Autoencoder Composite Scoring to Evaluate Prosthetic Performance in Individuals with Lower Limb Amputation
Abstract
1. Introduction
2. Methods
2.1. Study Design
2.2. Participants
2.3. Data and Statistical Analysis
2.4. Autoencoder Architecture
3. Results
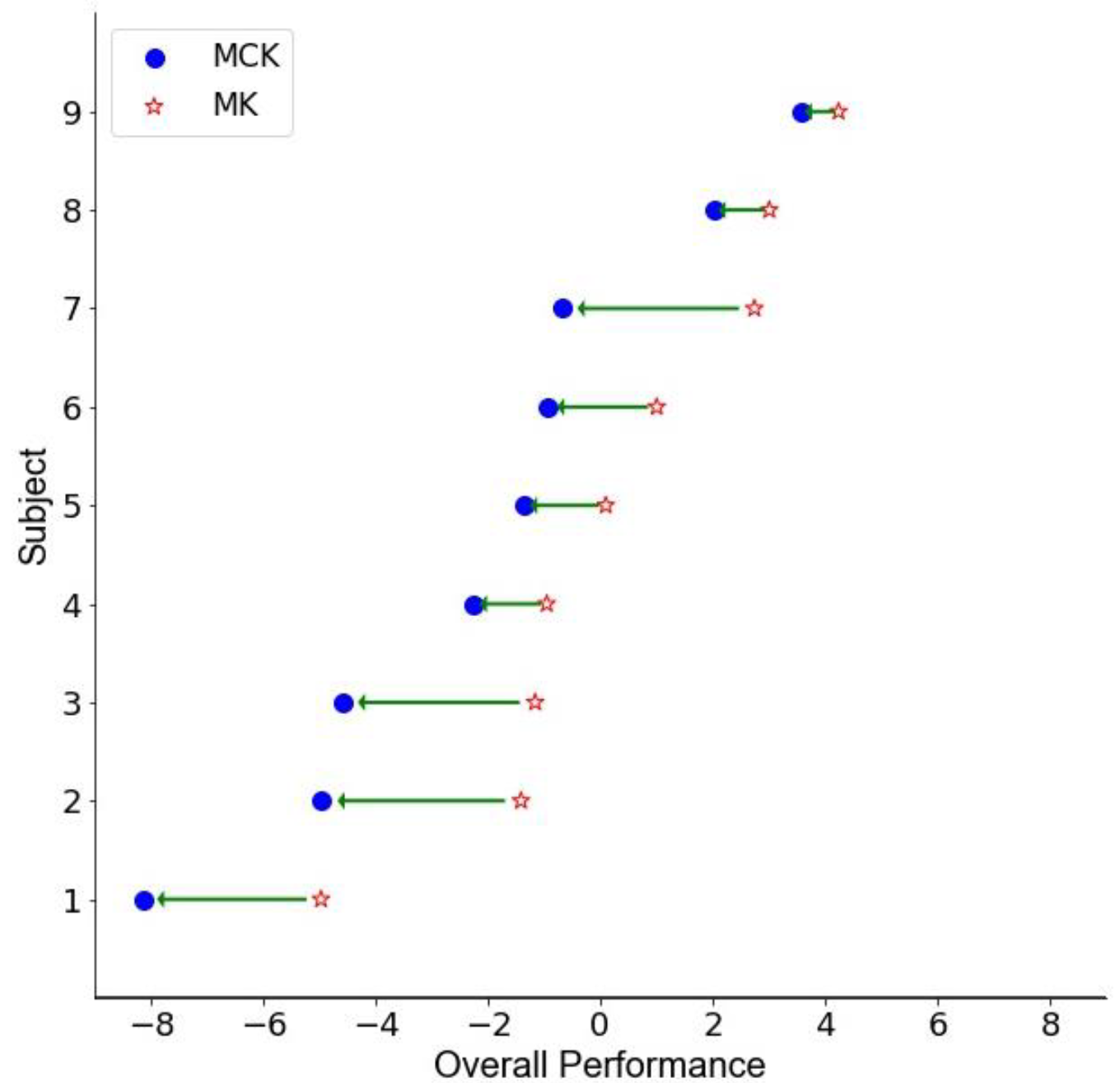
3.1. Performance and Self-Reported Measures
3.2. Autoencoder Composite Score
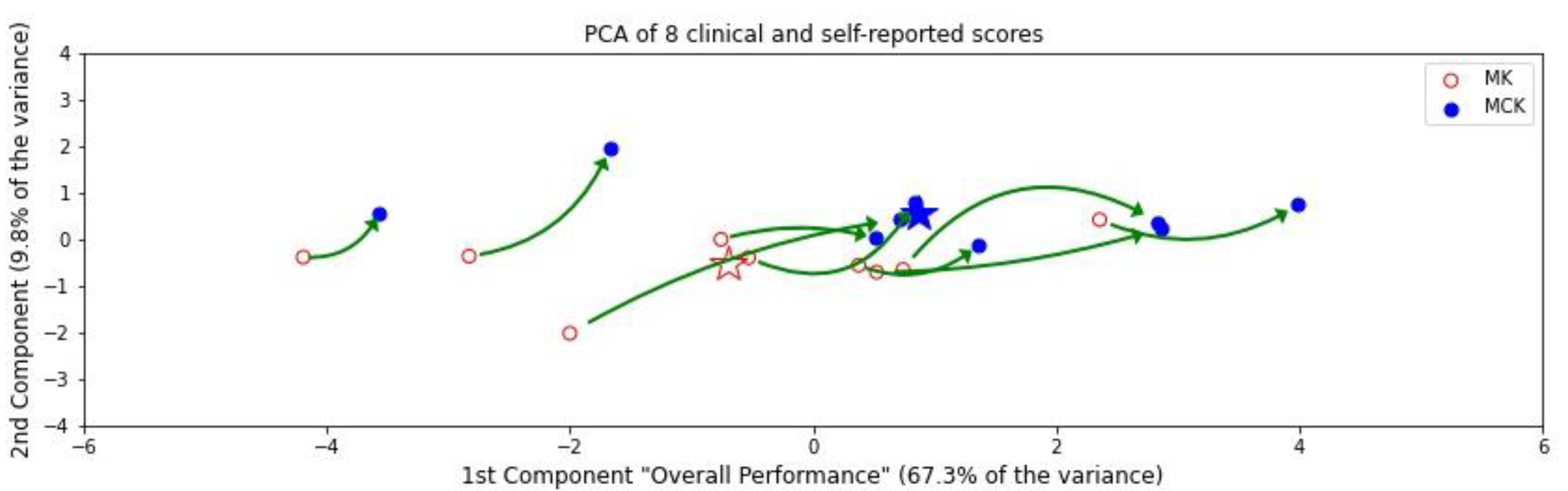
3.3. PCA Composite Score
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Üstün, T.B.; Chatterji, S.; Bickenbach, J.; Kostanjsek, N.; Schneider, M. The International Classification of Functioning, Disability and Health: A New Tool for Understanding Disability and Health. Disabil. Rehabil. 2003, 25, 565–571. [Google Scholar] [CrossRef] [PubMed]
- Renwick, R.; Nourhaghighi, N.; Manns, P.J.; Rudman, D.L. Quality of Life for People with Physical Disabilities: A New Instrument. Int. J. Rehabil. Res. 2003, 26, 279–287. [Google Scholar] [PubMed]
- Macduff, C. Respondent-Generated Quality of Life Measures: Useful Tools for Nursing or More Fool’s Gold? J. Adv. Nurs. 2000, 32, 375–382. [Google Scholar] [CrossRef] [PubMed]
- Wood-Dauphinee, S. Assessing Quality of Life in Clinical Research: From Where Have We Come and Where Are We Going? J. Clin. Epidemiol. 1999, 52, 355–363. [Google Scholar] [CrossRef]
- Xiao, T.; Albert, M.V. Big databig data in medical AI: How larger data sets lead to robust, automated learning for medicine. In Artificial Intelligence in Brain and Mental Health: Philosophical, Ethical & Policy Issues; Jotterand, F., Ienca, M., Eds.; Springer International Publishing: Cham, Switzerland, 2021; pp. 11–25. ISBN 9783030741884. [Google Scholar]
- Wang, L.; Sun, Y.; Li, Q.; Liu, T.; Yi, J. IMU-Based Gait Normalcy Index Calculation for Clinical Evaluation of Impaired Gait. IEEE J. Biomed. Health Inform 2021, 25, 3–12. [Google Scholar] [CrossRef]
- Domingos, P. A Few Useful Things to Know about Machine Learning. Commun. ACM 2012, 55, 78–87. [Google Scholar] [CrossRef]
- Simsek, M.; Obinikpo, A.A.; Kantarci, B. Deep learning in smart health: Methodologies, applications, challenges. In Connected Health in Smart Cities; El Saddik, A., Hossain, M.S., Kantarci, B., Eds.; Springer International Publishing: Cham, Switzerland, 2020; pp. 23–46. ISBN 9783030278441. [Google Scholar]
- Yang, Z.; Pou-Prom, C.; Jones, A.; Banning, M.; Dai, D.; Mamdani, M.; Oh, J.; Antoniou, T. Assessment of Natural Language Processing Methods for Ascertaining the Expanded Disability Status Scale Score from the Electronic Health Records of Patients With Multiple Sclerosis: Algorithm Development and Validation Study. JMIR Med. Inform. 2022, 10, e25157. [Google Scholar] [CrossRef]
- Nandigam, S.; Tabashum, T.; Xiao, T. Autoencoders and embeddings: How unsupervised structural learning enables fast and efficient goal-directed learning. In Bridging Human Intelligence and Artificial Intelligence; Albert, M.V., Lin, L., Spector, M.J., Dunn, L.S., Eds.; Springer International Publishing: Cham, Switzerland, 2022; pp. 47–63. ISBN 9783030847296. [Google Scholar]
- Kim, H.G.; Baddar, W.J.; Lim, H.-T.; Jeong, H.; Ro, Y.M. Measurement of exceptional motion in VR video contents for VR sickness assessment using deep convolutional autoencoder. In Proceedings of the 23rd ACM Symposium on Virtual Reality Software and Technology, Gothenburg, Sweden, 8–10 November 2017; pp. 1–7. [Google Scholar]
- Soni, M.H.; Patil, H.A. Novel deep autoencoder features for non-intrusive speech quality assessment. In Proceedings of the 2016 24th European Signal Processing Conference (EUSIPCO), Budapest, Hungary, 29 August–2 September 2016; pp. 2315–2319. [Google Scholar]
- Shaffie, A.; Soliman, A.; Ghazal, M.; Taher, F.; Dunlap, N.; Wang, B.; Van Berkel, V.; Gimelfarb, G.; Elmaghraby, A.; El-Baz, A. A novel autoencoder-based diagnostic system for early assessment of lung cancer. In Proceedings of the 2018 25th IEEE International Conference on Image Processing (ICIP), Athens, Greece, 7–10 October 2018; pp. 1393–1397. [Google Scholar]
- Wang, Y.; Yao, H.; Zhao, S. Auto-Encoder Based Dimensionality Reduction. Neurocomputing 2016, 184, 232–242. [Google Scholar] [CrossRef]
- Jolliffe, I.T.; Cadima, J. Principal Component Analysis: A Review and Recent Developments. Philos. Trans. A Math. Phys. Eng. Sci. 2016, 374, 20150202. [Google Scholar] [CrossRef]
- Gemperline, P. Principal Component Analysis. In Practical Guide to Chemometrics, 2nd ed.; CRC Press: Boca Raton, FL, USA, 2006; pp. 69–104. [Google Scholar]
- Plaut, E. From Principal Subspaces to Principal Components with Linear Autoencoders. arXiv 2018, arXiv:1804.10253. [Google Scholar]
- Baldi, P.; Hornik, K. Neural Networks and Principal Component Analysis: Learning from Examples without Local Minima. Neural Netw. 1989, 2, 53–58. [Google Scholar] [CrossRef]
- Shlens, J. A Tutorial on Principal Component Analysis. arXiv 2014, arXiv:1404.1100. [Google Scholar]
- Krakovska, O.; Christie, G.; Sixsmith, A.; Ester, M.; Moreno, S. Performance Comparison of Linear and Non-Linear Feature Selection Methods for the Analysis of Large Survey Datasets. PLoS ONE 2019, 14, e0213584. [Google Scholar] [CrossRef]
- Davis, C.; Fox, J. Sensitivity to Reward and Body Mass Index (BMI): Evidence for a Non-Linear Relationship. Appetite 2008, 50, 43–49. [Google Scholar] [CrossRef] [PubMed]
- Zeitzer, J.M.; Dijk, D.J.; Kronauer, R.; Brown, E.; Czeisler, C. Sensitivity of the Human Circadian Pacemaker to Nocturnal Light: Melatonin Phase Resetting and Suppression. J. Physiol. 2000, 526 Pt 3, 695–702. [Google Scholar] [CrossRef]
- Yu, S.; Príncipe, J.C. Understanding Autoencoders with Information Theoretic Concepts. Neural Netw. 2019, 117, 104–123. [Google Scholar] [CrossRef]
- Tschannen, M.; Bachem, O.; Lucic, M. Recent Advances in Autoencoder-Based Representation Learning. arXiv 2018, arXiv:1812.05069. [Google Scholar]
- Chan, C.K.; Hadjitheodorou, A.; Tsai, T.Y.-C.; Theriot, J.A. Quantitative Comparison of Principal Component Analysis and Unsupervised Deep Learning Using Variational Autoencoders for Shape Analysis of Motile Cells. bioRxiv 2020. [Google Scholar] [CrossRef]
- Franco, E.F.; Rana, P.; Cruz, A.; Calderón, V.V.; Azevedo, V.; Ramos, R.T.J.; Ghosh, P. Performance Comparison of Deep Learning Autoencoders for Cancer Subtype Detection Using Multi-Omics Data. Cancers 2021, 13, 2013. [Google Scholar] [CrossRef]
- Ziegler-Graham, K.; MacKenzie, E.J.; Ephraim, P.L.; Travison, T.G.; Brookmeyer, R. Estimating the Prevalence of Limb Loss in the United States: 2005 to 2050. Arch. Phys. Med. Rehabil. 2008, 89, 422–429. [Google Scholar] [CrossRef]
- Staff, N. Amputation Statistics by Cause, Limb Loss in the United States. Available online: file:///C:/Users/MDPI/Downloads/ACA%20Statistics.pdf (accessed on 15 September 2022).
- Owings, M.F.; Kozak, L.J. Ambulatory and Inpatient Procedures in the United States, 1996. Vital Health Stat. 13 1998, 13, 1–119. [Google Scholar]
- ADA. Statistics about Diabetes; ADA: Arlington, VA, USA, 2014. [Google Scholar]
- Skrepnek, G.H.; Mills, J.L.; Lavery, L.A.; Armstrong, D.G. Health Care Service and Outcomes among an Estimated 6.7 Million Ambulatory Care Diabetic Foot Cases in the US. Diabetes Care 2017, 40, 936–942. [Google Scholar] [CrossRef] [PubMed]
- Pandian, G. Rehabilitation of the patient with peripheral vascular disease and diabetic foot problems. In Rehabiliation Medicine: Principles and Practice, 3rd ed.; DeLisa, J.A., Gans, B.M., Eds.; Lippincott-Raven: Philadelphia, PA, USA, 1998. [Google Scholar]
- Li, G.; Liu, T.; Yi, J. Wearable Sensor System for Detecting Gait Parameters of Abnormal Gaits: A Feasibility Study. IEEE Sens. J. 2018, 18, 4234–4241. [Google Scholar] [CrossRef]
- Jun, K.; Lee, D.-W.; Lee, K.; Lee, S.; Kim, M.S. Feature Extraction Using an RNN Autoencoder for Skeleton-Based Abnormal Gait Recognition. IEEE Access 2020, 8, 19196–19207. [Google Scholar] [CrossRef]
- Tabashum, T.; Wang, S.-J.; Kruger, K.; Krzak, J.; Albert, M.V. Autoencoder-Derived Single Summary Metric to Assess Gait Quality. Arch. Phys. Med. Rehabil. 2021, 102, e95. [Google Scholar] [CrossRef]
- Patel, B.; Srikanthan, S.; Asanit, F.; Agu, E. Machine learning prediction of tbi from mobility, gait and balance patterns. In Proceedings of the 2021 IEEE/ACM Conference on Connected Health: Applications, Systems and Engineering Technologies (CHASE), Washington, DC, USA, 16–17 December 2021; pp. 11–22. [Google Scholar]
- Jayaraman, C.; Mummidisetty, C.K.; Albert, M.V.; Lipschutz, R.; Hoppe-Ludwig, S.; Mathur, G.; Jayaraman, A. Using a Microprocessor Knee (C-Leg) with Appropriate Foot Transitioned Individuals with Dysvascular Transfemoral Amputations to Higher Performance Levels: A Longitudinal Randomized Clinical Trial. J. NeuroEng. Rehabil. 2021, 18, 88. [Google Scholar] [CrossRef]
- Gehring, J.; Miao, Y.; Metze, F.; Waibel, A. Extracting deep bottleneck features using stacked auto-encoders. In Proceedings of the 2013 IEEE International Conference on Acoustics, Speech and Signal Processing, Vancouver, BC, Canada, 26–31 May 2013; pp. 3377–3381. [Google Scholar]
- Hinton, G.E.; Salakhutdinov, R.R. Reducing the Dimensionality of Data with Neural Networks. Science 2006, 313, 504–507. [Google Scholar] [CrossRef]
- Anaissi, A.; Zandavi, S.M. Multi-Objective Variational Autoencoder: An Application for Smart Infrastructure Maintenance. arXiv 2020, arXiv:2003.05070. [Google Scholar] [CrossRef]
- Baldi, P. Autoencoders, Unsupervised Learning, and Deep Architectures. J. Mach. Learn. Res. 2012, 27, 37–50. [Google Scholar]
- Liou, C.-Y.; Cheng, W.-C.; Liou, J.-W.; Liou, D.-R. Autoencoder for Words. Neurocomputing 2014, 139, 84–96. [Google Scholar] [CrossRef]
- Ghimire, S.; Deo, R.C.; Casillas-Pérez, D.; Salcedo-Sanz, S. Improved Complete Ensemble Empirical Mode Decomposition with Adaptive Noise Deep Residual Model for Short-Term Multi-Step Solar Radiation Prediction. Renew. Energy 2022, 190, 408–424. [Google Scholar] [CrossRef]
- Li, H.; Deng, J.; Feng, P.; Pu, C.; Arachchige, D.D.K.; Cheng, Q. Short-Term Nacelle Orientation Forecasting Using Bilinear Transformation and ICEEMDAN Framework. Front. Energy Res. 2021, 9, 780928. [Google Scholar] [CrossRef]
- Parmar, G.; Li, D.; Lee, K.; Tu, Z. Dual Contradistinctive Generative Autoencoder. arXiv 2020, arXiv:2011.10063. [Google Scholar]
- Ghimire, S.; Deo, R.C.; Wang, H.; Al-Musaylh, M.S.; Casillas-Pérez, D.; Salcedo-Sanz, S. Stacked LSTM Sequence-to-Sequence Autoencoder with Feature Selection for Daily Solar Radiation Prediction: A Review and New Modeling Results. Energies 2022, 15, 1061. [Google Scholar] [CrossRef]

| Layer | Layer Type | Number Nodes | Activation |
|---|---|---|---|
| 1 | Input | 8 | |
| 2 | Dense | 24 | relu |
| 3 | Dropout | 0.4 | |
| 4 | Dense | 1 | linear |
| 5 | Dropout | 0.4 | |
| 6 | Dense | 24 | relu |
| 7 | Output | 8 |
| Test | Conditions | |
|---|---|---|
| MK (Mean ± Std) | MCK (Mean ± Std) | |
| 6 MWT | 137.48 ± 85.63 | 145.43 ± 110.30 |
| 10 MWT | 22.27 ± 9.72 | 15.3 ± 7.24 |
| BERG | 37.11 ± 7.5 | 43.56 ± 13.20 |
| FSST | 17.37 ± 5.04 | 16.79 ± 11.17 |
| TUG | 27.47 ±14.96 | 25.32 ± 14.14 |
| AMP | 31.44 ± 7.04 | 35.67 ± 5.40 |
| MFES | 7.78 ± 1.14 | 9.33 ± 0.69 |
| PEQ-amb | 60.63 ± 18.75 | 81.92 ± 18.74 |
| AE | 0.29 ± 0.89 | −1.92 ± 1.13 |
| Test | Load Factors | Direction of Improvement in Scores |
|---|---|---|
| AMP | 0.395 | Higher |
| BERG | 0.388 | Higher |
| 6 MWT | 0.332 | Higher |
| PEQ-amb | 0.313 | Higher |
| MFES | 0.286 | Higher |
| FSST | −0.355 | Lower |
| 10 MWT | −0.362 | Lower |
| TUG | −0.383 | Lower |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tabashum, T.; Xiao, T.; Jayaraman, C.; Mummidisetty, C.K.; Jayaraman, A.; Albert, M.V. Autoencoder Composite Scoring to Evaluate Prosthetic Performance in Individuals with Lower Limb Amputation. Bioengineering 2022, 9, 572. https://doi.org/10.3390/bioengineering9100572
Tabashum T, Xiao T, Jayaraman C, Mummidisetty CK, Jayaraman A, Albert MV. Autoencoder Composite Scoring to Evaluate Prosthetic Performance in Individuals with Lower Limb Amputation. Bioengineering. 2022; 9(10):572. https://doi.org/10.3390/bioengineering9100572
Chicago/Turabian StyleTabashum, Thasina, Ting Xiao, Chandrasekaran Jayaraman, Chaithanya K. Mummidisetty, Arun Jayaraman, and Mark V. Albert. 2022. "Autoencoder Composite Scoring to Evaluate Prosthetic Performance in Individuals with Lower Limb Amputation" Bioengineering 9, no. 10: 572. https://doi.org/10.3390/bioengineering9100572
APA StyleTabashum, T., Xiao, T., Jayaraman, C., Mummidisetty, C. K., Jayaraman, A., & Albert, M. V. (2022). Autoencoder Composite Scoring to Evaluate Prosthetic Performance in Individuals with Lower Limb Amputation. Bioengineering, 9(10), 572. https://doi.org/10.3390/bioengineering9100572







