Abstract
Isoflavonoid synthase (IFS) is a critical enzyme for the biosynthesis of over 2400 isoflavonoids. Isoflavonoids are an important class of plant secondary metabolites that have a range of pharmaceutical and nutraceutical properties. With growing interest in isoflavonoids from both research and industrial perspectives, efforts are being forwarded to enhance isoflavonoid production in-planta and ex-planta; therefore, in-silico analysis and characterisation of available IFS protein sequences are needed. The present study is the first-ever attempt toward phylogenetic analysis and protein modelling of available IFS protein sequences. Phylogenetic analysis has shown that IFS amino acid sequences have 86.4% pairwise identity and 26.5% identical sites, and the sequences were grouped into six different clades. The presence of a β-hairpin and extra loop at catalytic sites of Trifolium pratense, Beta vulgaris and Medicago truncatula, respectively, compared with Glycyrrhiza echinata are critical structural differences that may affect catalytic function. Protein docking highlighted the preference of selected IFS for liquiritigenin compared with naringenin and has listed T. pratense as the most efficient candidate for heterologous biosynthesis of isoflavonoids. The in-silico characterisation of IFS represented in this study is vital in realising the new bioengineering endeavours and will help in the characterisation and selection of IFS candidate enzymes for heterologous biosynthesis of isoflavonoids.
1. Introduction
Isoflavonoid synthase (EC 1.14.14.87), a membrane-bound cytochrome P450 monooxygenase, is the key enzyme responsible for the biosynthesis of isoflavonoids [1,2]. Migration of the aryl ring from the C2 to C3 position is the first step toward synthesising the isoflavonoid core skeleton from flavonoid precursors (Figure 1) [3]. Being the first enzyme of the pathway, IFS directs the flow of intermediates from the flavonoid pathway to the isoflavonoid pathway and, therefore, defines a branch point in the synthesis of plant natural products [4]. Isoflavonoids are a unique sub-class of flavonoids that play an essential role in plant growth and development. In addition, the role of isoflavonoids in human health is well established, with recent studies highlighting their pharmacological properties ranging from hot flashes to cancer treatment [5,6,7,8]. It is envisaged that further characterisation will broaden the range of applications of isoflavonoids for human health.
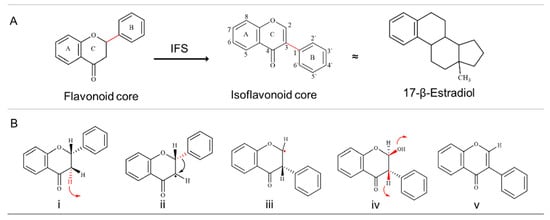
Figure 1.
Basic structure of isoflavonoid and aryl ring migration reaction mechanism. (A) B-ring is attached at the C3 position in isoflavonoids, making them different from flavonoids and similar to the human estrogen hormone (17-β-Estradiol). (B) The aryl ring migration reaction can be divided into five steps (Red colour represents the site of reactivity). (i) A H atom from the C3 position is removed (red arrow represents the removal of H), (ii) which is followed by the migration of the aryl ring from C2 to the C3 position. (iii) Due to aryl ring migration, C2 becomes electron-deficient, which is (iv) stabilised with the addition of the OH group, (v) A dehydration step that either occurs spontaneously or with the help of 2-hydroxyisoflavonid dehydratase (HIDH) completes the reaction. Step i to iv are catalysed by IFS. IFS: Isoflavonoid Synthase.
Isoflavonoids are mainly identified from Papilonoidae, a sub-family of Leguminosae; however, up to 59 non-leguminous plant families (i.e., Iridaceae, Rosaceae, and Liliaceae) have also been reported to synthesise isoflavonoids [9]. As legumes are primary isoflavonoid producers, it was initially believed that the IFS gene is present in legumes only [10,11,12]. Increasing evidence of isoflavonoid’s presence in non-leguminous plants was followed by the isolation of the IFS gene from sugar beet, proving that IFS is not limited to legumes only [10]. Isoflavonoids play multiple roles in plant growth and development, and their role in establishing plant-microbial interactions has been well documented [13]. Over 2400 isoflavonoids have been identified from over 300 plants [3]. As IFS is responsible for synthesising parent isoflavonoids, the widespread presence of isoflavonoids indicates the importance and diversity of the enzyme [14,15]. It is assumed that the aryl migration activity of IFS must have emerged through a rare gain-of-function event by mutations from ancestral CYP93 genes and have survived and become dominant in present-day legumes due to the ecophysiological importance of the products it makes [16].
Advancements in synthetic biology and metabolic engineering made it possible to replicate the complete biosynthetic pathway of any natural plant product to heterologous hosts (specifically in microorganisms) [17]. Heterologous biosynthesis of isoflavonoids in microorganisms, i.e., Saccharomyces cerevisiae, and Escherichia coli, has been achieved (Table 1). Most of these studies highlighted low isoflavonoid titer due to low IFS expression, functionality, or incompatibility with a co-expressed CPR partner. A better understanding of the IFS sequence and structure relationship, along with the addition of more chassis (Yarrowia lipolytica), co-culturing as well as cell-free biosynthesis, are a few avenues that can help develop highly efficient heterologous isoflavonoid production systems [18,19,20]. In addition to this, applications of protein engineering and directed evolution strategies can be employed to engineer IFS isoforms capable of synthesising new-to-nature isoflavonoids with improved pharmacokinetics at industrially acceptable titer, rate and yield [5,21].

Table 1.
Heterologous expression of IFS in microorganisms.
Isoflavonoids (biochanin-A, formononetin and genistein) are recently characterised as potential anti-cancer compounds, and demand for isoflavonoids is rising [3]. The market value of isoflavonoids was US $13.5 billion in 2018 and is expected to rise to US $30 billion by 2025 [30]. However, inefficient, traditional extraction methods, low plant yield, climate change, and priority to agricultural land use have questioned the availability of isoflavonoids to the general public [5,31]. Therefore, there is a drive for the biotechnology industry to produce isoflavonoids by using microorganisms sustainably and responsibly rather than using vital and valuable arable land [3]. Additionally, being a promising alternative, heterologous biosynthesis systems face low yield issues due to slow enzyme kinetics [23,27,28]. Therefore, the present study explores genetic variability in IFS protein sequences across different plant lineages and identifies key residues that define enzymes’ reactivity. Our results highlighted critical structural differences at catalytic sites that are assumed to play an important role in catalysis. Additionally, the preference of IFS for liquiritigenin (LQN) compared with naringenin (NGN) has been documented to help select the appropriate IFS isoform for heterologous isoflavonoid biosynthesis.
2. Materials and Methods
2.1. Sequence Retrieval
A BLASTP search was performed in the NCBI Non-redundant protein sequences (nr) database (https://blast.ncbi.nlm.nih.gov/Blast.cgi (accessed on 6 March 2022)) using Glycyrrhiza echinata IFS (One of the first identified and well-characterized IFS sequences) as the query sequence to retrieve available IFS protein sequences. Overall, 138 amino acid sequences representing 37 species having E-value = 0, and all key amino acids, i.e., Ser-310, Leu-371, Lys-375, and Arg-104 (amino acid numbers based on G. echinata sequence), were retrieved. P450 Engineering Database, University of Stuttgart (https://cyped.biocatnet.de/sequence-browser (accessed on 6 March 2022)), is also a valuable resource for the P450 family; however, it has fewer IFS entries as compared with NCBI. The detailed information (accession number etc.) about all the IFS sequences used in this study is listed in Table S1.
2.2. Sequence Alignment and Phylogenetic Analysis
The 139 IFS amino acid sequences (138 retrieved from NCBI and G. echinata sequence) were aligned using the MUSCLE alignment algorithm with default parameters. A phylogenetic tree was constructed using the Neighbor-Joining algorithm with default parameters on Geneious Prime version 2021.1.1, and the results were documented.
2.3. Protein Modelling Analysis
Protein homology models of selected IFS sequences were developed on the Alphafold homology modelling server and by using Salvia miltiorrhiza CYP76AH1 (PDB: 5YLW) structure as a template on the SWISS-MODEL protein structure homology modelling server (https://swissmodel.expasy.org (accessed on 10 April 2022)). Protein homology models were analyzed in PyMOL.
2.4. Protein Docking Analysis
For protein docking, MGLTools v1.5.7 has been used. Briefly, target .pdb files were developed as discussed under the Section 2.3. 3D conformers of LQN (PubChem CID: 114829) and NGN (PubChem CID: 932) ligands files were downloaded from the PubChem database (https://pubchem.ncbi.nlm.nih.gov/ (accessed 21 May 2022)) in .sdf format and were converted to .pdb format using BIOVIA Discovery Studio Visualizer. Target and ligand .pdbqt files were prepared using AutoDockTools v4.2. In the docking simulation, the grid box size was set to 70 Å × 40 Å × 40 Å with a space of 0.375 Å in the structure of enzymes. The Lamarckian genetic algorithm was used to predict the binding modes of the target and ligands. The docking parameters were set for a population size of 150. The GA runs were set to 25, and the maximum number of generations was set at 25,000 for each docking study. The molecular docking was executed in Cygwin, and the results were analysed in AutoDockTools v4.2. Finally, the docked complex structure of the ligands and the surrounding amino acid residues were analysed in PyMOL and BIOVIA Discovery Studio Visualizer.
3. Results
3.1. Phylogenetic Analysis of IFS Amino Acid Sequences
Multiple sequence alignment has shown that the selected IFS amino acid sequence has 86.4 % pairwise identity and 26.5% identical sites (Figure 2). High sequence similarity among P450 family members has been reported, which is very clear from our results [15]. Following alignment, a phylogenetic tree was developed which grouped selected IFS amino acid sequences into at least 6 different clades (Figure 3, Table S2).

Figure 2.
Sequence logos of the multiple sequence alignments of the 139 IFS amino acids sequences: The sequence logos are based on IFS (CYP93C subfamily) amino acids alignment using the Neighbor-Joining algorithm on Geneious Prime. The logos were generated using Weblogo (https://weblogo.berkeley.edu/logo.cgi (accessed on 26 March 2022)). The bit score indicates the information content for each position in the sequence. Hight of the letters designating the amino acid residues at each position represents the degree of conservation. The key conserved motifs are underlined; the black line indicates the P450 motifs, and the blue ones indicate substrate recognition sites (SRSs). Red stars and boxes represent key amino acids, and black triangles and boxes represent important amino acids that also have an important role in catalysis.
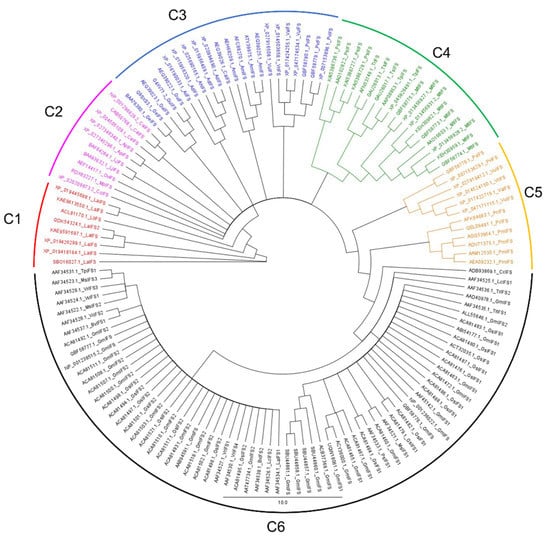
Figure 3.
Phylogenetic tree of IFS amino acid sequences. The phylogenetic tree is constructed using the Neighbour Joining (NJ) algorithm for 139 IFS amino acid sequences. Branch labels represent the NCBI accession number followed by species name initials and IFS isoform number. Colours are used to differentiate between clades. The scale bar represents amino acid substitutions per site.
3.1.1. Clade 1
There were 8 IFS sequences in clade 1; they all belong to the genus Lupinus and represent three species (L. albus, L. angustifolius, and L. luteus). These sequences have 84.0% pairwise identity, 62.9% identical sites, and 59.2% similarity with the G. echinata IFS sequence (used as a template for comparison with selected IFS sequences). Sequences of this group show variations at substrate recognition sites 2 (SRS2) and SRS3 positions (Figure S1). These sequences have Glu at position 237 instead of Asp; however, both amino acids are acidic, so this substitution might not affect the enzyme’s catalytic efficiency. Additionally, they have Lys instead of Arg at position 112 in SRS1 compared to G. echinata. Members of this clade also have a deletion at 426 positions compared with the G. echinata IFS sequence.
3.1.2. Clade 2
IFS amino acid sequences of Cicer arietinum, Cullen corylifolium, Abrus precatorius, Lotus japonicus, Onobrychis viciifolia and Mucuna pruriens are grouped in clade 2. These 10 sequences have 85.5% pairwise identity, 62.7% identical sites, and 61.6% similarity with the G. echinata sequence. O. viciifolia has an addition of 4 amino acids at positions 427–430 compared with G. echinata and other clade members.
3.1.3. Clade 3
Twenty-one IFS sequences representing the genus Glycyrrhiza, Arachis, Astragalus, Caragana, Vigna and Phaseolus are grouped in clade 3. These sequences have 86.0% pairwise identity, 65.6% identical sites, and 65.2% similarity with the G. echinata sequence. The addition of Glu at position 304 in P. vulgaris compared with other members and G. echinata IFS is unique to this clade.
3.1.4. Clade 4
IFS sequences from the genus Pisum, Trifolium, and Medicago are grouped in clade 4. These 19 IFS sequences have 89.6% pairwise identity, 67.9% identical sites, and 67.1% similarity with the G. echinata sequence. All four sequences of P. sativum and two sequences of G. echinata have the addition of Glu and Arg/Lys at positions 283–284 as compared with other clade members. Similarly, 3 of M. truncatula IFS seq. (QBF58773.1, QBF58774.1 and QBF58775.1) have the addition of Glu at position 326 compared with G. echinata and other clade members.
3.1.5. Clade 5
Clade 5 has 12 sequences representing 3 genera (Phaseolus, Vigna, and Pueraria). These sequences have 94.1% pairwise identity, 84.9% identical sites, and 77.1% similarity with G. echinata sequence. All these sequences have a deletion of two amino acids at positions 259 and 260 compared with the G. echinata sequence. Together with that, the addition of Glu at position 302 is unique to P. vulgaris IFS sequence (QBF58778.1) compared with the G. echinata and other clade members.
3.1.6. Clade 6
Sixty-nine IFS sequences representing 10 genera (Cajanus, Lens, Trifolium, Pisum, Lupinus, Beta, Vigna, Medicago, Vicia and Glycine) are grouped in clade 6. It is clear from Figure 3 that clade 6 is dominated by G. max and G. soja IFS sequences. These 69 sequences have 96.6% pairwise identity, 53.0% identical sites, and 51.1% similarity with the G. echinata template. G. max IFS seq. AAT47734.1 has the addition of 5 amino acids at position 232-236 and G. max IFS seq. QBF58777.1 and QBF58777.1 have a Glu addition at position 302 compared with the G. echinata and other clade members.
3.2. Protein Homology Modelling of Selected IFS Candidates
Being a P450 class enzyme, instability, scarcity, and the presence of many homologous proteins make solubilisation and purification challenging; that is why the crystal structure of IFS has not been solved yet [1]. However, a few key amino acids; Ser-310, Leu-371, Lys-375, and Arg-104 (amino acid numbers based on G. echinata sequence) involved in substrate selection and aryl ring migration have been identified (Figure 1) [14,32]. Based on these findings, it was concluded that the substitution of Ser-310 could suppress; however, Lys-375 is critical for the aryl migration activity of IFS, and Arg-104 is essential to maintain P450 architecture [14]. Many IFS encoding genes have been identified and characterised from various legumes and non-legume plants (Table S3). Following phylogenetic analysis, 38 representative IFS sequences (at least 1 IFS seq./species and 2 for G. max) were selected for homology modelling (Figure S2, Tables S4 and S5). Protein models developed on the SWISS-MODEL server have shown sequence identity ≥29.36% and coverage ≥ 0.82. For a few sequences, more than 2 models were developed, and a model having a higher GMGQ score was selected for further analysis (Table S4). On the other hand, protein models developed on Alphafold having the lowest E-value were selected for further analysis (Table S5).
Protein homology modelling has highlighted structural variations at the catalytic site for Astragalus mongholicus (AEH68209.1), Beta vulgaris (AAF34538.1), Lens culinaris (AAF34525.1), Medicago truncatula (AAO16603.1), Trifolium pratense (AAP06953.1) and Vigna angularis (XP_017424255.1) compared with G. echinata (BAA76380.1) (Figure S2 and Table S4). Following that, we developed homology models of the remaining IFS isoforms of these species; Beta vulgaris: AAF34537.1, Lens culinaris: AAF34526.1, Medicago truncatula: KEH30959.1, KEH30962.1, QBF58773.1, QBF58774.1, QBF58775.1, RDX63227.1, Trifolium pratense: XP_045826491.1 and Vigna angularis: XP_017422719.1 (Figure S3 and Table S6). However, a significant structural difference was only seen for B. vulgaris (AAF34538.1), T. pratense (AAP06953.1) and M. truncatula (QBF58774.1) IFS homology models compared with the G. echinata (BAA76380.1). On the other hand, protein models developed on the Alphafold server have not shown any extra β-hairpin or loop structure; instead, a new β-sheet (β4-sheet) has been seen in all models. β4-sheet is believed to be an integral part of the P450 structure and was also proposed for IFS by Sawada et al.; however, the β4-sheet is not present in the crystal structure of Salvia miltiorrhiza CYP76AH1 (PDB: 5YLW), which is quite surprising [14,32,33]. Therefore, the presence of β4-sheet can only be confirmed by solving the 3D model of IFS.
The presence of an extra β-hairpin near the active site in B. vulgaris (AAF34538.1) and T. pratense (AAP06953.1) and a long loop in M. truncatula (QBF58774.1) compared with the G. echinata (BAA76380.1) protein model is a significant difference in protein structures (Figure 4). The extra β-hairpin and loop structure are present over the heme unit, inside the active site and are surrounded by key amino acids. The orientation of Leu-371 and Lys-375 has been changed due to the extra β-hairpin, and these amino acids are tilted more towards the heme unit and Ser-310 (Figure S4). On the other hand, the orientation of Ser-310, Leu-371 and Lys-375 in models of B. vulgaris (AAF34538.1), T. pratense (AAP06953.1), M. truncatula (QBF58774.1) and G. echinata (BAA76380.1) developed on Alphafold are different compared with models developed on SWISS-MODEL (Figure 4). This change in the position of key amino acids might affect the enzyme’s catalytic efficiency.
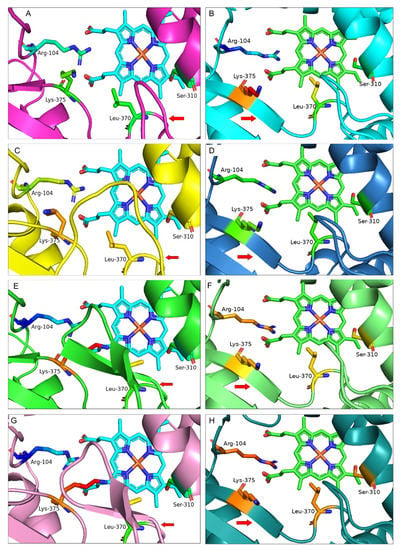
Figure 4.
Active site structure of IFS from (A,B) G. echinata (BAA76380.1) (C,D) M. truncatula (QBF58774.1) (E,F) B. vulgaris (AAF34538.1) and (G,H) T. pratense (AAP06953.1). Protein models on the left (A,C,E,G) are developed using Salvia miltiorrhiza CYP76AH1 (PDB: 5YLW) as a template on the SWISS-MODEL protein structure homology modelling server (https://swissmodel.expasy.org (accessed on 10 April 2022)). Protein models on the right (B,D,F,H) are developed on the Alphafold server. Key amino acids are shown as sticks; amino acid numbering is based on the G. echinata amino acid sequence. Red arrows indicate the presence of a long loop in M. truncatula and extra β-hairpin of B. vulgaris and T. pratense compared with the G. echinata protein model and the β4-sheet present in all models. The presence of an extra β-hairpin in the active site over the Heme unit might control/interfere with the entrance and selection of substrate and may have a role in enzyme efficiency.
Similarly, the effect of addition and deletion of amino acids, i.e., ER at position 260–261 in clade 4–6 (amino acid numbers are based on G. echinata sequence), was studied. The addition and deletion of amino acids are away from the active site, in loops structure present over the protein surface. These structural changes, along with the substitution of amino acids in SRS sequences, may affect substrate binding and interaction with CPR partners. Therefore, analysis of the effects of these variations is a potential area of research.
The amino acid sequence ‘KVSMEERAGLTVP’ (G. echinata amino acid 489–501) of G. echinata, B. vulgaris, M. truncatula and T. pratense at the extra β-hairpin site, and loop site was unchanged except for only two amino acids substitutions present in T. pratense (AAP06953.1) sequence (Figure S4). So, to check whether the extra β-hairpin are artefacts generated during the homology modelling, B. vulgaris, M. truncatula, T. pratense and G. echinata protein models were aligned with S. miltiorrhiza CYP76AH1 (PDB: 5YLW) structure in PyMOL (Figure S5). Key amino acid sites, I-helix, and all other structures except for the long loop of M. truncatula and extra β-hairpin of B. vulgaris and T. pratense were perfectly matched with the S. miltiorrhiza CYP76AH1 template. Additionally, the loop of M. truncatula and β-hairpin of B. vulgaris and T. pratense was not observed in any other IFS models. Thus, we concluded that the extra β-hairpin and loop might result from the addition and/or substitution of amino acids at other sites; therefore, further structural, and functional characterization of IFS is required.
Protein structure prediction models are becoming increasingly important in understanding sequence-structural relationships for proteins, like IFS, that do not have crystal structure solved [34,35]. Homology modelling is considered the most accurate of the available methods, and backbone prediction with sub-Angstrom Root Mean Square Distance (RMSD-Cα) has become a routine practice, and prediction of side chains has become increasingly accurate [34,35]. Protein models developed through homology modelling, even for challenging proteins, are now considered reliable [35]. Advancements in protein structure modelling have great potential because more reliable and accurate protein models will help better understand sequence-structure relationships.
3.3. Protein Docking of Selected IFS Candidates
Following protein modelling, the selected IFS; B. vulgaris (AAF34538.1), T. pratense (AAP06953.1), M. truncatula (QBF58774.1) and G. echinata (BAA76380.1) was docked with LQN and NGN as ligands (Figure 5, Table 2). The binding energy (ΔG) comparison highlighted the preference of IFS toward respective substrates [36,37]. All four IFS models have shown the lowest binding energy for LQN compared with NGN (Table 2). The difference in binding energy for LQN clearly explains the preference of IFS toward LQN in natural and heterologous hosts, as stated in the literature [38,39]. Protein docking analysis of IFS models developed on the SWISS-MODEL server has shown slightly lower binding energies compared with models developed using Alphafold (Table 2).
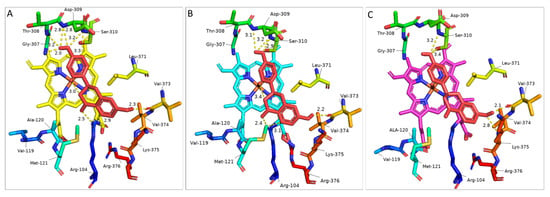
Figure 5.
Key amino acids along with Liquiritigenin (docked as a ligand) in selected IFS homology models: (A) T. pratense (AAP06953.1), (B) G. echinata (BAA76380.1) and (C) M. truncatula (QBF58774.1). Key amino acids and LQN structure are shown in the stick model. LQN is shown in red; key amino acids are shown in rainbow colours. Polar interactions are shown in yellow dotted lines, and the distance is shown in Å units.

Table 2.
Binding energy (ΔG) comparison of selected IFS docked with LQN and NGN.
Docking results have highlighted T. pratense as the most efficient candidate for conversion of LQN to isoflavonoids compared with other IFS models. On the other hand, the binding energies of T. pratense, M. truncatula and G. echinata for NGN were almost the same. These IFS may show similar conversion efficiency in heterologous hosts. As there is not much difference in binding energies, the interaction of ligand (LQN) with the enzyme at the catalytic site can help us explain and select an IFS candidate. Docking analysis has shown that LQN develops interaction with the Fe-O complex at the C3 position of LQN (for hydrogen abstraction (Figure 1)) to start the catalysis reaction. The distance between the Fe-O complex and C3 of LQN is 3Å for T. pratense, which is increased to 3.4Å for G. echinata (Figure 5). The Fe-O and C3 of LQN interaction are not seen in any of the 25 docked complexes of M. truncatula and B. vulgaris. It is believed that LQN would develop interaction with Fe-O; however, further refinement of models using molecular simulation algorithms can help find that interaction. Chemler et al. have compared 5 IFS, including T. pratense, G. echinata, and M. truncatula, in a yeast system, and they have ranked T. pratanse as the most efficient candidate for bioconversion of NGN [40]. Compared with other models, the low binding energy of T. pratense for LQN and NGN makes it the most efficient candidate for the biosynthesis of isoflavonoids in heterologous hosts (Table 1).
Docking results have highlighted a few more key amino acids that might play an important role in catalysis/aryl migration reaction and in stabilising the substrate in the active site over the heme unit. Interaction of the 7-OH group with Val-373 and 4′-OH with Gly-307, Thr-308 and Asp-309 of LQN (amino acid number based on G. echinata sequence) are significant findings. Val-119, Ala-120 and Met-121 as well as Arg-376, seem to be involved in heme stabilization. Previously, the interaction of Leu-371 with the O atom of the C-ring of LQN based on a homology model of GeIFS built using the crystal structure of bacterial BM3 as a template was reported; however, the authors concluded that Leu-371, along with Ser-310 and Lys-375 (amino acid number based on G. echinata sequence) specifically play a role in accommodation of substate and stabilisation of P450 architecture [14,32]. Together with that, the catalytic importance of the extended ring present over the catalytic site in the homology model of M. truncatula and extra β-hairpin in B. vulgaris and T. pratense have been highlighted by docking results. The orientation of ligands has been slightly changed in M. truncatula due to amino acid residues in the loop. In addition, the change in orientation of Lys-375 due to β-hairpin in B. vulgaris and T. pratense may be the reason for the higher catalysis rate of T. pratense [40]. However, in-depth protein docking analysis with more models/samples is required.
4. Discussion
In the present study, phylogenetic analysis and protein modelling has been performed on selected IFS amino acid sequences representing 37 species. Phylogenetic analysis has shown 86.4% pairwise identity and 26.5% identical sites among IFS amino acid sequences, highlighting a recent origin of IFS (CYP93C subfamily) from the CYP93B subfamily [15]. It is believed that CYP93C and CYP93B families share the same origin, further supported by the evidence that these subfamilies share the same substrate [41,42]. As subfamily CYP93C originated from CYP93B, Sawada et al. replaced key amino acids (Ser-310 and Lys-375) in the CYP93B subfamily enzyme; however, no aryl migration activity was seen [14,32]. The authors conclude that the key amino acids (Ser-310, Lys-375 and Leu-371, Arg-104) identified through mutational studies are not solely responsible for aryl-migration activity [14]. It was proposed that further amino acids in the IFS enzyme having a role in aryl migration reaction are assumed to be present, and overall protein architecture is an important factor that controls aryl-migration activity [14]. Together with that, the formation of the by-product (3-hydroxyflavone) also indicates the inefficiency of the aryl-migration apparatus, which means the answer to IFS enzymatic efficiency lies in amino acids sequence variability, and screening of different IFS isoforms can help to select highly efficient IFS candidate. In this regard, variations in amino acid sequences highlighted in the present study can help direct future studies for better IFS structure-function characterisation. It is envisaged that substitutions at distal sites might affect enzyme reactivity differently [43]. Therefore, further studies are required to better understand the function of amino acids identified in the present study.
IFS, a P450 enzyme, requires electrons from its partner enzyme, known as cytochrome P450 reductase (CPR), for catalysis [44]. That means IFS interacts with CPR and substrate molecules simultaneously, and anything that affects the stability of this interaction will influence enzyme reactivity and/or rate of catalysis. Literature indicates that the source of IFS and CPR strongly influence the rate of isoflavonoid biosynthesis [40]. That means amino acids involved in the electron transfer pathway present in CPR and IFS may have a preference for each other, and the overall architecture of both enzymes may have a role in electron transportation [14,40]. Therefore, amino acids that play a role in electron transfer and in, substrate selection, stabilisation and interaction with the heme unit and are involved in the recruitment of other enzymes require further insights [43,45,46]. Recent developments in molecular simulations can help advance our understanding specifically of IFS-CPR interaction, electron transfer pathway and substrate entrance channels [47]. Such studies will further strengthen our knowledge about aryl ring migration reaction and catalytic efficiency, which will help select appropriate IFS candidates for heterologous isoflavonoid biosynthesis.
With recent developments in synthetic biology and metabolic engineering, there is a drive for the biotechnology industry to enhance isoflavonoid production in-planta and ex-planta [48,49]. The potential to introduce exogenous isoflavonoid biosynthesis into cereal crops is an exciting avenue, but one in which the outcomes may be limited due to the negative public sentiment and regulatory barriers surrounding genetically modified crops [31]. The proof of concept evidence in onion and rice demonstrates that the approach is viable and likely beneficial to the agricultural industry [50,51]. While there is an excellent attraction toward the biofortification of crops through genetic engineering, societal perceptions and regulatory barriers will need to be addressed and overcome for this approach to be of value. On the other hand, microorganisms are a more attractive tool towards the generation of isoflavonoids due to their scalability, small footprint and rapid production [52]. As isoflavonoid products would be either produced through generally regarded as safe microorganisms, i.e., yeast or isolated and purified from the growth media, regulatory hurdles would be less restrictive [18]. Additionally, developing prokaryotic compatible IFS for scaling in bacterial systems has immense potential; however, this requires a large amount of research work, including protein engineering, to make IFS a cytosolic self-sufficient enzyme like prokaryotic P450 [44,53].
In conclusion, the structure-function characterisation of IFS for the highest isoflavonoid biosynthesis in plants and microorganisms, specifically in prokaryotic systems, has immense potential to transform future farming systems and the biotechnology industry.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/bioengineering9110609/s1, Figure S1: Weblogo of SRSs based on the amino acid sequence alignment across all clades. The bit score indicates the information content for each position in the sequence. Numbers 1–6 represent respective clades. Figure S2: Protein homology models of selected IFS sequences. Homology models on the left are developed using Salvia miltiorrhiza CYP76AH1 (PDB: 5YLW) as a template on the SWISS-MODEL protein structure homology modelling server (https://swissmodel.expasy.org (accessed on 20 April 2022)), and homology models on the right are developed using the Alphafold. Further details about homology models are given in Table S4 & S5. Figure S3: Protein homology models of Astragalus mongholicus, Beta vulgaris, Lens culinaris, Medicago truncatula, Trifolium pratense, Vigna angularis and G. echinata IFS sequences. Homology models are developed using Salvia miltiorrhiza CYP76AH1 (PDB: 5YLW) as a template on the SWISS-MODEL protein structure homology modelling server (https://swissmodel.expasy.org (accessed on 20 April 2022)). Further details about homology models are given in Table S6. Figure S4. Active site structure of IFS from (A) G. echinata (BAA76380.1) (B) M. truncatula (QBF58774.1) (C) B. vulgaris (AAF34538.1) and (D) T. pratense (AAP06953.1). Homology models are developed using Salvia miltiorrhiza CYP76AH1 (PDB: 5YLW) as a template on the SWISS-MODEL protein structure homology modelling server (https://swissmodel.expasy.org (accessed on 10 April 2022)). Key amino acids are shown as sticks; amino acid numbering is based on the G. echinata amino acid sequence. Red arrows indicate the presence of a long loop in M. truncatula and extra β-hairpin of B. vulgaris and T. pratense compared with the G. echinata protein model. The purple arrow indicates the change in orientation of Lys-375. The presence of an extra β-hairpin in the active site over the heme unit might control/interfere with the entrance and selection of substrate and may have a role in enzyme efficiency. The table represents the amino acid sequence of the loop/β-hairpin region (Amino acids numbers are based on the G. echinata sequence). Figure S5: Comparison of protein homology models (A) Template (PDB: 5YLW), (B) B. vulgaris, (C) T. pratense, (D) M. truncatula and (E) G. echinata, (F) homology models overlap with each other and (G) homology models overlapped with the template (PDB: 5ylw). Table S1. IFS amino acid sequences were used in this study. Table S2. IFS clade grouping based on phylogenetic analysis. Table S3. Functionally characterised Isoflavonoids synthase (IFS). Table S4: Protein modelling parameters of selected IFS sequences on SWISS-MODEL server. Table S5: Protein modelling parameters of selected IFS sequences on the Alphafold server. Table S6: Protein homology modelling parameters of selected IFS sequences from A. membranaceus, B. vulgaris, L. culinaris, M. truncatula, T. pratense, V. angularis and G. echinata. [1,2,10,11,12,14,32,54,55,56,57,58,59].
Author Contributions
M.S.: Data curation, Formal analysis, Investigation, Methodology, Validation, Visualization, Writing—original draft. S.R.S.: Investigation, Methodology, Supervision, Validation, Visualization, Writing—review & editing. P.K.: Conceptualisation, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Validation, Visualization, Writing—review & editing. All authors have read and agreed to the published version of the manuscript.
Funding
Not applicable.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
M.S. acknowledges the University of Western Australia for UIFS and UPA scholarships. P.K. is supported by the University of Western Australia with additional research funding from ExPlanta Pty Ltd. towards the Biosynthesis of Isoflavones-Formononetin (FMN) Proof of Concept project.
Conflicts of Interest
There is no conflict of interest.
References
- Akashi, T.; Aoki, T.; Ayabe, S. Cloning and Functional Expression of a Cytochrome P450 CDNA Encoding 2-Hydroxyisoflavanone Synthase Involved in Biosynthesis of the Isoflavonoid Skeleton in Licorice. Plant Physiol. 1999, 121, 821–828. [Google Scholar] [CrossRef] [PubMed]
- Steele, C.L.; Gijzen, M.; Qutob, D.; Dixon, R.A. Molecular Characterization of the Enzyme Catalyzing the Aryl Migration Reaction of Isoflavonoid Biosynthesis in Soybean. Arch. Biochem. Biophys. 1999, 367, 146–150. [Google Scholar] [CrossRef] [PubMed]
- Sajid, M.; Stone, S.R.; Kaur, P. Recent Advances in Heterologous Synthesis Paving Way for Future Green-Modular Bioindustries-A Review with Special Reference to Isoflavonoids. Front. Bioeng. Biotechnol. 2021, 9, 532. [Google Scholar] [CrossRef] [PubMed]
- Du, H.; Huang, Y.; Tang, Y. Genetic and Metabolic Engineering of Isoflavonoid Biosynthesis. Appl. Microbiol. Biotechnol. 2010, 86, 1293–1312. [Google Scholar] [CrossRef]
- Sajid, M.; Channakesavula, C.N.; Stone, S.R.; Kaur, P. Synthetic Biology towards Improved Flavonoid Pharmacokinetics. Biomolecules 2021, 11, 754. [Google Scholar] [CrossRef]
- Bustos-Salgado, P.; Andrade-Carrera, B.; Domínguez-Villegas, V.; Díaz-Garrido, N.; Rodríguez-Lagunas, M.J.; Badía, J.; Baldomà, L.; Mallandrich, M.; Calpena-Campmany, A.; Garduño-Ramírez, M.L. Screening Anti-Inflammatory Effects of Flavanones Solutions. Int. J. Mol. Sci. 2021, 22, 8878. [Google Scholar] [CrossRef]
- Kim, I.-S. Current Perspectives on the Beneficial Effects of Soybean Isoflavones and Their Metabolites for Humans. Antioxidants 2021, 10, 1064. [Google Scholar] [CrossRef]
- Gómez-Zorita, S.; González-Arceo, M.; Fernández-Quintela, A.; Eseberri, I.; Trepiana, J.; Portillo, M.P. Scientific Evidence Supporting the Beneficial Effects of Isoflavones on Human Health. Nutrients 2020, 12, 3853. [Google Scholar] [CrossRef]
- Lapčík, O. Isoflavonoids in Non-Leguminous Taxa: A Rarity or a Rule? Phytochemistry 2007, 68, 2909–2916. [Google Scholar] [CrossRef]
- Jung, W.; Yu, O.; Lau, S.-M.C.; O’Keefe, D.P.; Odell, J.; Fader, G.; McGonigle, B. Identification and Expression of Isoflavone Synthase, the Key Enzyme for Biosynthesis of Isoflavones in Legumes. Nat. Biotechnol. 2000, 18, 208–212. [Google Scholar] [CrossRef]
- Kim, B.G.; Kim, S.-Y.; Song, H.S.; Lee, C.; Hur, H.-G.; Kim, S.; Ahn, J.-H. Cloning and Expression of the Isoflavone Synthase Gene (IFS-Tp) from Trifolium Pratense. Mol. Cells (Springer Sci. Bus. Media BV) 2003, 15, 301–306. [Google Scholar]
- Shimada, N.; Akashi, T.; Aoki, T.; Ayabe, S. Induction of Isoflavonoid Pathway in the Model Legume Lotus Japonicus: Molecular Characterization of Enzymes Involved in Phytoalexin Biosynthesis. Plant Sci. 2000, 160, 37–47. [Google Scholar] [CrossRef]
- Tong, C.; Liu, X.; Wu, Y.; Zhao, Y.; Wang, J. Regulation of Endogenous Isoflavones on Alfalfa Nodulation and Nitrogen Fixation and Nitrogen Use Efficiency. Acta Prataculturae Sin. 2022, 31, 124. [Google Scholar]
- Sawada, Y.; Kinoshita, K.; Akashi, T.; Aoki, T.; Ayabe, S. Key Amino Acid Residues Required for Aryl Migration Catalysed by the Cytochrome P450 2-hydroxyisoflavanone Synthase. Plant J. 2002, 31, 555–564. [Google Scholar] [CrossRef] [PubMed]
- Du, H.; Ran, F.; Dong, H.-L.; Wen, J.; Li, J.-N.; Liang, Z. Genome-Wide Analysis, Classification, Evolution, and Expression Analysis of the Cytochrome P450 93 Family in Land Plants. PLoS ONE 2016, 11, e0165020. [Google Scholar]
- Stafford, H.A.; Ibrahim, R.K. Recent Advances in Phytochemistry; Plenum Press: New York, NY, USA, 1992; Volume 26, ISBN 0306442310. [Google Scholar]
- Huccetogullari, D.; Luo, Z.W.; Lee, S.Y. Metabolic Engineering of Microorganisms for Production of Aromatic Compounds. Microb. Cell Factories 2019, 18, 1–29. [Google Scholar] [CrossRef]
- Cravens, A.; Payne, J.; Smolke, C.D. Synthetic Biology Strategies for Microbial Biosynthesis of Plant Natural Products. Nat. Commun. 2019, 10, 1–12. [Google Scholar] [CrossRef]
- Lv, Y.; Marsafari, M.; Koffas, M.; Zhou, J.; Xu, P. Optimizing Oleaginous Yeast Cell Factories for Flavonoids and Hydroxylated Flavonoids Biosynthesis. ACS Synth. Biol. 2019, 8, 2514–2523. [Google Scholar] [CrossRef]
- Zang, Y.; Zha, J.; Wu, X.; Zheng, Z.; Ouyang, J.; Koffas, M.A. In Vitro Naringenin Biosynthesis from P-Coumaric Acid Using Recombinant Enzymes. J. Agric. Food Chem. 2019, 67, 13430–13436. [Google Scholar] [CrossRef]
- Wang, Y.; Xue, P.; Cao, M.; Yu, T.; Lane, S.T.; Zhao, H. Directed Evolution: Methodologies and Applications. Chem. Rev. 2021, 121, 12384–12444. [Google Scholar] [CrossRef]
- Tian, L.; Dixon, R.A. Engineering Isoflavone Metabolism with an Artificial Bifunctional Enzyme. Planta 2006, 224, 496–507. [Google Scholar] [CrossRef] [PubMed]
- Trantas, E.; Panopoulos, N.; Ververidis, F. Metabolic Engineering of the Complete Pathway Leading to Heterologous Biosynthesis of Various Flavonoids and Stilbenoids in Saccharomyces Cerevisiae. Metab. Eng. 2009, 11, 355–366. [Google Scholar] [CrossRef]
- Katsuyama, Y.; Miyahisa, I.; Funa, N.; Horinouchi, S. One-Pot Synthesis of Genistein from Tyrosine by Coincubation of Genetically Engineered Escherichia Coli and Saccharomyces Cerevisiae Cells. Appl. Microbiol. Biotechnol. 2007, 73, 1143–1149. [Google Scholar] [CrossRef] [PubMed]
- Leonard, E.; Koffas, M.A. Engineering of Artificial Plant Cytochrome P450 Enzymes for Synthesis of Isoflavones by Escherichia Coli. Appl. Environ. Microbiol. 2007, 73, 7246–7251. [Google Scholar] [CrossRef] [PubMed]
- Trantas, E.A.; Koffas, M.A.G.; Xu, P.; Ververidis, F. When Plants Produce Not Enough or at All: Metabolic Engineering of Flavonoids in Microbial Hosts. Front. Plant Sci. 2015, 6, 1–16. [Google Scholar] [CrossRef]
- Kim, B.-G. Biological Synthesis of Genistein in Escherichia Coli. J. Microbiol. Biotechnol. 2020, 30, 770–776. [Google Scholar] [CrossRef]
- Liu, Q.; Liu, Y.; Li, G.; Savolainen, O.; Chen, Y.; Nielsen, J. De Novo Biosynthesis of Bioactive Isoflavonoids by Engineered Yeast Cell Factories. Nat. Commun. 2021, 12, 1–15. [Google Scholar] [CrossRef]
- Meng, Y.; Liu, X.; Zhang, L.; Zhao, G.-R. Modular Engineering of Saccharomyces Cerevisiae for De Novo Biosynthesis of Genistein. Microorganisms 2022, 10, 1402. [Google Scholar] [CrossRef]
- Ahuja, K.; Mamtani, K. Industry Analysis Report, Regional Outlook, Application Development Potential, Price Trends, Competitive Market Share & Forecast, 2019–2025. 2019. Available online: https://www.gminsights.com/industry-analysis/isoflavones-market (accessed on 10 February 2022).
- Sajid, M.; Stone, S.R.; Kaur, P. Transforming Traditional Nutrition Paradigms with Synthetic Biology Driven Microbial Production Platforms. Curr. Res. Biotechnol. 2021, 3, 260–268. [Google Scholar] [CrossRef]
- Sawada, Y.; Ayabe, S. Multiple Mutagenesis of P450 Isoflavonoid Synthase Reveals a Key Active-Site Residue. Biochem. Biophys. Res. Commun. 2005, 330, 907–913. [Google Scholar] [CrossRef]
- Gu, M.; Wang, M.; Guo, J.; Shi, C.; Deng, J.; Huang, L.; Huang, L.; Chang, Z. Crystal Structure of CYP76AH1 in 4-PI-Bound State from Salvia Miltiorrhiza. Biochem. Biophys. Res. Commun. 2019, 511, 813–819. [Google Scholar] [CrossRef] [PubMed]
- Hameduh, T.; Haddad, Y.; Adam, V.; Heger, Z. Homology Modeling in the Time of Collective and Artificial Intelligence. Comput. Struct. Biotechnol. J. 2020, 18, 3494. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A. Highly Accurate Protein Structure Prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Wasukan, N.; Kuno, M.; Maniratanachote, R. Molecular Docking as a Promising Predictive Model for Silver Nanoparticle-Mediated Inhibition of Cytochrome P450 Enzymes. J. Chem. Inf. Model. 2019, 59, 5126–5134. [Google Scholar] [CrossRef]
- Fan, Y.; Tao, Y.; Liu, G.; Wang, M.; Wang, S.; Li, L. Interaction Study of Engeletin toward Cytochrome P450 3A4 and 2D6 by Multi-Spectroscopy and Molecular Docking. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2022, 264, 120311. [Google Scholar] [CrossRef]
- Uchida, K.; Akashi, T.; Aoki, T. Functional Expression of Cytochrome P450 in Escherichia Coli: An Approach to Functional Analysis of Uncharacterized Enzymes for Flavonoid Biosynthesis. Plant Biotechnol. 2015, 32, 205–213. [Google Scholar] [CrossRef]
- Kaducová, M.; Eliašová, A.; Trush, K.; Bačovčinová, M.; Sklenková, K.; Pal’ove-Balang, P. Accumulation of Isoflavonoids in Lotus Corniculatus after UV-B Irradiation. Theor. Exp. Plant Physiol. 2022, 34, 53–62. [Google Scholar] [CrossRef]
- Chemler, J.A.; Lim, C.G.; Daiss, J.L.; Koffas, M.A. A Versatile Microbial System for Biosynthesis of Novel Polyphenols with Altered Estrogen Receptor Binding Activity. Chem. Biol. 2010, 17, 392–401. [Google Scholar] [CrossRef][Green Version]
- Fliegmann, J.; Furtwängler, K.; Malterer, G.; Cantarello, C.; Schüler, G.; Ebel, J.; Mithöfer, A. Flavone Synthase II (CYP93B16) from Soybean (Glycine max L.). Phytochemistry 2010, 71, 508–514. [Google Scholar] [CrossRef]
- Du, Y.; Chu, H.; Chu, I.K.; Lo, C. CYP93G2 Is a Flavanone 2-Hydroxylase Required for C-Glycosylflavone Biosynthesis in Rice. Plant Physiol. 2010, 154, 324–333. [Google Scholar] [CrossRef]
- Waki, T.; Yoo, D.; Fujino, N.; Mameda, R.; Denessiouk, K.; Yamashita, S.; Motohashi, R.; Akashi, T.; Aoki, T.; Ayabe, S. Identification of Protein–Protein Interactions of Isoflavonoid Biosynthetic Enzymes with 2-Hydroxyisoflavanone Synthase in Soybean (Glycine max (L.) Merr.). Biochem. Biophys. Res. Commun. 2016, 469, 546–551. [Google Scholar] [CrossRef] [PubMed]
- Finnigan, J.D.; Young, C.; Cook, D.J.; Charnock, S.J.; Black, G.W. Cytochromes P450 (P450s): A Review of the Class System with a Focus on Prokaryotic P450s. Adv. Protein Chem. Struct. Biol. 2020, 122, 289–320. [Google Scholar] [PubMed]
- Anguraj Vadivel, A.K.; McDowell, T.; Renaud, J.B.; Dhaubhadel, S. A Combinatorial Action of GmMYB176 and GmbZIP5 Controls Isoflavonoid Biosynthesis in Soybean (Glycine max). Commun. Biol. 2021, 4, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Mameda, R.; Waki, T.; Kawai, Y.; Takahashi, S.; Nakayama, T. Involvement of Chalcone Reductase in the Soybean Isoflavone Metabolon: Identification of GmCHR5, Which Interacts with 2-Hydroxyisoflavanone Synthase. Plant J. 2018, 96, 56–74. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, G.; Nandekar, P.P.; Wade, R.C. An Electron Transfer Competent Structural Ensemble of Membrane-Bound Cytochrome P450 1A1 and Cytochrome P450 Oxidoreductase. Commun. Biol. 2021, 4, 1–13. [Google Scholar] [CrossRef]
- Rani, D.; Vimolmangkang, S. Trends in the Biotechnological Production of Isoflavonoids in Plant Cell Suspension Cultures. Phytochem. Rev. 2022, 21, 1–20. [Google Scholar] [CrossRef]
- Rani, D.; Meelaph, T.; De-Eknamkul, W.; Vimolmangkang, S. Yeast Extract Elicited Isoflavonoid Accumulation and Biosynthetic Gene Expression in Pueraria Candollei Var. Mirifica Cell Cultures. Plant Cell Tissue Organ Cult. (PCTOC) 2020, 141, 661–667. [Google Scholar] [CrossRef]
- Sreevidya, V.S.; Srinivasa Rao, C.; Sullia, S.B.; Ladha, J.K.; Reddy, P.M. Metabolic Engineering of Rice with Soybean Isoflavone Synthase for Promoting Nodulation Gene Expression in Rhizobia. J. Exp. Bot. 2006, 57, 1957–1969. [Google Scholar] [CrossRef]
- Malla, A.; Shanmugaraj, B.; Srinivasan, B.; Sharma, A.; Ramalingam, S. Metabolic Engineering of Isoflavonoid Biosynthesis by Expressing Glycine Max Isoflavone Synthase in Allium Cepa L. for Genistein Production. Plants 2021, 10, 52. [Google Scholar] [CrossRef]
- Yang, D.; Park, S.Y.; Park, Y.S.; Eun, H.; Lee, S.Y. Metabolic Engineering of Escherichia Coli for Natural Product Biosynthesis. Trends Biotechnol. 2020, 38, 745–765. [Google Scholar] [CrossRef]
- Li, Z.; Jiang, Y.; Guengerich, F.P.; Ma, L.; Li, S.; Zhang, W. Engineering Cytochrome P450 Enzyme Systems for Biomedical and Biotechnological Applications. J. Biol. Chem. 2020, 295, 833–849. [Google Scholar] [CrossRef]
- Siminszky, B.; Corbin, F.T.; Ward, E.R.; Fleischmann, T.J.; Dewey, R.E. Expression of a Soybean Cytochrome P450 Monooxygenase CDNA in Yeast and Tobacco Enhances the Metabolism of Phenylurea Herbicides. Proc. Natl. Acad. Sci. USA 1999, 96, 1750–1755. [Google Scholar] [CrossRef] [PubMed]
- Wiriyaampaiwong, P.; Thanonkeo, S.; Thanonkeo, P. Molecular Characterization of Isoflavone Synthase Gene from Pueraria candollei var. mirifica. Afr. J. Agric. Res. 2012, 7, 4489–4498. [Google Scholar] [CrossRef]
- Pičmanová, M.; Reňák, D.; Feciková, J.; Růžička, P.; Mikšátková, P.; Lapčík, O.; Honys, D. Functional Expression and Subcellular Localization of Pea Polymorphic Isoflavone Synthase CYP93C18. Biol. Plant. 2013, 57, 635–645. [Google Scholar] [CrossRef]
- Misra, P.; Pandey, A.; Tewari, S.K.; Nath, P.; Trivedi, P.K. Characterization of Isoflavone Synthase Gene from Psoralea Corylifolia: A Medicinal Plant. Plant Cell Rep. 2010, 29, 747–755. [Google Scholar] [CrossRef]
- He, X.; Blount, J.W.; Ge, S.; Tang, Y.; Dixon, R.A. A Genomic Approach to Isoflavone Biosynthesis in Kudzu (Pueraria lobata). Planta 2011, 233, 843–855. [Google Scholar] [CrossRef]
- Kaur, N.; Murphy, J.B. Cloning, Characterization, and Functional Analysis of Cowpea Isoflavone Synthase (IFS) Homologs. J. Plant Mol. Biol. Biotechnol. PMBB 2010, 1. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).