdam, The Netherlands, 2021; pp. 135–163. ISBN 9780128214718. [Google Scholar]
- Hill, W.G.; Robertson, A. Linkage Disequilibrium in Finite Populations. Theor. Appl. Genet. 1968, 38, 226–231. [Google Scholar] [CrossRef]
- Shi, Y.Y.; He, L. SHEsis, a Powerful Software Platform for Analyses of Linkage Disequilibrium, Haplotype Construction, and Genetic Association at Polymorphism Loci. Cell Res. 2005, 15, 97–98. [Google Scholar] [CrossRef]
- Browning, B.L.; Zhou, Y.; Browning, S.R. A One-Penny Imputed Genome from Next-Generation Reference Panels. Am. J. Hum. Genet. 2018, 103, 338–348. [Google Scholar] [CrossRef]
- Mao, Y.; Zhu, X.; Xing, S.; Zhang, M.; Zhang, H.; Wang, X.; Karrow, N.; Yang, L.; Yang, Z. Polymorphisms in the Promoter Region of the Bovine Lactoferrin Gene Influence Milk Somatic Cell Score and Milk Production Traits in Chinese Holstein Cows. Res. Vet. Sci. 2015, 103, 107–112. [Google Scholar] [CrossRef]
- Romero, J.; Benavides, E.; Meza, C. Assessing Financial Impacts of Subclinical Mastitis on Colombian Dairy Farms. Front. Vet. Sci. 2018. [Google Scholar] [CrossRef]
- Rutitzky, L.I.; Lopes da Rosa, J.R.; Stadecker, M.J. Severe CD4 T Cell-Mediated Immunopathology in Murine Schistosomiasis Is Dependent on IL-12p40 and Correlates with High Levels of IL-17. J. Immunol. 2005, 175, 3920–3926. [Google Scholar] [CrossRef]
- Straus, D.S. TNFα and IL-17 Cooperatively Stimulate Glucose Metabolism and Growth Factor Production in Human Colorectal Cancer Cells. Mol. Cancer 2013, 12, 78. [Google Scholar] [CrossRef]
- Saraiva, A.M.; Alves e Silva, M.R.M.; de Fátima Correia Silva, J.; da Costa, J.E.; Gollob, K.J.; Dutra, W.O.; Moreira, P.R. Evaluation of IL17A Expression and of IL17A, IL17F and IL23R Gene Polymorphisms in Brazilian Individuals with Periodontitis. Hum. Immunol. 2013, 74, 207–214. [Google Scholar] [CrossRef]
- Tao, W.; Mallard, B. Differentially Expressed Genes Associated with Staphylococcus Aureus Mastitis of Canadian Holstein Cows. Vet. Immunol. Immunopathol. 2007, 120, 201–211. [Google Scholar] [CrossRef]
- Gilbert, F.B.; Cunha, P.; Jensen, K.; Glass, E.J.; Foucras, G.; Robert-Granié, C.; Rupp, R.; Rainard, P. Differential Response of Bovine Mammary Epithelial Cells to Staphylococcus Aureus or Escherichia Coli Agonists of the Innate Immune System. Vet. Res. 2013, 44, 40. [Google Scholar] [CrossRef]
- Bougarn, S.; Cunha, P.; Gilbert, F.B.; Harmache, A.; Foucras, G.; Rainard, P. Staphylococcal-Associated Molecular Patterns Enhance Expression of Immune Defense Genes Induced by IL-17 in Mammary Epithelial Cells. Cytokine 2011, 56, 749–759. [Google Scholar] [CrossRef]
- Tassi, R.; McNeilly, T.N.; Fitzpatrick, J.L.; Fontaine, M.C.; Reddick, D.; Ramage, C.; Lutton, M.; Schukken, Y.H.; Zadoks, R.N. Strain-Specific Pathogenicity of Putative Host-Adapted and Nonadapted Strains of Streptococcus Uberis in Dairy Cattle. J. Dairy Sci. 2013, 96, 5129–5145. [Google Scholar] [CrossRef]
- Roussel, P.; Cunha, P.; Porcherie, A.; Petzl, W.; Gilbert, F.B.; Riollet, C.; Zerbe, H.; Rainard, P.; Germon, P. Investigating the Contribution of IL-17A and IL-17F to the Host Response during Escherichia Coli Mastitis. Vet. Res. 2015, 46, 56. [Google Scholar] [CrossRef]
- Blackburn, P.S. The Variation in the Cell Count of Cow’s Milk throughout Lactation and from One Lactation to the Next. J. Dairy Res. 1966, 33, 193–198. [Google Scholar] [CrossRef]
- Porcherie, A.; Gilbert, F.B.; Germon, P.; Cunha, P.; Trotereau, A.; Rossignol, C.; Winter, N.; Berthon, P.; Rainard, P. IL-17A Is an Important Effector of the Immune Response of the Mammary Gland to Escherichia Coli Infection. J. Immunol. 2016, 196, 803–812. [Google Scholar] [CrossRef]
- Gaffen, S.L.; Moutsopoulos, N.M. Regulation of Host-Microbe Interactions at Oral Mucosal Barriers by Type 17 Immunity. Sci. Immunol. 2020, 5. [Google Scholar] [CrossRef]
| Item | Percentage |
|---|---|
| Ingredient, % of DM | |
| Alfalfa hay | 25.31 |
| Corn silage | 28.50 |
| Oat hay | 6.16 |
| Ground corn | 17.48 |
| Soybean meal | 5.26 |
| Cottonseed meal | 4.06 |
| Distillers dried grains with solubles | 5.30 |
| Barely | 5.18 |
| Limestone | 0.32 |
| NaHCO3 | 0.36 |
| NaCl | 0.31 |
| CaHPO4 | 0.56 |
| Premix | 1.20 |
| Composition, % of DM | |
| Crude protein | 15.02 |
| Ether extract | 3.96 |
| Neutral detergent fiber | 41.11 |
| Acid detergent fiber | 22.04 |
| Calcium | 0.80 |
| Phosphorus | 0.44 |
| NEL2 Mcal/kg | 6.29 |
| Primer. | (5′→3′) Primer of Sequences | Size of Production (bp) | Position of Production | Annealing Temperature (°C) |
|---|---|---|---|---|
| P1 | F: GGAGTGTGGTGGAGGGTAAAA | 778 | −2060~−1282 | 57 |
| R: CCTATTCCCAAACCTACTGCCA | ||||
| P2 | F: AGTTGAATCACTTTGCTTTACAGT | 845 | −1413~−568 | 55 |
| R: ACATCTACTCTGCCTGAGGAAC | ||||
| P3 | F: TCACCACCTTTCTGCAGTCTC | 775 | −748~27 | 57.5 |
| R: TGAACTTGTGCTCGCTGTGA | ||||
| P4 | F: GGGGCGGTTTTTCTTTGACC | 457 | −143~314 | 58 |
| R: TGTGTGGTTTAGCCCCAGTC | ||||
| P5 | F: GCCATGGTCCTAATGTCACT | 505 | 1063–1568 | 56 |
| R: TGGCTCTTCCAGGTTTGACA | ||||
| P6 | F: AGGAATTCACTTTCTTCCTGGCTT | 759 | 2747–3506 | 57 |
| R: TGCTGTCTCTCTTGTAATGCCT |
| SNP Locus | Genotype | Genotype Frequency | Sample Number | Allele | Allele Frequency | χ2 Value for the H-W Test | Pearson’s p Test |
|---|---|---|---|---|---|---|---|
| IL17A (-398) | AA | 0.247 | 239 | A | 0.514 | 4.478 | 0.034 |
| AT | 0.534 | 516 | T | 0.486 | |||
| TT | 0.219 | 212 | |||||
| IL17A (-1578) | AA | 0.109 | 89 | A | 0.374 | 13.742 | 0.000 |
| AG | 0.529 | 431 | G | 0.626 | |||
| GG | 0.362 | 295 | |||||
| IL17A (-1835) | AA | 0.026 | 21 | A | 0.126 | 6.778 | 0.009 |
| AG | 0.200 | 162 | G | 0.874 | |||
| GG | 0.774 | 627 | |||||
| IL17A (3164) | CC | 0.398 | 364 | C | 0.637 | 1.139 | 0.286 |
| CT | 0.479 | 438 | T | 0.363 | |||
| TT | 0.123 | 113 | |||||
| IL17A (3409) | CC | 0.383 | 363 | C | 0.488 | 316.527 | 0.000 |
| CG | 0.211 | 200 | G | 0.512 | |||
| GG | 0.406 | 385 |
| Haplotype | IL17A -1578A>G | IL17A -1835G>A | IL17A -398T>A | IL17A 3164T>C | IL17A 3409G>C | Sample | Estimated Frequency |
|---|---|---|---|---|---|---|---|
| 1 | G | G | A | C | C | 384 | 0.303 |
| 2 | A | G | T | T | G | 356 | 0.281 |
| 3 | G | G | A | C | G | 111 | 0.087 |
| 4 | G | G | T | T | G | 80 | 0.063 |
| 5 | G | G | T | C | C | 78 | 0.062 |
| 6 | A | G | A | C | C | 61 | 0.048 |
| 7 | G | A | A | C | C | 48 | 0.038 |
| 8 | A | A | T | T | G | 24 | 0.019 |
| 9 | G | A | T | C | C | 24 | 0.019 |
| 10 | A | G | T | C | C | 18 | 0.015 |
| 11 | G | A | A | C | G | 16 | 0.013 |
| 12 | G | A | T | C | G | 16 | 0.012 |
| 13 | A | A | A | C | C | 16 | 0.012 |
| 14 | G | G | T | C | G | 14 | 0.011 |
| 15 | A | G | A | C | G | 10 | 0.008 |
| 16 | A | A | T | C | C | 4 | 0.003 |
| 17 | A | A | A | C | G | 4 | 0.003 |
| 18 | A | A | T | C | G | 2 | 0.002 |
| 19 | G | A | T | T | G | 1 | 0.001 |
| Total | 1267 | 1 |
| SNP Locus | Genotypes | Record Number | Tested Day Milk Yield (kg) | Milk Fat Percentage (%) | Milk Protein Percentage (%) | Somatic Cell Score |
|---|---|---|---|---|---|---|
| IL17A-1578A>G | AA | 827 | 34.35 ± 0.37 b | 3.57 ± 0.03 | 3.21 ± 0.01 | 2.91 ± 0.08 a |
| GA | 3984 | 35.03 ± 0.17 a | 3.65 ± 0.01 | 3.23 ± 0.01 | 2.75 ± 0.03 b | |
| GG | 2661 | 34.82 ± 0.21 ab | 3.67 ± 0.02 | 3.26 ± 0.01 | 2.69 ± 0.04 b | |
| Total | 7472 | 34.97 ± 0.11 | 3.64 ± 0.01 | 3.24 ± 0.00 | 2.76 ± 0.02 | |
| F value | 5.583 ** | 1.622 | 0.385 | 3.486 * | ||
| IL17A-1835G>A | AA | 209 | 36.14 ± 0.75 | 3.71 ± 0.06 | 3.21 ± 0.02 | 3.19 ± 0.15 |
| GA | 1445 | 35.28 ± 0.28 | 3.64 ± 0.02 | 3.21 ± 0.01 | 2.67 ± 0.06 | |
| GG | 5894 | 35.09 ± 0.14 | 3.63 ± 0.01 | 3.23 ± 0.00 | 2.80 ± 0.03 | |
| F value | 1.538 | 1.098 | 1.146 | 1.846 | ||
| IL17A-398T>A | AA | 2247 | 35.36 ± 0.24 a | 3.63 ± 0.02 | 3.25 ± 0.01 | 2.71 ± 0.04 |
| TA | 4859 | 35.01 ± 0.15 ab | 3.63 ± 0.01 | 3.22 ± 0.01 | 2.80 ± 0.03 | |
| TT | 1813 | 34.62 ± 0.25 b | 3.69 ± 0.02 | 3.23 ± 0.01 | 2.73 ± 0.05 | |
| F value | 5.692 ** | 1.988 | 0.105 | 1.188 | ||
| IL17A 3164T>C | CC | 3487 | 35.40 ± 0.19 a | 3.62 ± 0.02 | 3.24 ± 0.01 | 2.71 ± 0.04 |
| TC | 4041 | 35.14 ± 0.17 a | 3.63 ± 0.01 | 3.21 ± 0.01 | 2.84 ± 0.03 | |
| TT | 986 | 34.37 ± 0.34 b | 3.67 ± 0.03 | 3.24 ± 0.01 | 2.74 ± 0.06 | |
| F value | 3.948 ** | 0.592 | 1.129 | 0.199 | ||
| IL17A 3409G>C | CC | 3480 | 35.38 ± 0.19 a | 3.63 ± 0.02 | 3.24 ± 0.01 | 2.70 ± 0.04 |
| GC | 1972 | 35.08 ± 0.24 ab | 3.63 ± 0.02 | 3.22 ± 0.01 | 2.69 ± 0.05 | |
| GG | 3267 | 34.82 ± 0.19 b | 3.65 ± 0.02 | 3.22 ± 0.01 | 2.88 ± 0.04 | |
| F value | 4.711 ** | 0.46 | 0.195 | 0.262 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ghulam Mohyuddin, S.; Liang, Y.; Ni, W.; Adam Idriss Arbab, A.; Zhang, H.; Li, M.; Yang, Z.; Karrow, N.A.; Mao, Y. Polymorphisms of the IL-17A Gene Influence Milk Production Traits and Somatic Cell Score in Chinese Holstein Cows. Bioengineering 2022, 9, 448. https://doi.org/10.3390/bioengineering9090448
Ghulam Mohyuddin S, Liang Y, Ni W, Adam Idriss Arbab A, Zhang H, Li M, Yang Z, Karrow NA, Mao Y. Polymorphisms of the IL-17A Gene Influence Milk Production Traits and Somatic Cell Score in Chinese Holstein Cows. Bioengineering. 2022; 9(9):448. https://doi.org/10.3390/bioengineering9090448
Chicago/Turabian StyleGhulam Mohyuddin, Sahar, Yan Liang, Wei Ni, Abdelaziz Adam Idriss Arbab, Huiming Zhang, Mingxun Li, Zhangping Yang, Niel A. Karrow, and Yongjiang Mao. 2022. "Polymorphisms of the IL-17A Gene Influence Milk Production Traits and Somatic Cell Score in Chinese Holstein Cows" Bioengineering 9, no. 9: 448. https://doi.org/10.3390/bioengineering9090448
APA StyleGhulam Mohyuddin, S., Liang, Y., Ni, W., Adam Idriss Arbab, A., Zhang, H., Li, M., Yang, Z., Karrow, N. A., & Mao, Y. (2022). Polymorphisms of the IL-17A Gene Influence Milk Production Traits and Somatic Cell Score in Chinese Holstein Cows. Bioengineering, 9(9), 448. https://doi.org/10.3390/bioengineering9090448






