RTF-RCNN: An Architecture for Real-Time Tomato Plant Leaf Diseases Detection in Video Streaming Using Faster-RCNN
Abstract
1. Introduction
- To detect tomato plant leaf disease automatically, a faster R-CNN model is proposed;
- The suggested model uses both images and video to detect tomato plant leaf disease;
- In terms of accuracy, loss, Precision, Recall, and F-Measure, the suggested methodologies are compared to existing models such as Alex net and generic CNN.
2. Related Works
3. Materials and Methods
3.1. Region Convolutional Neural Networks (R-CNN)
- The main limitation of R-CNN is slow training. Its training phase increases if there are more areas or objects to detect or classify;
- Secondly, as it takes a long time to train, the R-CNN cannot be considered a real-time detector as its detection process takes more time for simulation.
3.2. Fast R-CNN
- Firstly, it takes the amount of 20 s for every single test image. And it is a slow detection process;
- Secondly, it is still not accurate for real-time data detection;
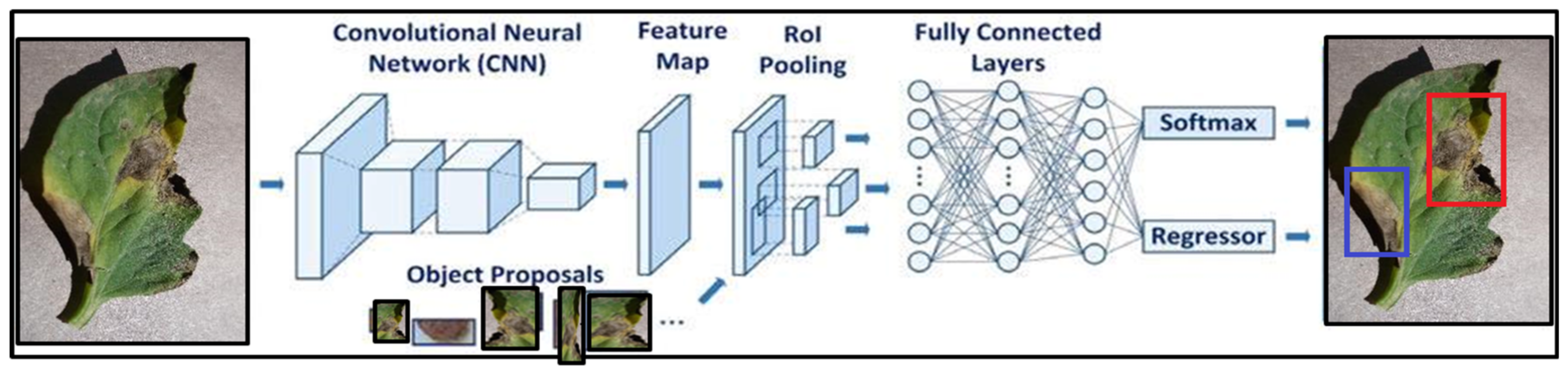
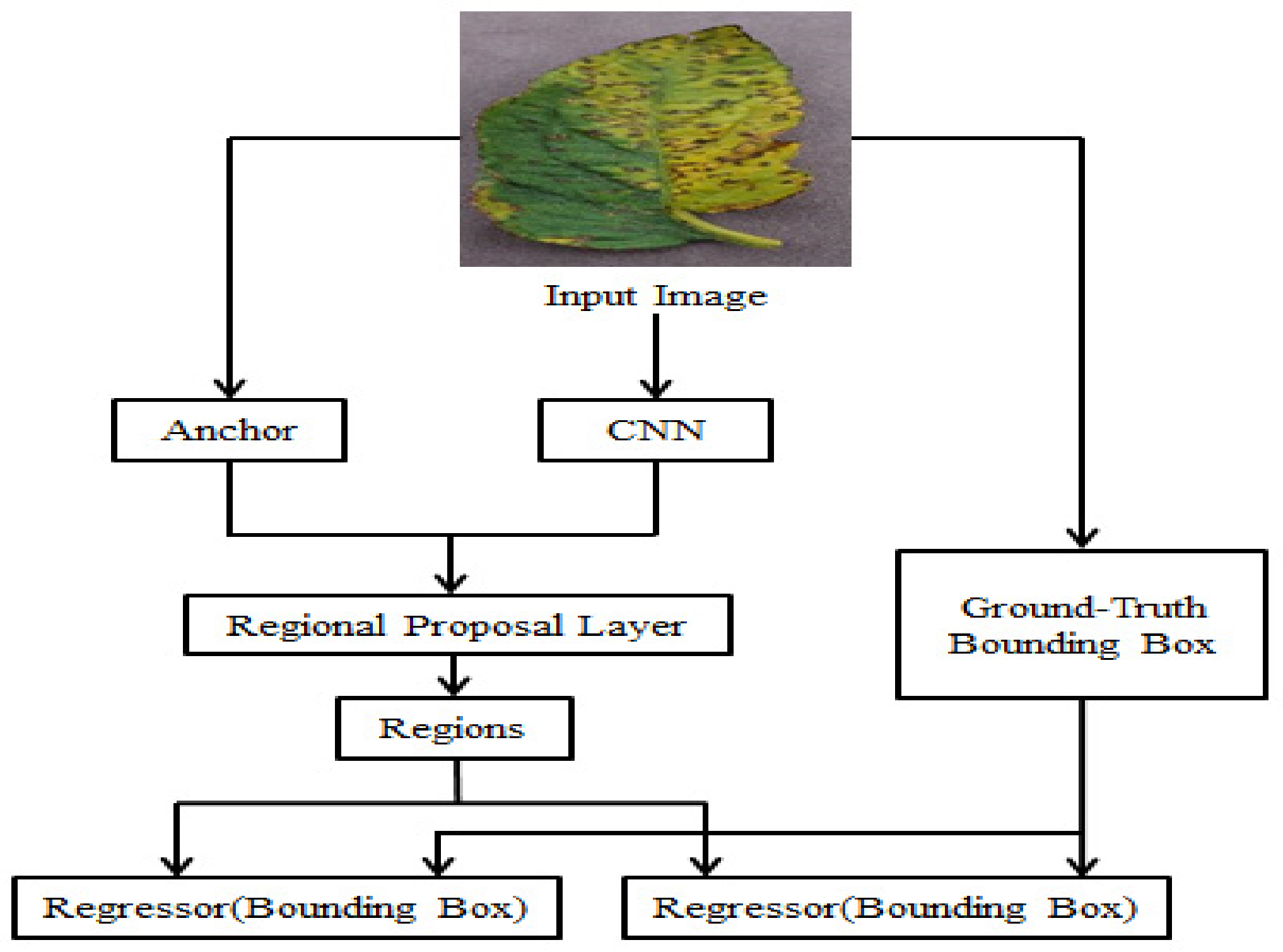
3.3. Faster R-CNN
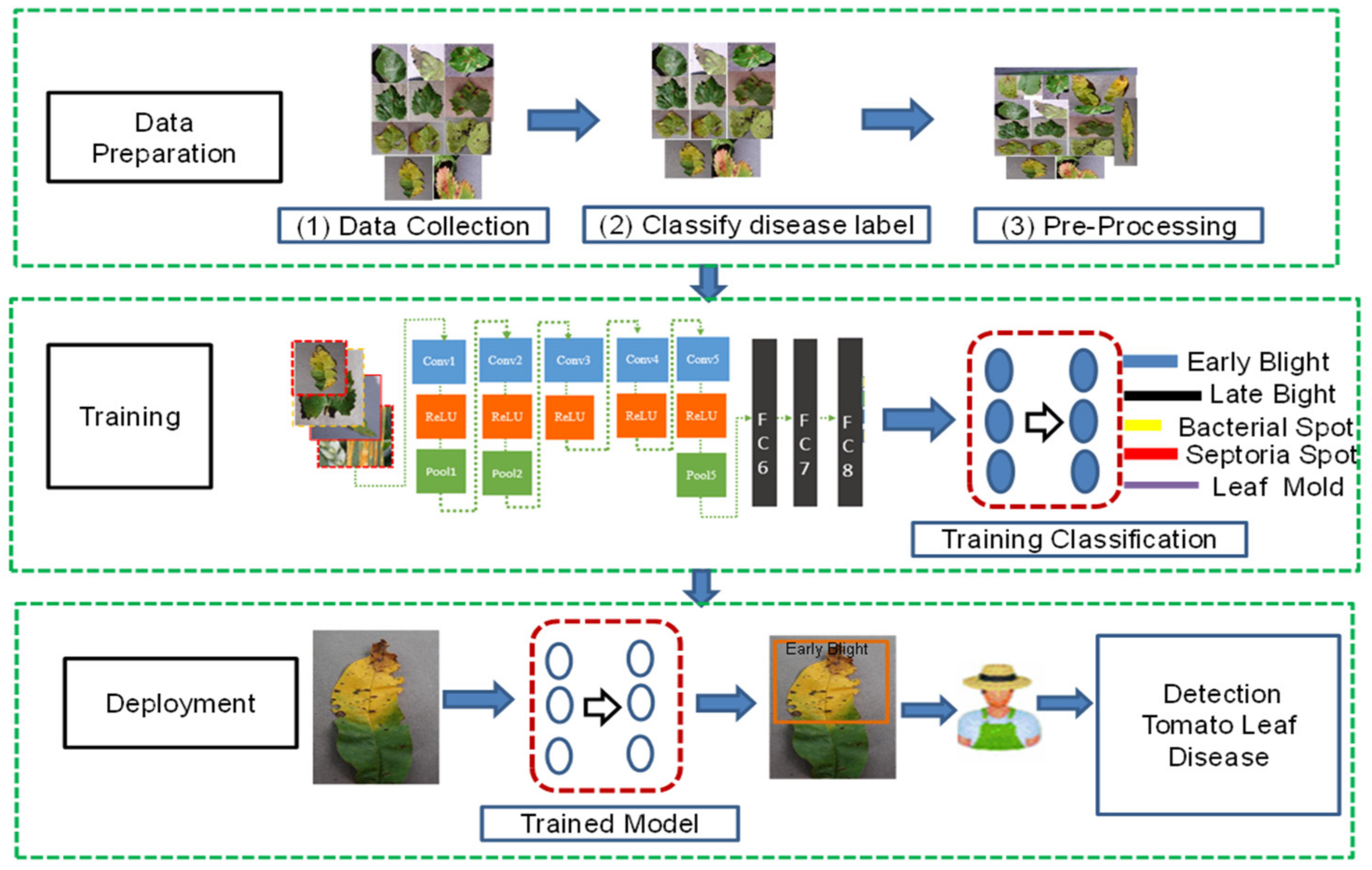
3.4. Proposed Research Framework
4. Data Collection
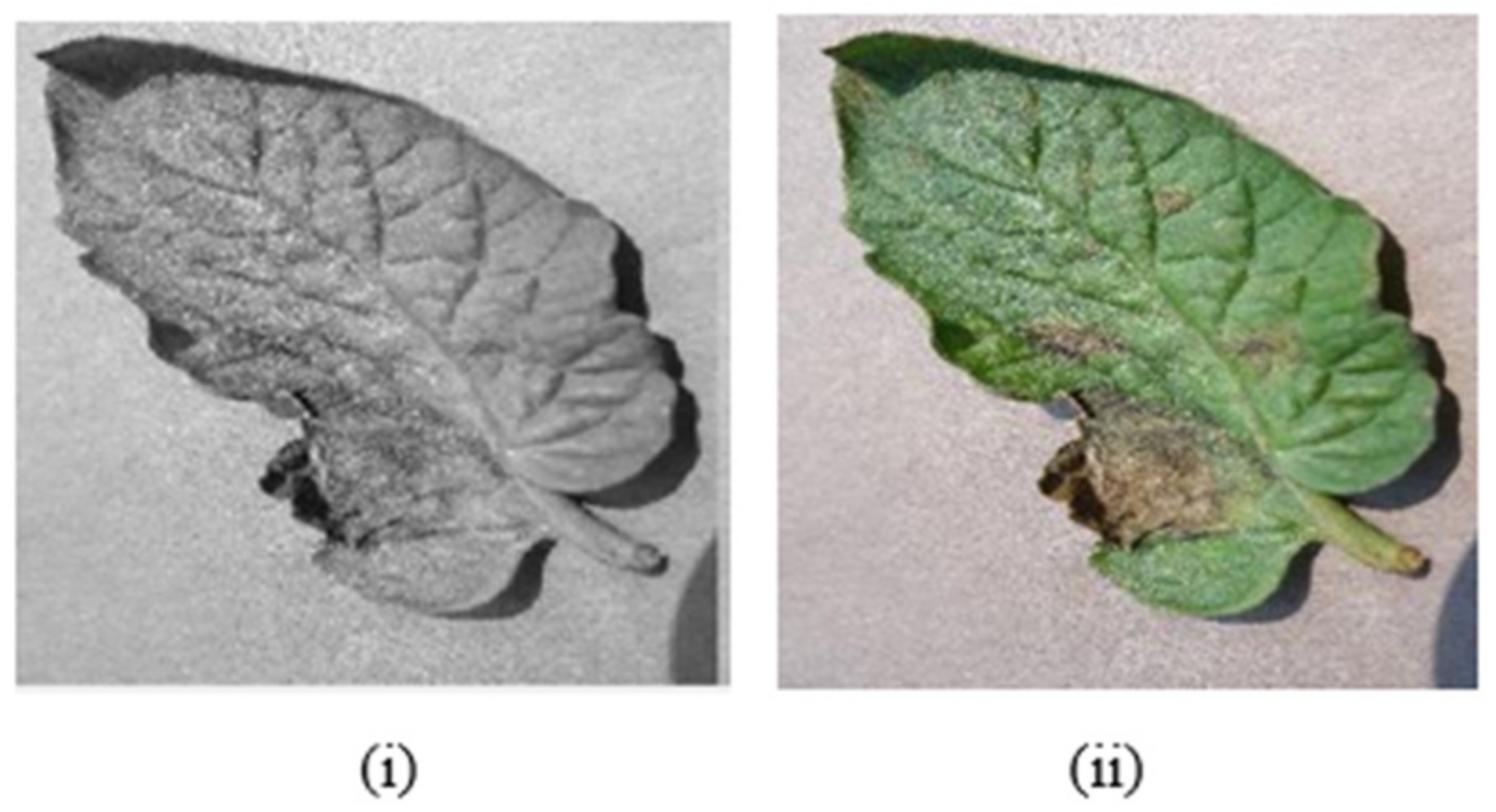
5. Data Preprocessing
5.1. Resizing Images
5.2. Image Enhancement
5.3. Noise Removal
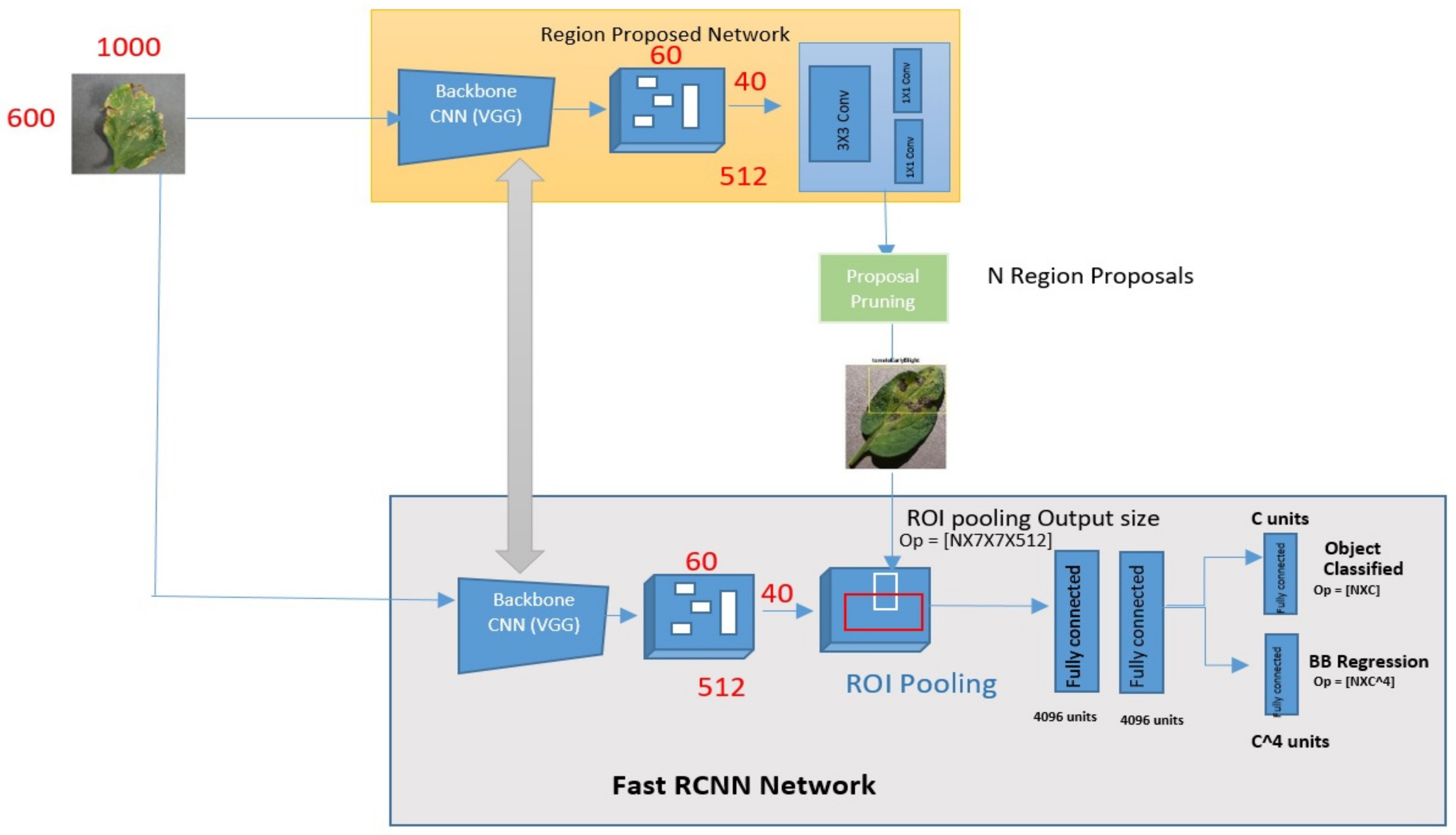
5.4. Proposed Faster R-CNN
- Taking the corresponding region from a backbone feature map to a proposal;
- By partitioning the region into a fixed number of sub-images;
- Using max-pooling on sub-windows, you can get a fixed-size output.
| Algorithm 1 Proposed model algorithm |
| Input: input images/video to the Faster R-CNN model Output: display the result of the tomato plant leaf with detected the affected part. Start Step 1: initialize the structure of the proposed model and initial parameters Step 2: Load the input data Step 3: Label session of Label Data Step 4: Save Session of Label Data Step 5: Load Label Session data for Training Step 6: Determine the Total number of Images Path in Training Step 7: Initialize proposed Faster R_CNN model Step 8: read (Size of Label Session) For 1 to N Calculate error For End Step 9: For K to Epochs Number Apply in Proposed Method CON=>ReLU=>CON CON=>ReLU=>POOL Set Fully Connected (FC)=> SofMax Return Netwrok Architecture Constructed For End Step 10: Visualization and Process results Post End |
6. Experiments and Results
6.1. Preliminaries
6.2. Results
- A faster R-CNN has been proposed;
- Alex net and;
- CNN.
6.3. Convolutional Neural Network (CNN) Model
6.4. Alex Net Model
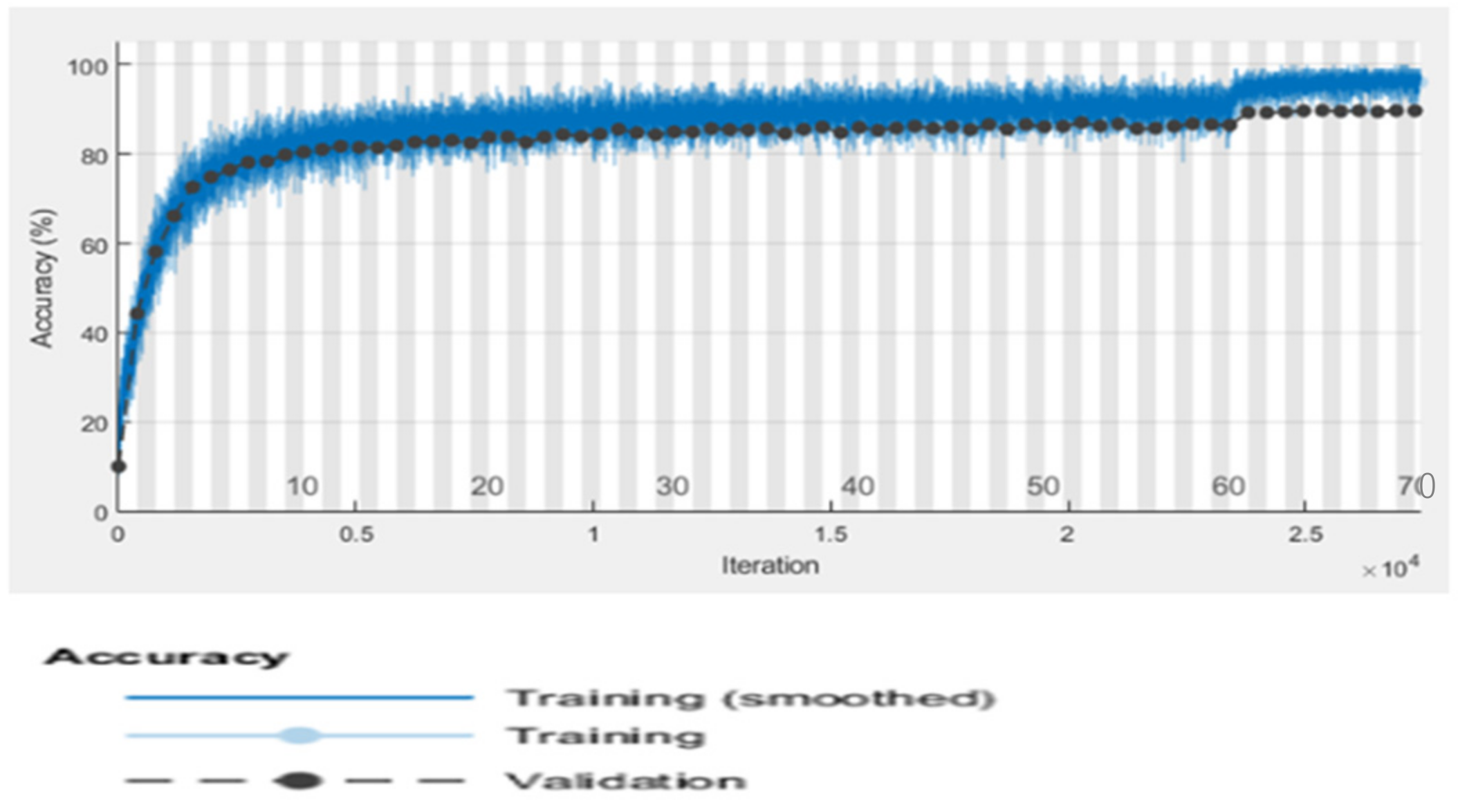
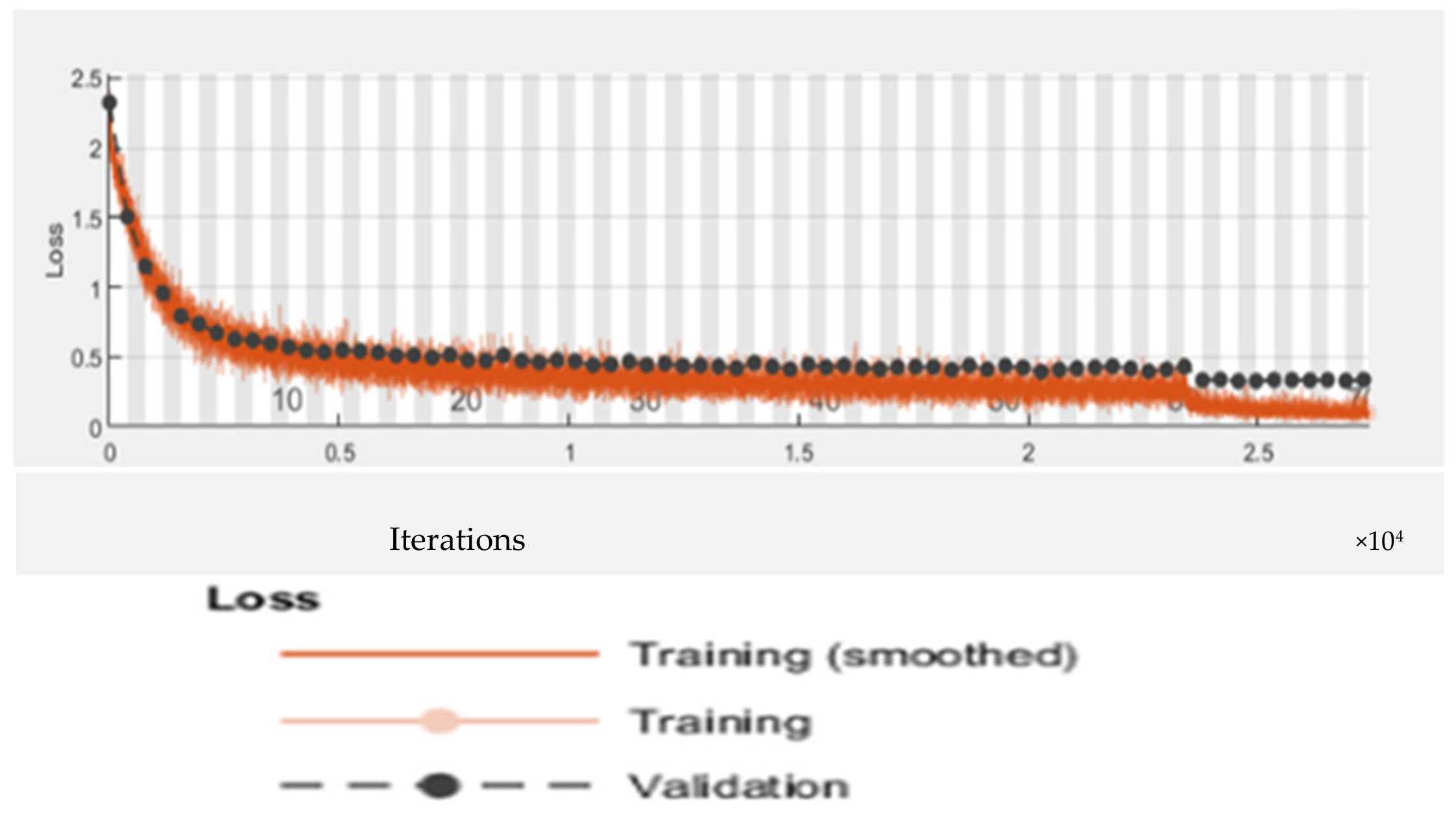
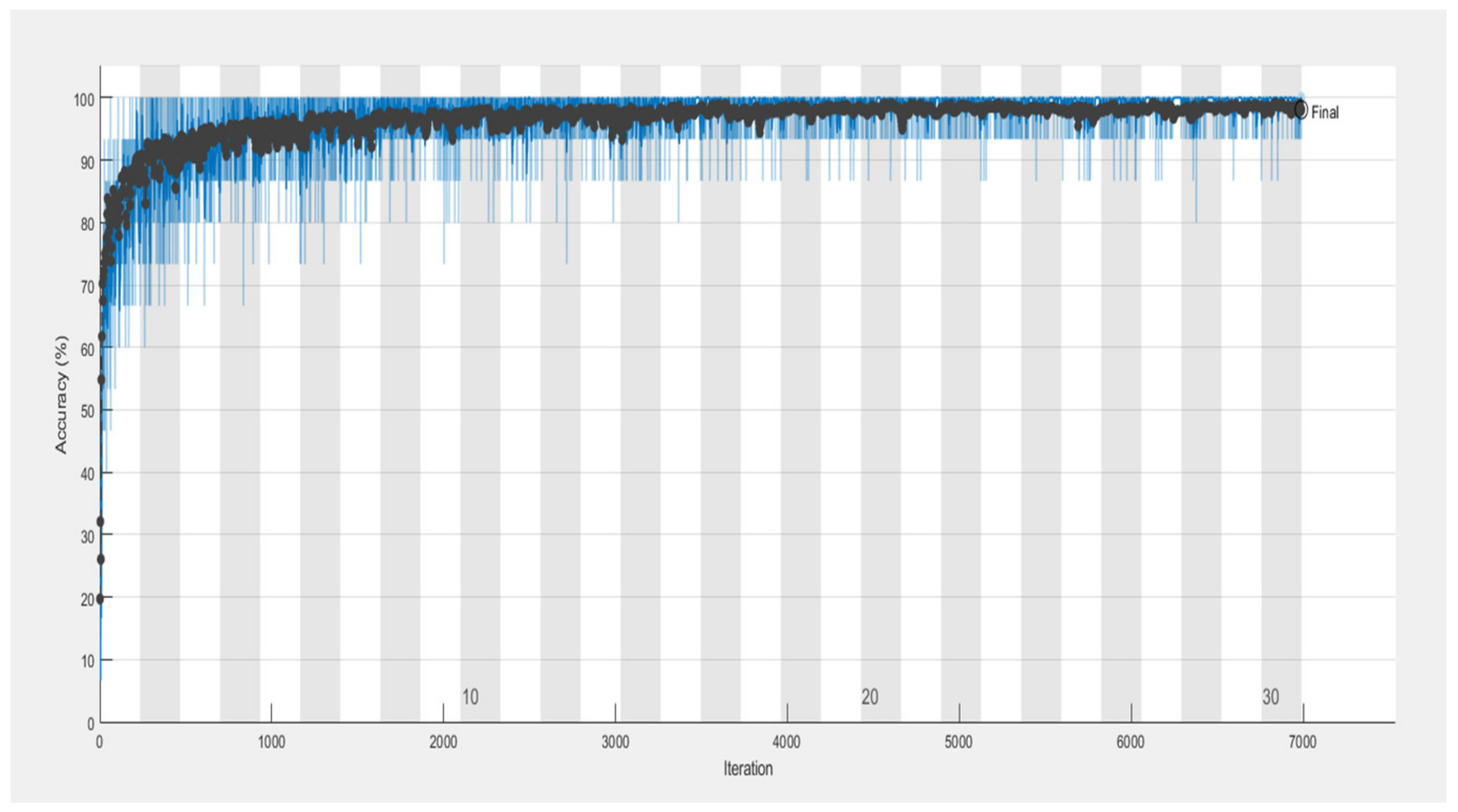
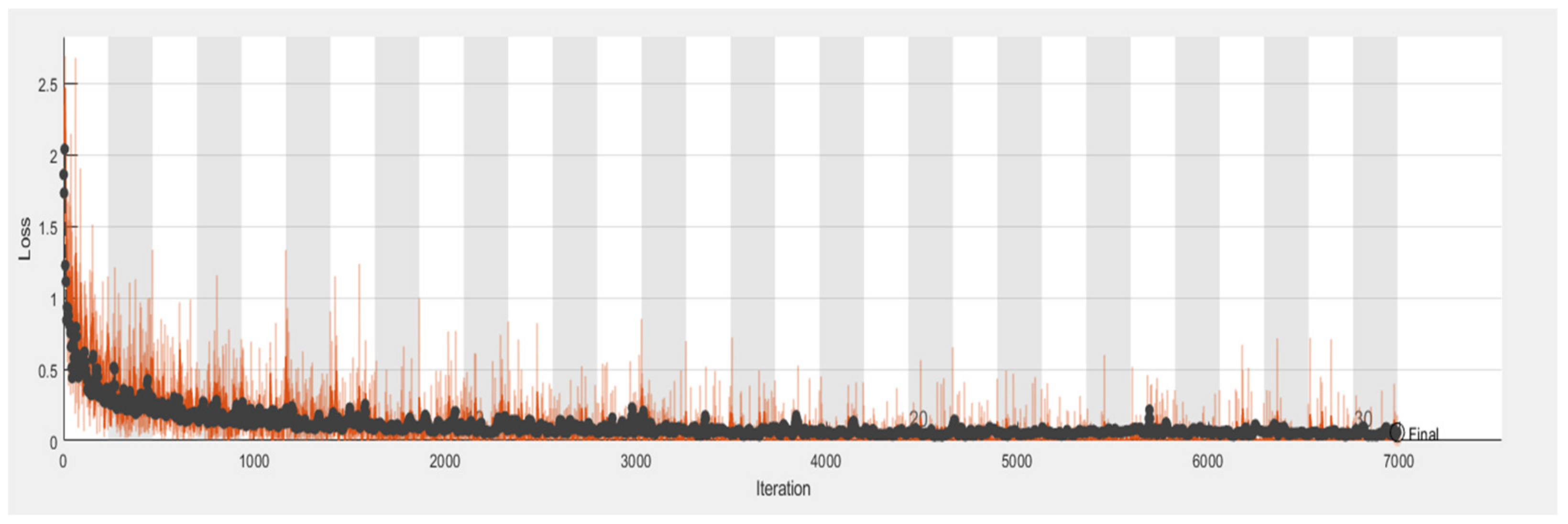
6.5. Proposed RTF-RCNN
6.6. Accuracy, Loss, Precision, Recall and F-Measure Comparison Performance
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Blancard, D. Diagnosis of parasitic and nonparasitic diseases. In Tomato Diseases; CRC Press: Boca Raton, FL, USA, 2012; pp. 35–412. [Google Scholar]
- Brahimi, M.; Boukhalfa, K.; Moussaoui, A.J.A.A.I. Deep learning for tomato diseases: Classification and symptoms visualization. Appl. Artif. Intell. 2017, 31, 299–315. [Google Scholar] [CrossRef]
- Al-Hiary, H.; Bani-Ahmad, S.; Reyalat, M.; Braik, M.; Alrahamneh, Z. Fast and accurate detection and classification of plant diseases. Int. J. Comput. Appl. 2011, 17, 31–38. [Google Scholar] [CrossRef]
- Dandawate, Y.; Kokare, R. An automated approach for classification of plant diseases towards development of futuristic Decision Support System in Indian perspective. In Proceedings of the International Conference on Advances in Computing, Communications and Informatics, Kochi, India, 10–13 August 2015; pp. 794–799. [Google Scholar]
- Lu, Y.; Yi, S.; Zeng, N.; Liu, Y.; Zhang, Y.J.N. Identification of rice diseases using deep convolutional neural networks. Neurocomputing 2017, 267, 378–384. [Google Scholar] [CrossRef]
- Akhtar, A.; Khanum, A.; Khan, S.A.; Shaukat, A. Automated plant disease analysis (APDA): Performance comparison of machine learning techniques. In Proceedings of the 11th International Conference on Frontiers of Information Technology, Washington, DC, USA, 16–18 December 2013; pp. 60–65. [Google Scholar]
- LeCun, Y.; Bengio, Y.; Hinton, G.J.N. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef] [PubMed]
- DeChant, C.; Wiesner-Hanks, T.; Chen, S.; Stewart, E.L.; Yosinski, J.; Gore, M.A.; Nelson, R.J.; Lipson, H. Automated identification of northern leaf blight-infected maize plants from field imagery using deep learning. Phytopathology 2017, 107, 1426–1432. [Google Scholar] [CrossRef] [PubMed]
- Fujita, E.; Kawasaki, Y.; Uga, H.; Kagiwada, S.; Iyatomi, H. Basic investigation on a robust and practical plant diagnostic system. In Proceedings of the 15th IEEE International Conference on Machine Learning and Applications, Anaheim, CA, USA, 18–20 December 2016; pp. 989–992. [Google Scholar]
- Kawasaki, Y.; Uga, H.; Kagiwada, S.; Iyatomi, H. Basic study of automated diagnosis of viral plant diseases using convolutional neural networks. In Proceedings of the International Symposium on Visual Computing, Las Vegas, NV, USA, 14–16 December 2015; pp. 638–645. [Google Scholar]
- Mohanty, S.P.; Hughes, D.P.; Salathé, M. Using deep learning for image-based plant disease detection. Front. Plant Sci. 2016, 7, 1419. [Google Scholar] [CrossRef] [PubMed]
- Nachtigall, L.G.; Araujo, R.M.; Nachtigall, G.R. Classification of apple tree disorders using convolutional neural networks. In Proceedings of the 28th International Conference on Tools with Artificial Intelligence, San Jose, CA, USA, 6–8 November 2016; pp. 472–476. [Google Scholar]
- Sladojevic, S.; Arsenovic, M.; Anderla, A.; Culibrk, D.; Stefanovic, D. Deep neural networks based recognition of plant diseases by leaf image classification. Comput. Intell. Neurosci. 2016, 2016, 3289801. [Google Scholar] [CrossRef] [PubMed]
- Gould, S.; Fulton, R.; Koller, D. Decomposing a scene into geometric and semantically consistent regions. In Proceedings of the IEEE 12th International Conference on Computer Vision, Kyoto, Japan, 29 September–2 October 2009; pp. 1–8. [Google Scholar]
- Hughes, D.; Salathé, M. An open access repository of images on plant health to enable the development of mobile disease diagnostics. arXiv Preprint 2015, arXiv:1511.08060. [Google Scholar]
- Liu, B.; Zhang, Y.; He, D.; Li, Y. Identification of apple leaf diseases based on deep convolutional neural networks. Symmetry 2018, 10, 11. [Google Scholar] [CrossRef]
- Ferentinos, K.P. Deep learning models for plant disease detection and diagnosis. Comput. Electron. Agric. 2018, 145, 311–318. [Google Scholar] [CrossRef]
- Rangarajan, A.K.; Purushothaman, R.; Ramesh, A. Tomato crop disease classification using pre-trained deep learning algorithm. Procedia Comput. Sci. 2018, 133, 1040–1047. [Google Scholar] [CrossRef]
- Coulibaly, S.; Kamsu-Foguem, B.; Kamissoko, D.; Traore, D. Deep neural networks with transfer learning in millet crop images. Comput. Ind. 2019, 108, 115–120. [Google Scholar] [CrossRef]
- Sangeetha, R.; Rani, M. Tomato Leaf Disease Prediction Using Transfer Learning. In Proceedings of the International Advanced Computing Conference, Kandlakoya, India, 16–17 December 2020; pp. 3–18. [Google Scholar]
- Jiang, D.; Li, F.; Yang, Y.; Yu, S. A tomato leaf diseases classification method based on deep learning. In Proceedings of the Chinese Control and Decision Conference, Hefei, China, 22–24 August 2020; pp. 1446–1450. [Google Scholar]
- Agarwal, M.; Gupta, S.K.; Biswas, K.K. Development of Efficient CNN model for Tomato crop disease identification. Sustain. Comput. Inform. Syst. 2020, 28, 100407. [Google Scholar] [CrossRef]
- Kaur, P.; Gautam, V. Research patterns and trends in classification of biotic and abiotic stress in plant leaf. Mater. Today Proc. 2021, 45, 4377–4382. [Google Scholar] [CrossRef]
- Kaur, P.; Gautam, V. Plant Biotic Disease Identification and Classification Based on Leaf Image: A Review. In Proceedings of the 3rd International Conference on Computing Informatics and Networks, Delhi, India, 29–30 July 2020; pp. 597–610. [Google Scholar]
- Suryanarayana, G.; Chandran, K.; Khalaf, O.I.; Alotaibi, Y.; Alsufyani, A.; Alghamdi, S.A. Accurate Magnetic Resonance Image Super-Resolution Using Deep Networks and Gaussian Filtering in the Stationary Wavelet Domain. IEEE Access 2021, 9, 71406–71417. [Google Scholar] [CrossRef]
- Wu, Y.; Xu, L.; Goodman, E.D. Tomato Leaf Disease Identification and Detection Based on Deep Convolutional Neural Network. Intell. Autom. Soft Comput. 2021, 28, 561–576. [Google Scholar] [CrossRef]
- Kaushik, M.; Prakash, P.; Ajay, R.; Veni, S. Tomato Leaf Disease Detection using Convolutional Neural Network with Data Augmentation. In Proceedings of the 5th International Conference on Communication and Electronics Systems, Coimbatore, India, 10–12 June 2020; pp. 1125–1132. [Google Scholar]
- Ashok, S.; Kishore, G.; Rajesh, V.; Suchitra, S.; Sophia, S.G.; Pavithra, B. Tomato Leaf Disease Detection Using Deep Learning Techniques. In Proceedings of the 5th International Conference on Communication and Electronics Systems, Coimbatore, India, 10–12 June 2020; pp. 979–983. [Google Scholar]
- Girshick, R.; Donahue, J.; Darrell, T.; Malik, J. Rich feature hierarchies for accurate object detection and semantic segmentation. In Proceedings of the IEEE International Conference on Computer Vision and Pattern Recognition, Columbus, OH, USA, 23–28 June 2014; pp. 580–587. [Google Scholar]
- Girshick, R. Fast r-cnn. In Proceedings of the International Conference on Computer Vision, Santiago, Chile, 20–23 June 2015; pp. 1440–1448. [Google Scholar]
- Ren, S.; He, K.; Girshick, R.; Sun, J. Faster r-cnn: Towards real-time object detection with region proposal networks. Adv. Neural Inf. Process. Syst. 2015, 28, 91–99. [Google Scholar] [CrossRef] [PubMed]
- Goodfellow, I.; Bengio, Y.; Courville, A. Deep Learning; MIT Press: Cambridge, MA, USA, 2016. [Google Scholar]
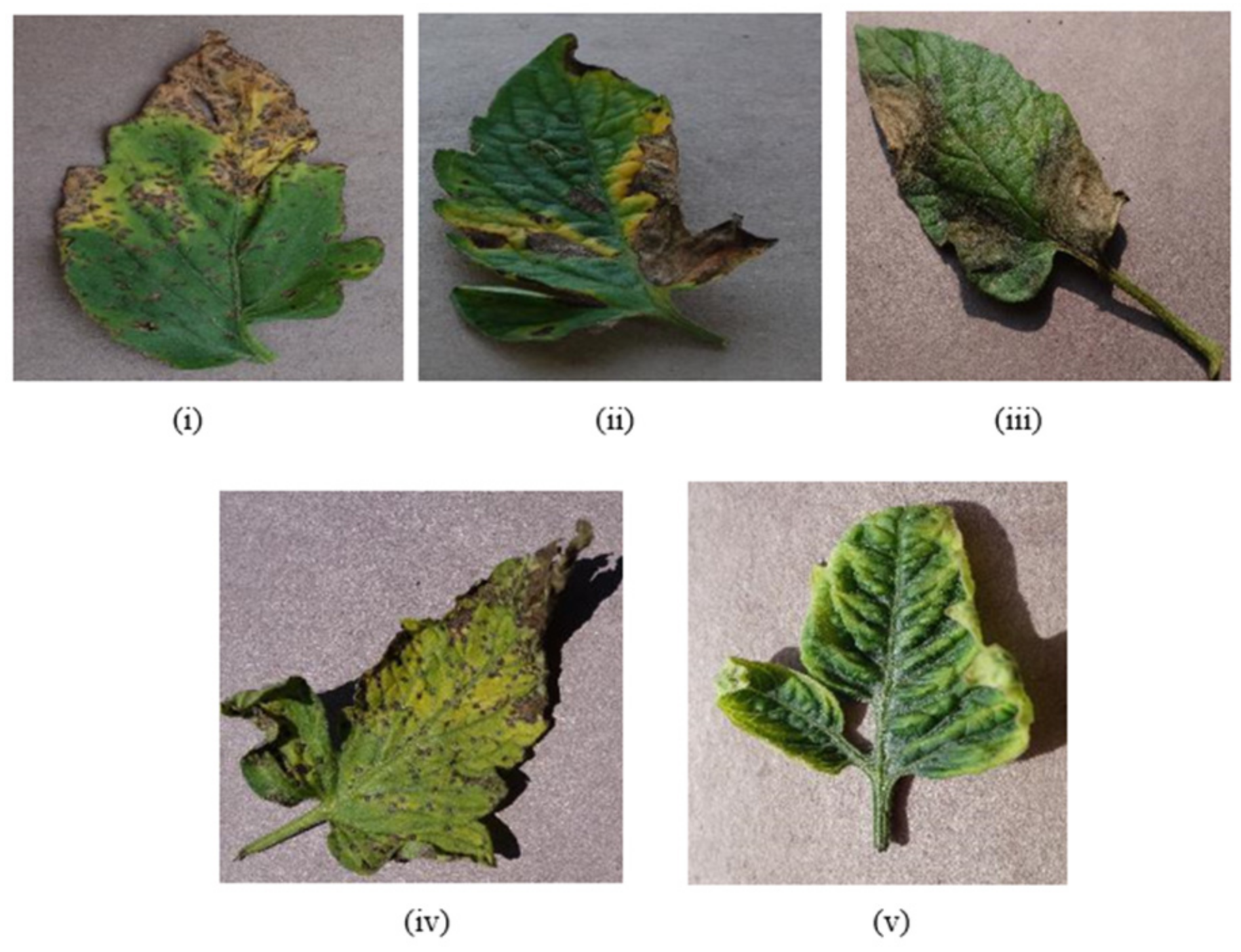




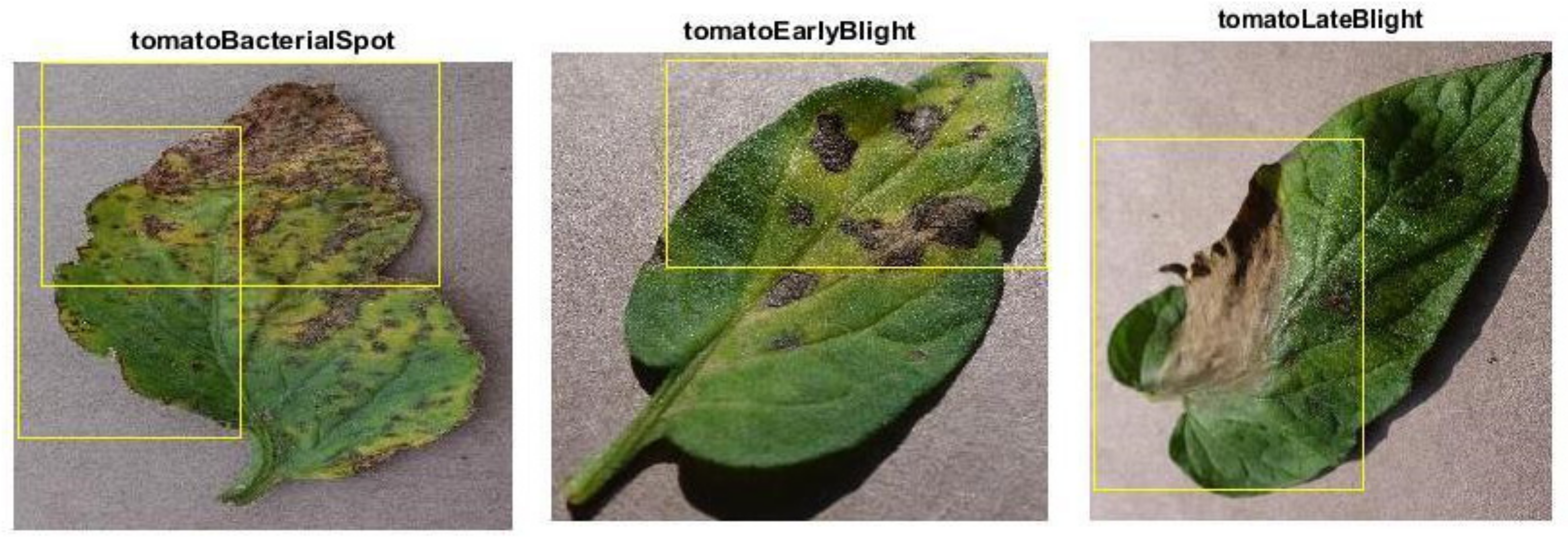



| Classes of Tomato Diseases | Number of Images |
|---|---|
| Yellow Leaf Curl Virus Tomato | 2500 |
| Septoria Leaf Spot Tomato | 2500 |
| Late Tomato Blight | 2500 |
| Bacterial Tomato Spot | 2500 |
| Early Tomato Blight | 2500 |
| Layers | Type | Number Kernel | Kernel Size | Stride |
|---|---|---|---|---|
| 0 | Input | 3 | 32 × 32 | - |
| 1 | Convolution | 32 | 3 × 3 | 1 |
| 2 | Relu | - | - | - |
| 3 | Convolution | 32 | 3 × 3 | 1 |
| 4 | Relu | - | - | - |
| 5 | Max pooling | - | 3 × 3 | 2 |
| 6 | Fully Connected | 64 | - | - |
| 7 | Relu | - | - | - |
| 8 | Fully Connected | 5 | - | - |
| 9 | SoftMax | - | - | - |
| Layers | Type | Number Kernel | Kernel Size | Stride |
|---|---|---|---|---|
| 1 | Input | 3 | 32 × 32 | - |
| 2 | Convolution | 32 | 3 × 3 | 1 |
| 3 | Relu | - | - | - |
| 4 | Convolution | 32 | 3 × 3 | 1 |
| 5 | Relu | - | - | - |
| 6 | Convolution | 32 | 3 × 3 | 1 |
| 7 | Relu | - | - | - |
| 8 | Max pooling | - | 3 × 3 | 2 |
| 9 | Fully Connected | 64 | - | - |
| 10 | Fully Connected | 64 | - | - |
| 11 | SoftMax | - | - | - |
| 12 | Classification | - | - | - |
| Layers | Type | Number Kernel | Kernel Size | Stride |
|---|---|---|---|---|
| 1 | Input | - | 227 × 227 | - |
| 2 | Convolution | 96 | 3 × 3 | 1 |
| 3 | Relu | - | - | - |
| 4 | Channel normalization | - | - | - |
| 5 | Pooling | - | - | - |
| 6 | Convolution | 256 | 5 × 5 | 1 |
| 7 | Relu | - | - | - |
| 8 | Channel normalization | - | - | - |
| 9 | Pooling | - | - | - |
| 10 | Convolution | 384 | 3 × 3 | 1 |
| 11 | Relu | - | - | - |
| 12 | Convolution | 384 | 3 × 3 | 1 |
| 13 | Relu | - | - | - |
| 14 | Convolution | 256 | 3 × 3 | 1 |
| 15 | Relu | - | - | - |
| 16 | Pooling | - | - | -. |
| 17 | Fully Connected | - | - | - |
| 18 | Relu | - | - | - |
| 19 | Dropout | - | - | - |
| 21 | Fully Connected | - | - | - |
| 21 | Relu | - | - | - |
| 22 | Dropout | - | - | - |
| 23 | Fully Connected | - | - | - |
| 24 | SoftMax | - | - | - |
| 25 | Classification | - | - | - |
| Name | Parameters |
|---|---|
| Algorithm | CNN, Alex net, Faster R-CNN |
| Convolutional Layers | Relu |
| Fully Connected Layers | SoftMax |
| Maximum Number of Epochs | 30 |
| Data Set | 12,500 images |
| Training Data | 70% |
| Testing Data | 30% |
| Environment | MATLAB with Deep Learning |
| Evaluation Parameter | Accuracy, MSE Loss, Precession, Recall and F-Measure |
| Epochs | Loss | Accuracy | Epochs | Loss | Accuracy |
|---|---|---|---|---|---|
| 1 | 0.5621 | 0.7031 | 3 | 0.5571 | 0.7078 |
| 5 | 0.5501 | 0.7079 | 7 | 0.5490 | 0.7150 |
| 9 | 0.5431 | 0.7191 | 11 | 0.5521 | 0.7067 |
| 13 | 0.5771 | 0.7009 | 15 | 0.5921 | 0.7001 |
| 17 | 0.5831 | 0.7021 | 19 | 0.5710 | 0.7123 |
| 21 | 0.5322 | 0.7698 | 23 | 0.5201 | 0.7876 |
| 25 | 0.4901 | 0.8108 | 27 | 0.4690 | 0.8543 |
| 29 | 0.4321 | 0.8908 | 30 | 0.4165 | 0.9221 |
| Name | Recall | Precision | F-Measure |
|---|---|---|---|
| Macro | 0.69 | 0.65 | 0.66 |
| Average | 0.66 | ||
| Micro | 0.71 | 0.70 | 0.70 |
| Average | 70.50 | ||
| Epochs | Loss | Accuracy | Epochs | Loss | Accuracy |
|---|---|---|---|---|---|
| 1 | 0.4621 | 0.7531 | 3 | 0.4571 | 0.7678 |
| 5 | 0.4501 | 0.7679 | 7 | 0.4490 | 0.7750 |
| 9 | 0.4231 | 0.7791 | 11 | 0.4221 | 0.7867 |
| 13 | 0.4171 | 0.8009 | 15 | 0.4121 | 0.8101 |
| 17 | 0.3931 | 0.8221 | 19 | 0.3710 | 0.8323 |
| 21 | 0.3622 | 0.8498 | 23 | 0.3601 | 0.8976 |
| 25 | 0.3401 | 0.9098 | 27 | 0.3390 | 0.9200 |
| 29 | 0.3121 | 0.9480 | 30 | 0.3065 | 0.9532 |
| Name | Precision | F-Measure | Recall |
|---|---|---|---|
| Macro | 0.69 | 0.71 | 0.74 |
| Average | 0.71 | ||
| Micro | 0.73 | 0.74 | 0.76 |
| Average | 0.74 | ||
| Epoch | Loss | Accuracy | Epoch | Loss | Accuracy |
|---|---|---|---|---|---|
| 1 | 0.7721 | 0.7631 | 17 | 0.5731 | 0.8750 |
| 3 | 0.7571 | 0.7678 | 19 | 0.5210 | 0.8984 |
| 5 | 0.7401 | 0.7979 | 21 | 0.4922 | 0.9024 |
| 7 | 0.7090 | 0.8330 | 23 | 0.4401 | 0.9146 |
| 9 | 0.6991 | 0.8491 | 25 | 0.4001 | 0.9298 |
| 11 | 0.6891 | 0.8533 | 27 | 0.3590 | 0.9500 |
| 13 | 0.6371 | 0.9009 | 29 | 0.3121 | 0.9608 |
| 15 | 0.6221 | 0.8739 | 30 | 0.2765 | 0.9742 |
| Name | Precision | Recall | F-Measure |
|---|---|---|---|
| Macro Average | 0.72 | 0.78 | 0.74 |
| Micro Average | 0.75 | 0.80 | 0.77 |
| Model | Accuracy | Loss | Precision | Recall | F-Measure |
|---|---|---|---|---|---|
| RTF-RCNN | 0.9742 | 0.2765 | 0.75 | 0.80 | 0.77 |
| Alex Net | 0.9532 | 0.3065 | 0.73 | 0.76 | 0.74 |
| CNN | 0.9221 | 0.4165 | 0.70 | 0.71 | 0.70 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alruwaili, M.; Siddiqi, M.H.; Khan, A.; Azad, M.; Khan, A.; Alanazi, S. RTF-RCNN: An Architecture for Real-Time Tomato Plant Leaf Diseases Detection in Video Streaming Using Faster-RCNN. Bioengineering 2022, 9, 565. https://doi.org/10.3390/bioengineering9100565
Alruwaili M, Siddiqi MH, Khan A, Azad M, Khan A, Alanazi S. RTF-RCNN: An Architecture for Real-Time Tomato Plant Leaf Diseases Detection in Video Streaming Using Faster-RCNN. Bioengineering. 2022; 9(10):565. https://doi.org/10.3390/bioengineering9100565
Chicago/Turabian StyleAlruwaili, Madallah, Muhammad Hameed Siddiqi, Asfandyar Khan, Mohammad Azad, Abdullah Khan, and Saad Alanazi. 2022. "RTF-RCNN: An Architecture for Real-Time Tomato Plant Leaf Diseases Detection in Video Streaming Using Faster-RCNN" Bioengineering 9, no. 10: 565. https://doi.org/10.3390/bioengineering9100565
APA StyleAlruwaili, M., Siddiqi, M. H., Khan, A., Azad, M., Khan, A., & Alanazi, S. (2022). RTF-RCNN: An Architecture for Real-Time Tomato Plant Leaf Diseases Detection in Video Streaming Using Faster-RCNN. Bioengineering, 9(10), 565. https://doi.org/10.3390/bioengineering9100565







