Heparan Sulfate Glycosaminoglycan Is Predicted to Stabilize Inflammatory Infiltrate Formation and RANKL/OPG Ratio in Severe Periodontitis in Humans
Abstract
1. Introduction
2. Materials and Methods
2.1. Samples Procurement
2.2. Immunohistochemistry and Immunofluorescence Staining
2.3. Image Acquisition and Processing
2.4. Histomorphometry and IF Signal Quantification
2.5. Statistical Analysis and Modeling
3. Results
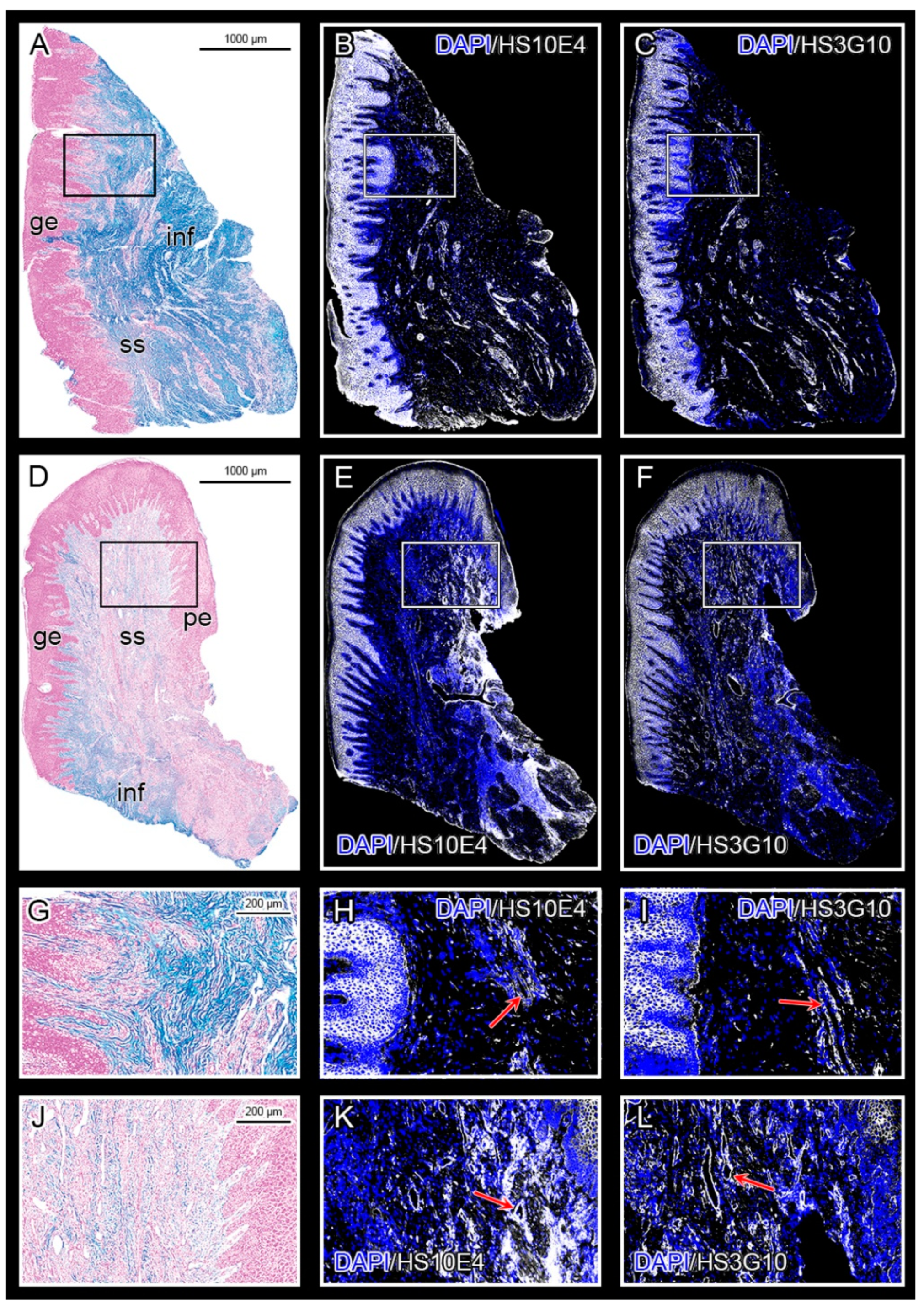
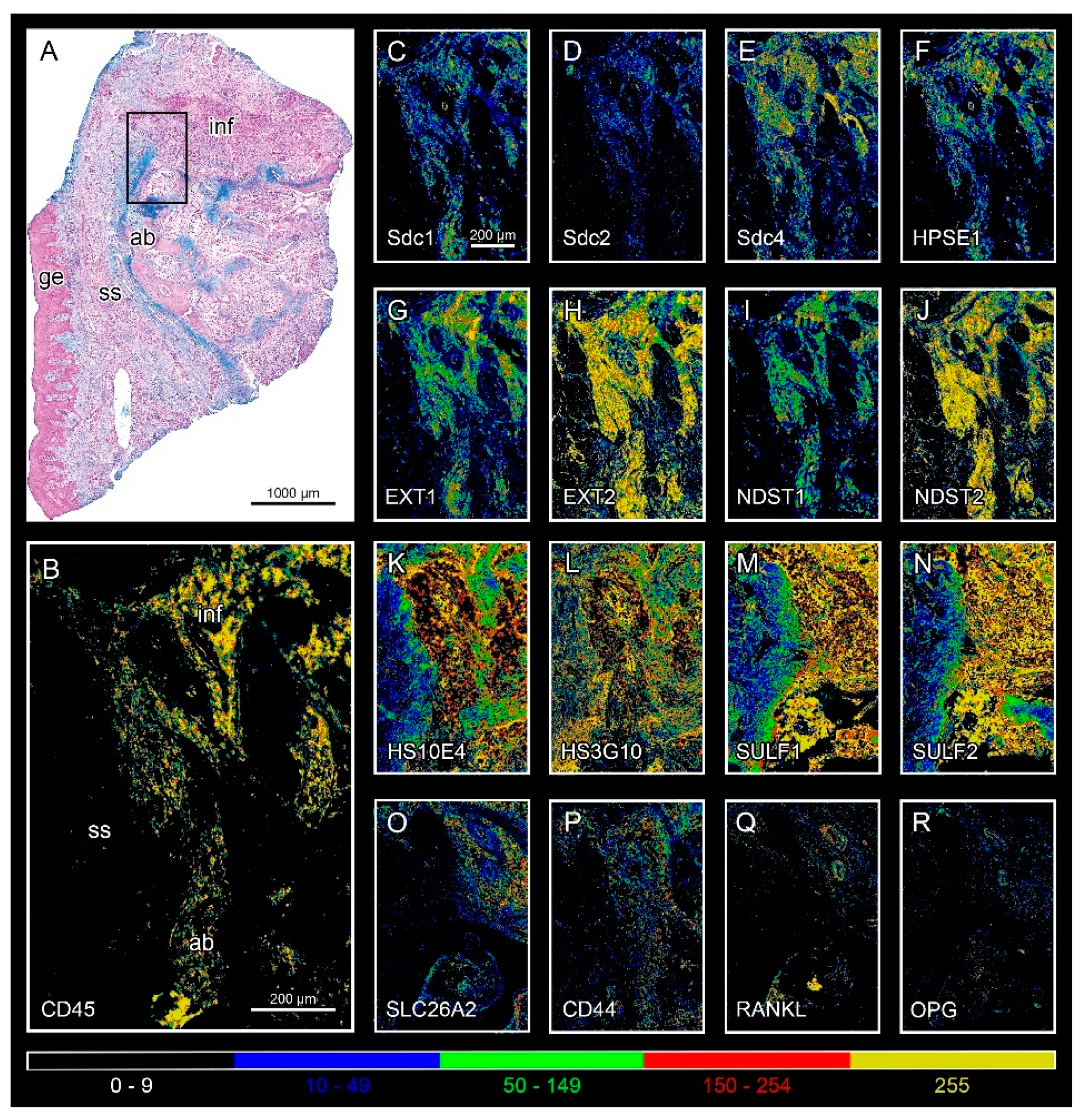
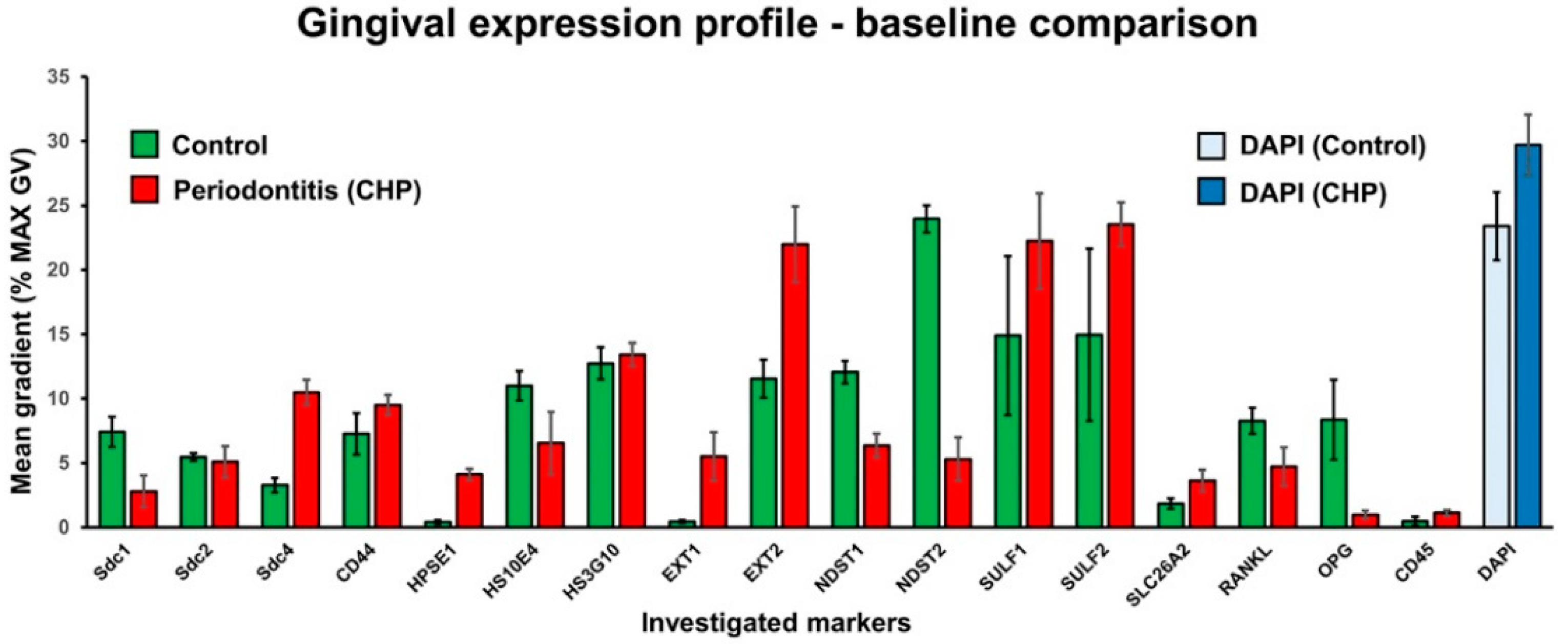
3.1. Expression of HS GAG and Associated Factors in Healthy Gingiva and in Gingiva of Patients Affected by Periodontitis
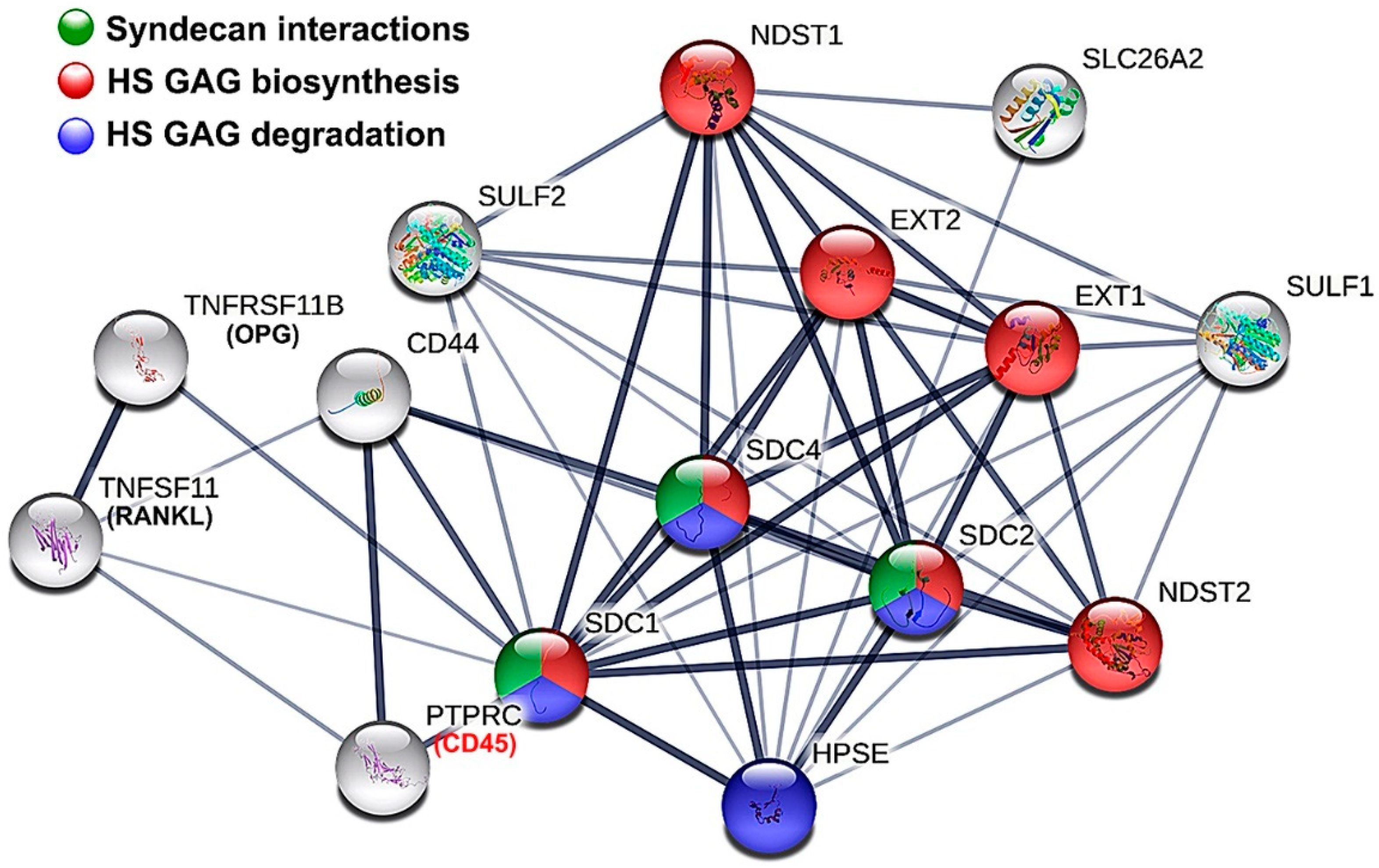
3.2. Parameters of Regression Equation System
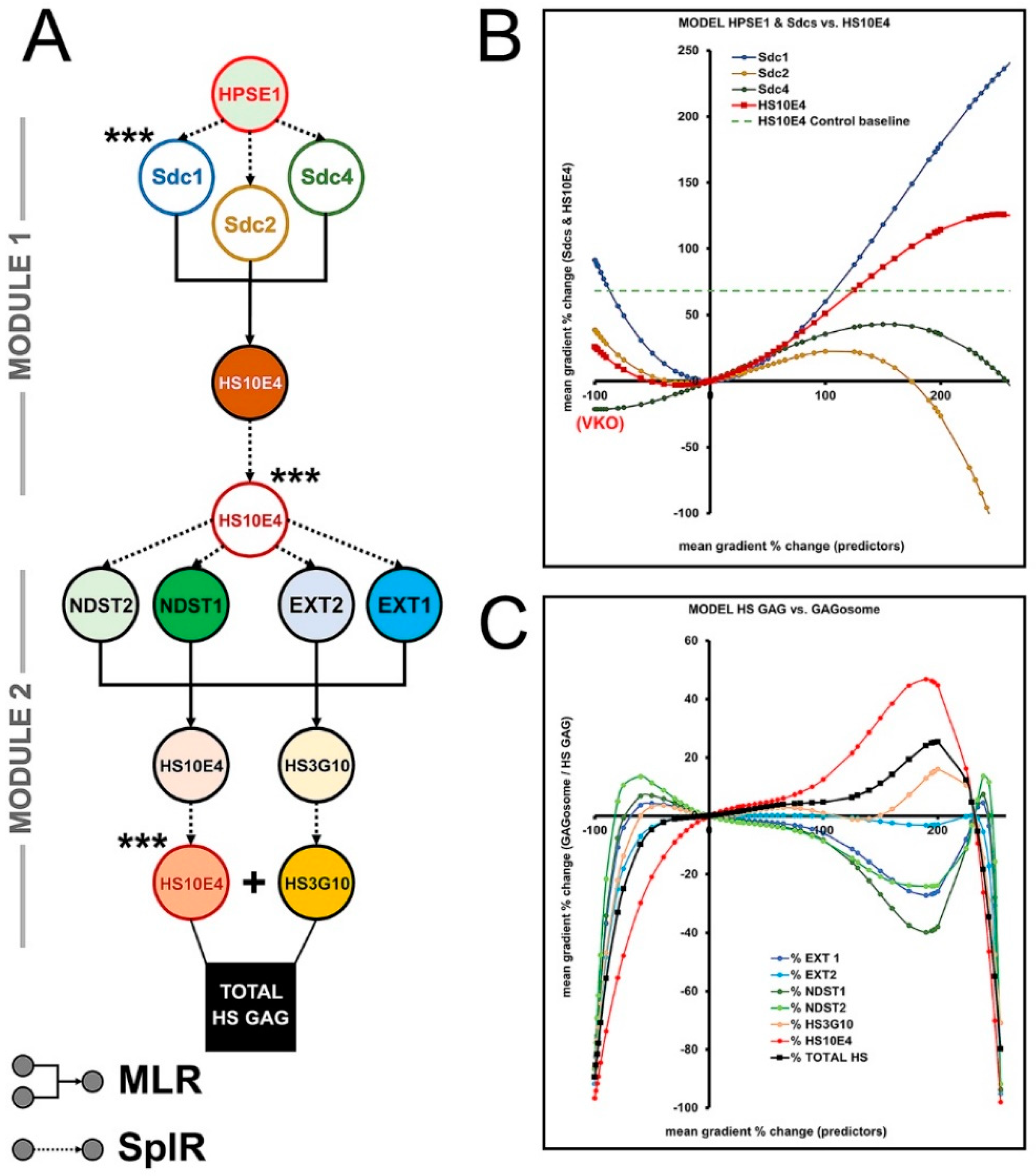
3.3. The Effects of HPSE1 and Sdcs on the Expression/ECM Content of HS GAG—Module 1
3.4. Biosynthesis of HS GAG and Feedback Loop between HS GAG and GAGosome Enzymes—Module 2
3.5. The Effects of SULF1-2 and SLC26A2 on Changes in Sulfation of HS GAG—Module 3
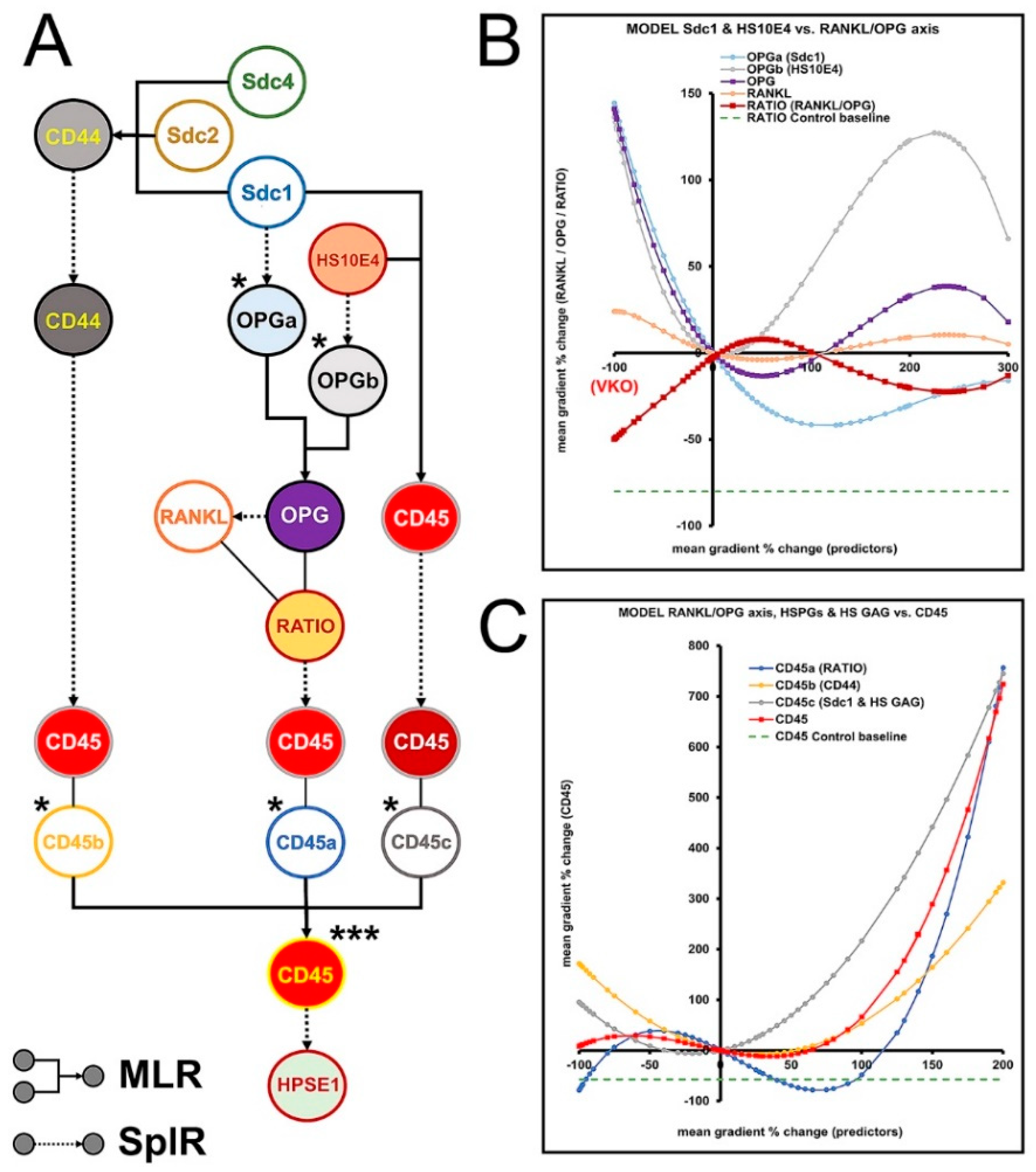
3.6. Modulation of RANKL/OPG Ratio and CD45 Expression by HS GAG—Module 4
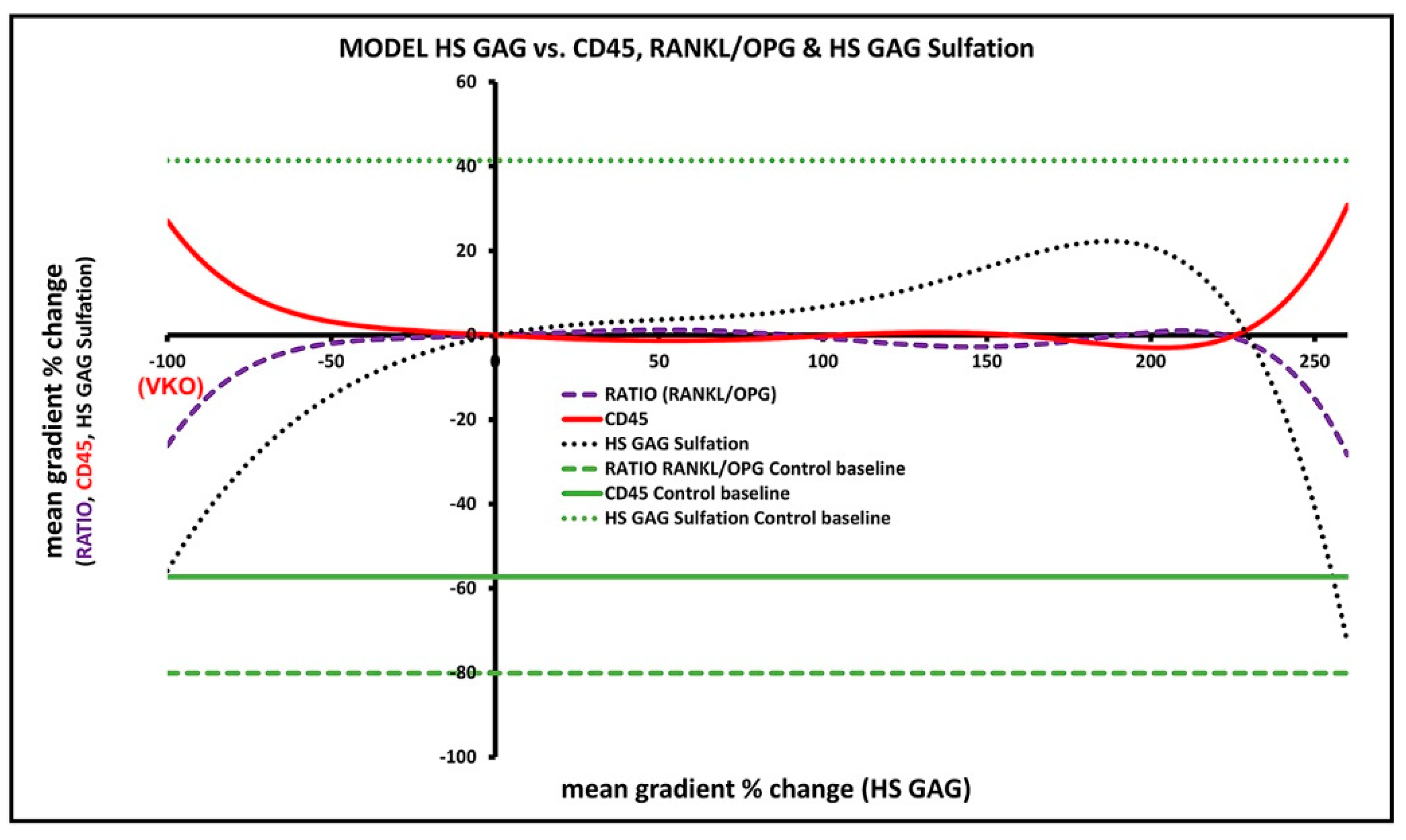
3.7. Predicted Effects of Native HS GAG on Gingival Inflammatory Infiltrate and Alveolar Bone Formation/Degradation from the Overall Model
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Slots, J. Periodontitis: Facts, fallacies and the future. Periodontology 2000 2017, 75, 7–23. [Google Scholar] [CrossRef] [PubMed]
- Tonetti, M.S.; Greenwell, H.; Kornman, K.S. Staging and grading of periodontitis: Framework and proposal of a new classification and case definition. J. Periodontol. 2018, 89 (Suppl. 1), S159–S172. [Google Scholar] [CrossRef] [PubMed]
- Tonetti, M.S.; Jepsen, S.; Jin, L.; Otomo-Corgel, J. Impact of the global burden of periodontal diseases on health, nutrition and wellbeing of mankind: A call for global action. J. Clin. Periodontol. 2017, 44, 456–462. [Google Scholar] [CrossRef] [PubMed]
- Alexopoulou, A.N.; Multhaupt, H.A.; Couchman, J.R. Syndecans in wound healing, inflammation and vascular biology. Int. J. Biochem. Cell Biol. 2007, 39, 505–528. [Google Scholar] [CrossRef] [PubMed]
- Bonnans, C.; Chou, J.; Werb, Z. Remodelling the extracellular matrix in development and disease. Nat. Rev. Mol. Cell Biol. 2014, 15, 786–801. [Google Scholar] [CrossRef] [PubMed]
- Stepp, M.A.; Pal-Ghosh, S.; Tadvalkar, G.; Pajoohesh-Ganji, A. Syndecan-1 and Its Expanding List of Contacts. Adv. Wound Care 2015, 4, 235–249. [Google Scholar] [CrossRef] [PubMed]
- Teng, Y.H.-F.; Aquino, R.S.; Park, P.W. Molecular functions of syndecan-1 in disease. Matrix Biol. 2012, 31, 3–16. [Google Scholar] [CrossRef] [PubMed]
- Gopal, S. Syndecans in Inflammation at a Glance. Front. Immunol. 2020, 11, 227. [Google Scholar] [CrossRef]
- Szatmári, T.; Mundt, F.; Heidari-Hamedani, G.; Zong, F.; Ferolla, E.; Alexeyenko, A.; Hjerpe, A.; Dobra, K. Novel Genes and Pathways Modulated by Syndecan-1: Implications for the Proliferation and Cell-Cycle Regulation of Malignant Mesothelioma Cells. PLoS ONE 2012, 7, e48091. [Google Scholar] [CrossRef] [PubMed]
- Eszatmári, T.; Edobra, K. The Role of Syndecan-1 in Cellular Signaling and its Effects on Heparan Sulfate Biosynthesis in Mesenchymal Tumors. Front. Oncol. 2013, 3, 310. [Google Scholar] [CrossRef]
- Talsma, D.T.; Katta, K.; Ettema, M.A.B.; Kel, B.; Kusche-Gullberg, M.; Daha, M.R.; Stegeman, C.A.; van den Born, J.; Wang, L. Endothelial heparan sulfate deficiency reduces inflammation and fibrosis in murine diabetic nephropathy. Lab. Investig. 2018, 98, 427–438. [Google Scholar] [CrossRef] [PubMed]
- Sanderson, R.D.; Elkin, M.; Rapraeger, A.C.; Ilan, N.; Vlodavsky, I. Heparanase regulation of cancer, autophagy and inflammation: New mechanisms and targets for therapy. FEBS J. 2016, 284, 42–55. [Google Scholar] [CrossRef] [PubMed]
- Na Ge, X.; Bastan, I.; Gil Ha, S.; Greenberg, Y.G.; Esko, J.D.; Rao, S.P.; Sriramarao, P. Regulation of eosinophil recruitment and allergic airway inflammation by heparan sulfate proteoglycan (HSPG) modifying enzymes. Exp. Lung Res. 2018, 44, 98–112. [Google Scholar] [CrossRef]
- Andreas, D. Implications of Heparan Sulfate and Heparanase in Inflammatory Diseases. Ph.D. Dissertation, Uppsala University, Uppsala, Sweden, 2017. [Google Scholar]
- Sanz, M.; Herrera, D.; Kebschull, M.; Chapple, I.; Jepsen, S.; Berglundh, T.; Sculean, A.; Tonetti, M.S.; Aass, A.M.; Aimetti, M.; et al. Treatment of stage I–III periodontitis—The EFP S3 level clinical practice guideline. J. Clin. Periodontol. 2020, 47 (Suppl. 22), 4–60. [Google Scholar] [CrossRef] [PubMed]
- Coyac, B.R.; Detzen, L.; Doucet, P.; Baroukh, B.; Llorens, A.; Bonnaure-Mallet, M.; Gosset, M.; Barritault, D.; Colombier, M.-L.; Saffar, J.-L. Periodontal reconstruction by heparan sulfate mimetic-based matrix therapy in Porphyromonas gingivalis-infected mice. Heliyon 2018, 4, e00719. [Google Scholar] [CrossRef]
- Papapanou, P.N.; Sanz, M.; Buduneli, N.; Dietrich, T.; Feres, M.; Fine, D.H.; Flemmig, T.F.; Garcia, R.; Giannobile, W.V.; Graziani, F.; et al. Periodontitis: Consensus report of workgroup 2 of the 2017 World Workshop on the Classification of Periodontal and Peri-Implant Diseases and Conditions. J. Periodontol. 2018, 89 (Suppl. 1), S173–S182. [Google Scholar] [CrossRef]
- Duplancic, R.; Roguljic, M.; Puhar, I.; Vecek, N.; Dragun, R.; Vukojevic, K.; Saraga-Babic, M.; Kero, D. Syndecans and Enzymes for Heparan Sulfate Biosynthesis and Modification Differentially Correlate With Presence of Inflammatory Infiltrate in Periodontitis. Front. Physiol. 2019, 10, 1248. [Google Scholar] [CrossRef]
- World Medical Association. World Medical Association Declaration of Helsinki: Ethical principles for medical research involving human subjects. JAMA 2013, 310, 2191–2194. [Google Scholar] [CrossRef]
- Kero, D.; Cigic, L.; Mikic, I.M.; Galic, T.; Cubela, M.; Vukojevic, K.; Saraga-Babic, M. Involvement of IGF-2, IGF-1R, IGF-2R and PTEN in development of human tooth germ—An immunohistochemical study. Organogenesis 2016, 12, 152–167. [Google Scholar] [CrossRef]
- Kero, D.; Vukojevic, K.; Stazic, P.; Sundov, D.; Brakus, S.M.; Saraga-Babic, M. Regulation of proliferation in developing human tooth germs by MSX homeodomain proteins and cyclin-dependent kinase inhibitor p19INK4d. Organogenesis 2017, 13, 141–155. [Google Scholar] [CrossRef]
- Duplancic, R.; Kero, D. Novel approach for quantification of multiple immunofluorescent signals using histograms and 2D plot profiling of whole-section panoramic images. Sci. Rep. 2021, 11, 8619. [Google Scholar] [CrossRef] [PubMed]
- Kero, D.; Ćavar, I. Correlation of the expression of hyaluronan and CD44 with the presence of gingival inflammatory infiltrate in advanced generalized periodontitis. St Open 2020, 1, 1–16. [Google Scholar] [CrossRef]
- Kero, D.; Bilandzija, T.S.; Arapovic, L.L.; Vukojevic, K.; Saraga-Babić, M. Syndecans and Enzymes Involved in Heparan Sulfate Biosynthesis and Degradation Are Differentially Expressed During Human Odontogenesis. Front. Physiol. 2018, 9, 732. [Google Scholar] [CrossRef] [PubMed]
- von Mering, C.; Huynen, M.; Jaeggi, D.; Schmidt, S.; Bork, P.; Snel, B. STRING: A database of predicted functional associations between proteins. Nucleic Acids Res. 2003, 31, 258–261. [Google Scholar] [CrossRef]
- Yaddanapudi, L.N. The American Statistical Association statement on P- values explained. J. Anaesthesiol. Clin. Pharmacol. 2016, 32, 421–423. [Google Scholar] [CrossRef] [PubMed]
- Jin, H.; Zhou, S.; Yang, S.; Cao, H.-M. Heparanase overexpression down-regulates syndecan-1 expression in a gallbladder carcinoma cell line. J. Int. Med Res. 2017, 45, 662–672. [Google Scholar] [CrossRef] [PubMed]
- David, G.; Zimmermann, P. Heparanase tailors syndecan for exosome production. Mol. Cell. Oncol. 2016, 3, e1047556. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Yu, S.; Lv, H.; Zhang, H.; Jiang, Y.; Hong, Y.; Xia, R.; Zhang, Q.; Ju, W.; Jiang, L.; Ou, G.; et al. Heparanase-1-induced shedding of heparan sulfate from syndecan-1 in hepatocarcinoma cell facilitates lymphatic endothelial cell proliferation via VEGF-C/ERK pathway. Biochem. Biophys. Res. Commun. 2017, 485, 432–439. [Google Scholar] [CrossRef] [PubMed]
- Esko, J.D.; Selleck, S.B. Order Out of Chaos: Assembly of Ligand Binding Sites in Heparan Sulfate. Annu. Rev. Biochem. 2002, 71, 435–471. [Google Scholar] [CrossRef]
- Carlsson, P.; Presto, J.; Spillmann, D.; Lindahl, U.; Kjellén, L. Heparin/Heparan Sulfate Biosynthesis: Processive formation of N-sulfated domains. J. Biol. Chem. 2008, 283, 20008–20014. [Google Scholar] [CrossRef]
- McCormick, C.; LeDuc, Y.; Martindale, D.; Mattison, K.; Esford, L.; Dyer, A.; Tufaro, F. The putative tumour suppressor EXT1 alters the expression of cell-surface heparan sulfate. Nat. Genet. 1998, 19, 158–161. [Google Scholar] [CrossRef] [PubMed]
- Presto, J.; Thuveson, M.; Carlsson, P.; Busse, M.; Wilén, M.; Eriksson, I.; Kusche-Gullberg, M.; Kjellén, L. Heparan sulfate biosynthesis enzymes EXT1 and EXT2 affect NDST1 expression and heparan sulfate sulfation. Proc. Natl. Acad. Sci. USA 2008, 105, 4751–4756. [Google Scholar] [CrossRef]
- Deligny, A.; Dierker, T.; Dagälv, A.; Lundequist, A.; Eriksson, I.; Nairn, A.; Moremen, K.W.; Merry, C.; Kjellén, L. NDST2 (N-Deacetylase/N-Sulfotransferase-2) Enzyme Regulates Heparan Sulfate Chain Length. J. Biol. Chem. 2016, 291, 18600–18607. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Wang, F.; Sheng, J. “Coding” and “Decoding”: Hypothesis for the regulatory mechanism involved in heparan sulfate biosynthesis. Carbohydr. Res. 2016, 428, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Buresh-Stiemke, R.A.; Malinowski, R.L.; Keil, K.P.; Vezina, C.M.; Oosterhof, A.; Van Kuppevelt, T.H.; Marker, P.C. Distinct expression patterns of Sulf1 and Hs6st1 spatially regulate heparan sulfate sulfation during prostate development. Dev. Dyn. 2012, 241, 2005–2013. [Google Scholar] [CrossRef]
- Kim, H.-J.; Kim, H.-S.; Hong, Y.-H. Sulfatase 1 and sulfatase 2 as novel regulators of macrophage antigen presentation and phagocytosis. Yeungnam Univ. J. Med. 2021, 38, 326–336. [Google Scholar] [CrossRef]
- Hästbacka, J.; Superti-Furga, A.; Wilcox, W.R.; Rimoin, D.L.; Cohn, D.H.; Lander, E.S. Atelosteogenesis type II is caused by mutations in the diastrophic dysplasia sulfate-transporter gene (DTDST): Evidence for a phenotypic series involving three chondrodysplasias. Am. J. Hum. Genet. 1996, 58, 255–262. [Google Scholar]
- Alper, S.L.; Sharma, A.K. The SLC26 gene family of anion transporters and channels. Mol. Asp. Med. 2013, 34, 494–515. [Google Scholar] [CrossRef]
- Kelly, T.; Suva, L.J.; Nicks, K.M.; MacLeod, V.; Sanderson, R.D. Tumor-derived syndecan-1 mediates distal cross-talk with bone that enhances osteoclastogenesis. J. Bone Miner. Res. 2010, 25, 1295–1304. [Google Scholar] [CrossRef]
- Benad-Mehner, P.; Thiele, S.; Rachner, T.D.; Göbel, A.; Rauner, M.; Hofbauer, L.C. Targeting syndecan-1 in breast cancer inhibits osteoclast functions through up-regulation of osteoprotegerin. J. Bone Oncol. 2013, 3, 18–24. [Google Scholar] [CrossRef]
- Timmen, M.; Hidding, H.; Götte, M.; El Khassawna, T.; Kronenberg, D.; Stange, R. The heparan sulfate proteoglycan Syndecan-1 influences local bone cell communication via the RANKL/OPG axis. Sci. Rep. 2020, 10, 20510. [Google Scholar] [CrossRef] [PubMed]
- Agere, S.A.; Kim, E.Y.; Akhtar, N.; Ahmed, S. Syndecans in chronic inflammatory and autoimmune diseases: Pathological insights and therapeutic opportunities. J. Cell. Physiol. 2017, 233, 6346–6358. [Google Scholar] [CrossRef] [PubMed]
- Morla, S. Glycosaminoglycans and Glycosaminoglycan Mimetics in Cancer and Inflammation. Int. J. Mol. Sci. 2019, 20, 1963. [Google Scholar] [CrossRef] [PubMed]
- Misra, S.; Hascall, V.C.; Markwald, R.R.; Ghatak, S. Interactions between Hyaluronan and Its Receptors (CD44, RHAMM) Regulate the Activities of Inflammation and Cancer. Front. Immunol. 2015, 6, 201. [Google Scholar] [CrossRef] [PubMed]
- Nedvetzki, S.; Gonen, E.; Assayag, N.; Reich, R.; Williams, R.O.; Thurmond, R.L.; Huang, J.-F.; Neudecker, B.A.; Wang, F.-S.; Turley, E.A.; et al. RHAMM, a receptor for hyaluronan-mediated motility, compensates for CD44 in inflamed CD44-knockout mice: A different interpretation of redundancy. Proc. Natl. Acad. Sci. USA 2004, 101, 18081–18086. [Google Scholar] [CrossRef]
- Man, M.; Elias, P.M.; Man, W.; Wu, Y.; Bourguignon, L.Y.W.; Feingold, K.R.; Man, M.-Q. The role of CD44 in cutaneous inflammation. Exp. Dermatol. 2009, 18, 962–968. [Google Scholar] [CrossRef] [PubMed]
- Escartin, Q.; Lallam-Laroye, C.; Baroukh, B.; Morvan, F.O.; Caruelle, J.P.; Godeau, G.; Barritault, D.; Saffar, J.-L. A new approach to treat tissue destruction in periodontitis with chemically modified dextran polymers. FASEB J. 2003, 17, 644–651. [Google Scholar] [CrossRef] [PubMed]
- Lallam-Laroye, C.; Escartin, Q.; Zlowodzki, A.-S.; Barritault, D.; Caruelle, J.-P.; Baroukh, B.; Saffar, J.-L.; Colombier, M.-L. Periodontitis destructions are restored by synthetic glycosaminoglycan mimetic. J. Biomed. Mater. Res. Part A 2006, 79, 675–683. [Google Scholar] [CrossRef] [PubMed]
- Lallam-Laroye, C.; Baroukh, B.; Doucet, P.; Barritault, D.; Saffar, J.-L.; Colombier, M.-L. ReGeneraTing Agents Matrix Therapy Regenerates a Functional Root Attachment in Hamsters with Periodontitis. Tissue Eng. Part A 2011, 17, 2359–2367. [Google Scholar] [CrossRef] [PubMed]
- De Vries, T.J.; Andreotta, S.; Loos, B.G.; Nicu, E.A. Genes Critical for Developing Periodontitis: Lessons from Mouse Models. Front. Immunol. 2017, 8, 1395. [Google Scholar] [CrossRef] [PubMed]
- Marchesan, J.; Girnary, M.S.; Jing, L.; Miao, M.Z.; Zhang, S.; Sun, L.; Morelli, T.; Schoenfisch, M.H.; Inohara, N.; Offenbacher, S.; et al. An experimental murine model to study periodontitis. Nat. Protoc. 2018, 13, 2247–2267. [Google Scholar] [CrossRef] [PubMed]
- Martinon, P.; Fraticelli, L.; Giboreau, A.; Dussart, C.; Bourgeois, D.; Carrouel, F. Nutrition as a Key Modifiable Factor for Periodontitis and Main Chronic Diseases. J. Clin. Med. 2021, 10, 197. [Google Scholar] [CrossRef] [PubMed]
- Khocht, A.; Orlich, M.; Paster, B.; Bellinger, D.; Lenoir, L.; Irani, C.; Fraser, G. Cross-sectional comparisons of subgingival microbiome and gingival fluid inflammatory cytokines in periodontally healthy vegetarians versus non-vegetarians. J. Periodontal Res. 2021, 56, 1079–1090. [Google Scholar] [CrossRef] [PubMed]
- Silva, N.; Abusleme, L.; Bravo, D.; Dutzan, N.; Garcia-Sesnich, J.; Vernal, R.; Hernández, M.; Gamonal, J. Host response mechanisms in periodontal diseases. J. Appl. Oral Sci. 2015, 23, 329–355. [Google Scholar] [CrossRef] [PubMed]
- Schubert, W.; Bonnekoh, B.; Pommer, A.J.; Philipsen, L.; Böckelmann, R.; Malykh, Y.; Gollnick, H.; Friedenberger, M.; Bode, M.; Dress, A.W.M. Analyzing proteome topology and function by automated multidimensional fluorescence microscopy. Nat. Biotechnol. 2006, 24, 1270–1278. [Google Scholar] [CrossRef] [PubMed]
- Park, C.C.; Georgescu, W.; Polyzos, A.; Pham, C.; Ahmed, K.M.; Zhang, H.; Costes, S.V. Rapid and automated multidimensional fluorescence microscopy profiling of 3D human breast cultures. Integr. Biol. 2013, 5, 681–691. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kero, D.; Saraga-Babic, M. Odontogenesis—A Masterful Orchestration of Functional Redundancy or What Makes Tooth Bioengineering an Intrinsically Difficult Concept. Int. J. Stem Cell Res. Ther. 2016, 1, 7. [Google Scholar] [CrossRef][Green Version]
- Zhang, S.; Yu, N.; Arce, R.M. Periodontal inflammation: Integrating genes and dysbiosis. Periodontology 2000 2020, 82, 129–142. [Google Scholar] [CrossRef]
- Chanalaris, A.; Clarke, H.; Guimond, S.E.; Vincent, T.L.; Turnbull, J.E.; Troeberg, L. Heparan Sulfate Proteoglycan Synthesis Is Dysregulated in Human Osteoarthritic Cartilage. Am. J. Pathol. 2019, 189, 632–647. [Google Scholar] [CrossRef] [PubMed]
- Bishop, J.R.; Schuksz, M.; Esko, J.D. Heparan sulphate proteoglycans fine-tune mammalian physiology. Nature 2007, 446, 1030–1037. [Google Scholar] [CrossRef]

| Primary Antibody | Dilution | Description/Function |
|---|---|---|
| Mouse monoclonal anti-Sdc1 (ab34164) * a | 1:100 | HSPGs; type 1 transmembrane proteins; cell surface receptors for HS GAG which participate in cell proliferation, cell migration and cell-matrix interactions; members of syndecan protein family |
| Rabbit polyclonal anti-Sdc2 (ab191062) * b | 1:200 | |
| Rabbit polyclonal anti-Sdc4 (ab24511) * b | 1:100 | |
| Rabbit polyclonal anti-EXT1 (ab126305) * b | 1:100 | Exostosins; endoplasmic reticulum resident type 2 transmembrane glycosyltransferases; involved in biosynthesis of HS GAG (chain elongation step); members of the GAGosome complex |
| Rabbit polyclonal anti-EXT2 (ab102843) * b | 1:50 | |
| Rabbit polyclonal anti-NDST1 (ab129248) * b | 1:50 | Bifunctional enzymes involved in biosynthesis of HS GAG; catalyze N-deacetylation and N-sulfation of glucosamine monosaccharides of HS GAG (initial sulfation step); members of the GAGosome complex |
| Rabbit polyclonal anti-NDST2 (ab151141) * b | 1:100 | |
| Mouse monoclonal anti-HS3G10 (370260-1) † c | 1:100 | HS GAG marker; reacts with HS GAG 3G10 epitope generated after the digestion of HS GAG with heparinase III enzyme; binds to non-sulfated regions of HS GAG; does not react with other classes of GAGs |
| Mouse monoclonal anti-HS10E4 (370255-1) † c | 1:100 | HS GAG marker; reacts with HS GAG 10E4 epitope which includes N-sulfated glucosamines; reactivity abolished after treatment with heparinase III enzyme; does not react with other classes of GAGs |
| Rabbit polyclonal anti-HPSE1 (ab85543) * b | 1:200 | Heparanase 1 enzyme (endoglycosidase); degrades HS GAG at cell surface and within ECM into shorter oligosaccharide fragments; facilitates shedding of syndecans’ extracellular domains |
| Rabbit polyclonal anti-SULF1 (ab32763) * b | 1:200 | Heparan sulfate 6-O-endo-sulfatases; selectively remove 6-O-sulfate groups from HS chains of HSPGs (HS GAG desulfation); modulate activity of HS GAG by altering binding sites for signaling molecules |
| Mouse monoclonal [2B4] anti-SULF2 (ab113405) * c | 1:25 | |
| Rabbit polyclonal anti-SLC26A2 (ab238591) * b | 1:200 | Transmembrane glycoprotein from the solute carrier protein family; involved in the transport of cellular sulfate ions essential for sulfation of HS GAG. |
| Rabbit polyclonal anti-CD44 (ab157107) * b | 1:500 | Cell-surface receptor for hyaluronan; also binds HS GAG and interacts with Sdcs, collagens, matrix metalloproteinases; involved in cell- interactions, cell adhesion and migration |
| Mouse monoclonal anti-CD45 (PTPRC) (ab8216) * c | 1:200 | Protein tyrosine phosphatase; type 1 transmembrane protein present in differentiated hematopoietic cells of myeloid and lymphoid lineage (common leukocyte antigen) |
| Rabbit polyclonal anti-RANKL (TNFSF11) (LS-B1425-0.05) ‡ b | 1:200 | Receptor activator of nuclear kappa-Β ligand (TNF receptor superfamily); type II membrane protein involved in bone remodeling and immune reactions; promotes differentiation and activation of osteoclasts and specific populations of immune cells |
| Rabbit polyclonal anti-OPG (TNFRSF11B) (ab73400) * b | 1:500 | Osteoclastogenesis inhibitory factor (TNF receptor superfamily); decoy receptor for RANKL; inhibits osteoclastogenesis and bone resorption |
| Model Parameters | ||||||||
|---|---|---|---|---|---|---|---|---|
| Predictors | Outcomes | R2 | Df † | FIT ‡ | Knots | MODULE § | ||
| Marker | Mean * | Std. Error | ||||||
| HPSE1 | Sdc1 | 2.80123 | 0.03626 | 0.99912 | 1 | SplR | 545 | 1 |
| Sdc2 | 5.07486 | 0.00304 | 0.99999 | 1 | 579 | |||
| Sdc4 | 10.49152 | 0.00395 | 0.99998 | 1 | 570 | |||
| Sdcs | HS10E4 | 6.54086 | 0.32258 | 0.98241 | 3 | MLR | N/A | |
| HS10E4 | 6.54091 | 0.00663 | 0.99999 | 1 | SplR | 358 | ||
| HS10E4 | EXT1 | 5.50899 | 0.00479 | 0.99999 | 1 | 226 | 2 | |
| EXT2 | 21.97313 | 0.01919 | 0.99996 | 1 | 188 | |||
| NDST1 | 6.346521 | 0.00357 | 0.99998 | 1 | 357 | |||
| NDST2 | 5.29767 | 0.00963 | 0.99997 | 1 | 217 | |||
| EXTs, NDSTs | HS10E4 | 6.54091 | 0.31644 | 0.99795 | 4 | MLR | N/A | |
| HS3G10 | 13.41192 | 0.42029 | 0.99902 | 4 | ||||
| HS10E4 | HS10E4 | 6.54103 | 0.02772 | 0.99987 | 1 | SplR | 185 | |
| HS3G10 | HS3G10 | 13.41494 | 0.01222 | 0.99981 | 1 | 451 | ||
| HPSE1, Sdcs, EXTs, NDSTs | SULF1 | 22.24313 | 0.55832 | 0.97743 | 7 | MLR | N/A | 3 |
| SULF2 | 23.54117 | 0.34153 | 0.95915 | 7 | ||||
| HPSE1, NDST1 | SLC26A2 | 3.62778 | 0.53376 | 0.58391 | 2 | |||
| SULF1 | SULF1 | 22.24361 | 0.01709 | 0.99998 | 1 | SplR | 642 | |
| SULF2 | SULF2 | 23.54061 | 0.01529 | 0.99992 | 1 | 730 | ||
| SLC26A2 | SLC26A2 | 3.62767 | 0.00372 | 0.99998 | 1 | 646 | ||
| SULFs, SLC26A2 | HS10E4 | 6.54181 | 0.37448 | 0.97629 | 3 | MLR | N/A | |
| HS10E4 | HS10E4 | 6.54103 | 0.00194 | 0.99999 | 1 | SplR | 1104 | |
| Sdc1 | OPG | 0.97662 | 0.00158 | 0.99998 | 1 | SplR | 454 | 4 |
| HS10E4 | 0.97665 | 0.00209 | 0.99996 | 1 | 437 | |||
| OPGa, OPGb | 0.97671 | 0.00125 | 0.99998 | 2 | MLR | N/A | ||
| OPG | RANKL | 4.72004 | 0.00279 | 0.99999 | 1 | SplR | 495 | |
| RANKL/OPG RATIO | CD45 | 1.11445 | 0.00105 | 0.99999 | 1 | 776 | ||
| Sdcs | CD44 | 9.51209 | 0.64461 | 0.32535 | 3 | MLR | N/A | |
| CD44 | CD44 | 9.51218 | 0.00318 | 0.99998 | 1 | SplR | 712 | |
| CD44 | CD45 | 1.11444 | 0.00188 | 0.99993 | 1 | 888 | ||
| Sdc1, HS10E4 | CD45 | 1.11445 | 0.13179 | 0.66525 | 2 | MLR | N/A | |
| CD45 | CD45 | 1.11445 | 0.00076 | 0.99998 | 1 | SplR | 683 | |
| CD45a, CD45b, CD45c | CD45 | 1.11445 | 0.00049 | 0.99999 | 3 | MLR | N/A | |
| CD45 | HPSE1 | 4.10009 | 0.00481 | 0.99989 | 1 | SplR | 841 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Duplancic, R.; Roguljic, M.; Bozic, D.; Kero, D. Heparan Sulfate Glycosaminoglycan Is Predicted to Stabilize Inflammatory Infiltrate Formation and RANKL/OPG Ratio in Severe Periodontitis in Humans. Bioengineering 2022, 9, 566. https://doi.org/10.3390/bioengineering9100566
Duplancic R, Roguljic M, Bozic D, Kero D. Heparan Sulfate Glycosaminoglycan Is Predicted to Stabilize Inflammatory Infiltrate Formation and RANKL/OPG Ratio in Severe Periodontitis in Humans. Bioengineering. 2022; 9(10):566. https://doi.org/10.3390/bioengineering9100566
Chicago/Turabian StyleDuplancic, Roko, Marija Roguljic, Darko Bozic, and Darko Kero. 2022. "Heparan Sulfate Glycosaminoglycan Is Predicted to Stabilize Inflammatory Infiltrate Formation and RANKL/OPG Ratio in Severe Periodontitis in Humans" Bioengineering 9, no. 10: 566. https://doi.org/10.3390/bioengineering9100566
APA StyleDuplancic, R., Roguljic, M., Bozic, D., & Kero, D. (2022). Heparan Sulfate Glycosaminoglycan Is Predicted to Stabilize Inflammatory Infiltrate Formation and RANKL/OPG Ratio in Severe Periodontitis in Humans. Bioengineering, 9(10), 566. https://doi.org/10.3390/bioengineering9100566







