SCRaMbLE: A Study of Its Robustness and Challenges through Enhancement of Hygromycin B Resistance in a Semi-Synthetic Yeast
Abstract
1. Introduction
2. Results and Discussion
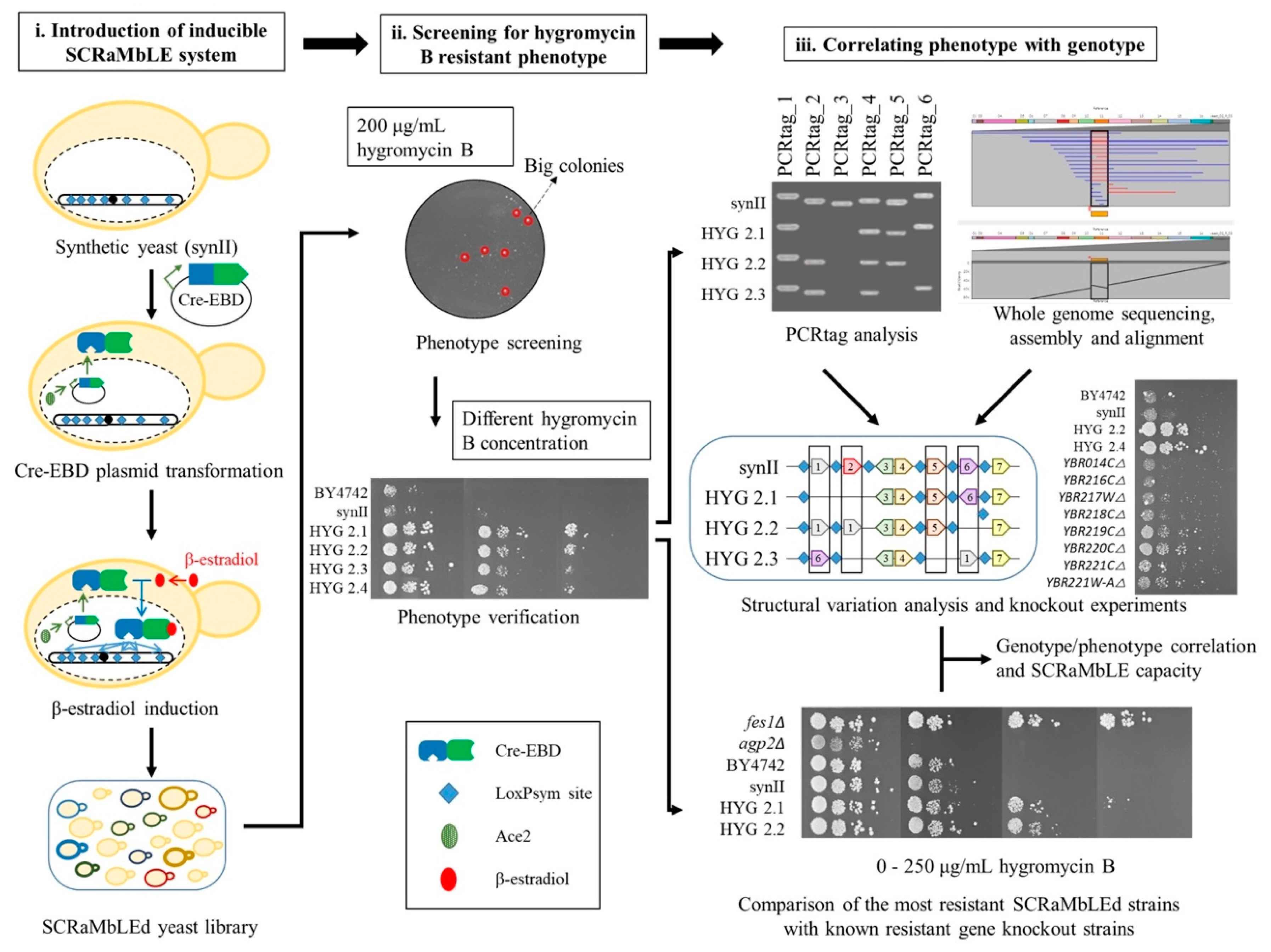
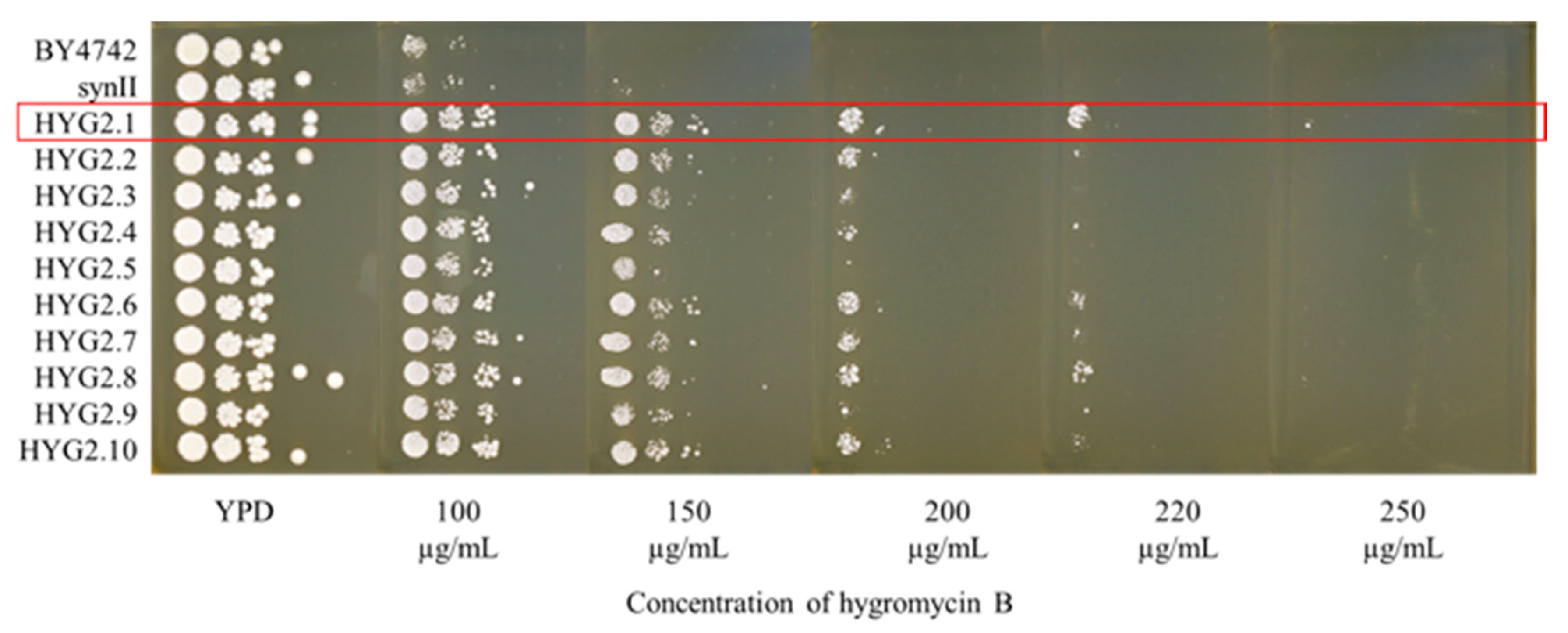
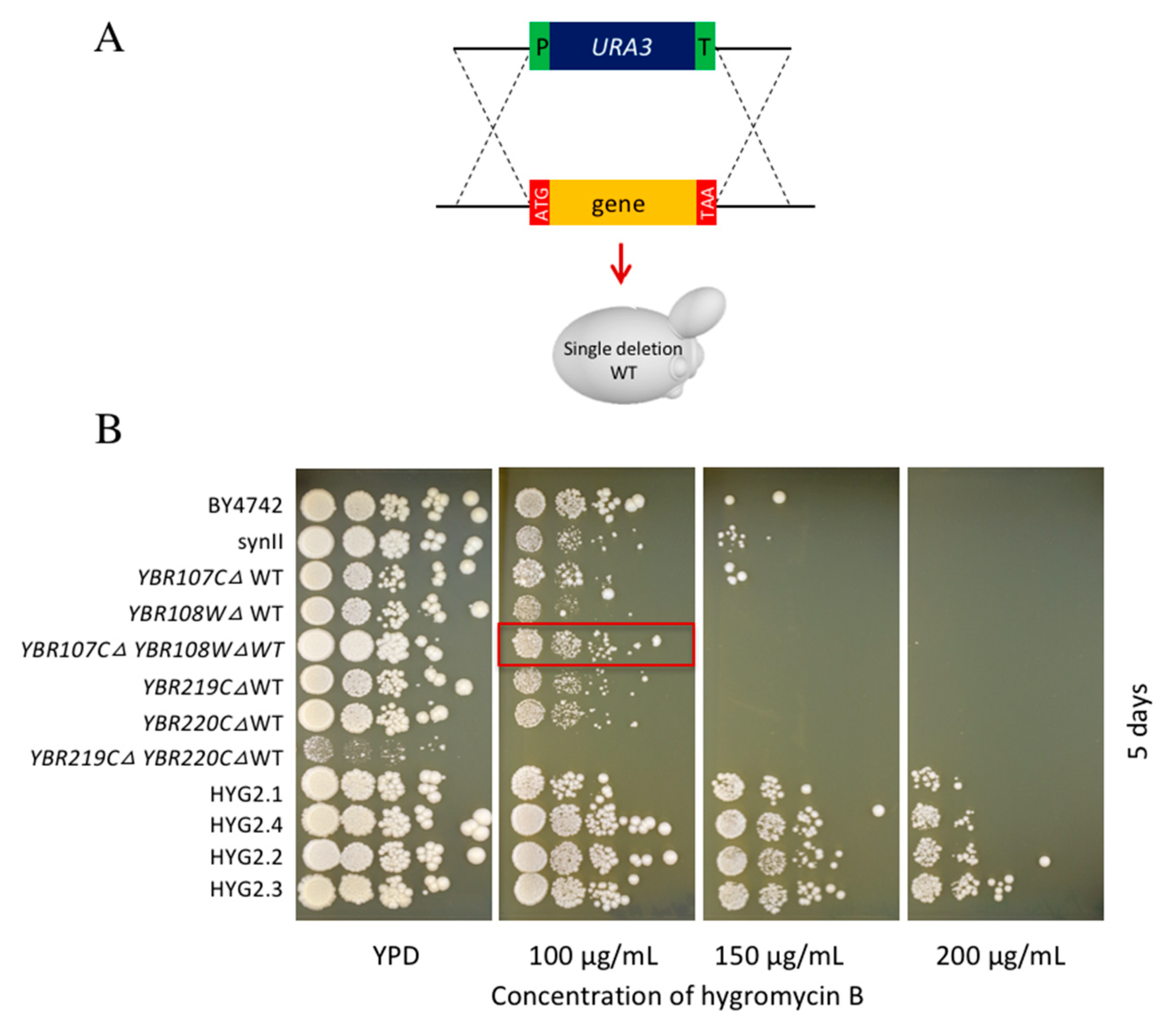
2.1. Generating a Hygromycin B Resistant Strain Using SCRaMbLE
2.2. PCRTag Analysis for SCRaMbLEd Hygromycin B Resistant Strains
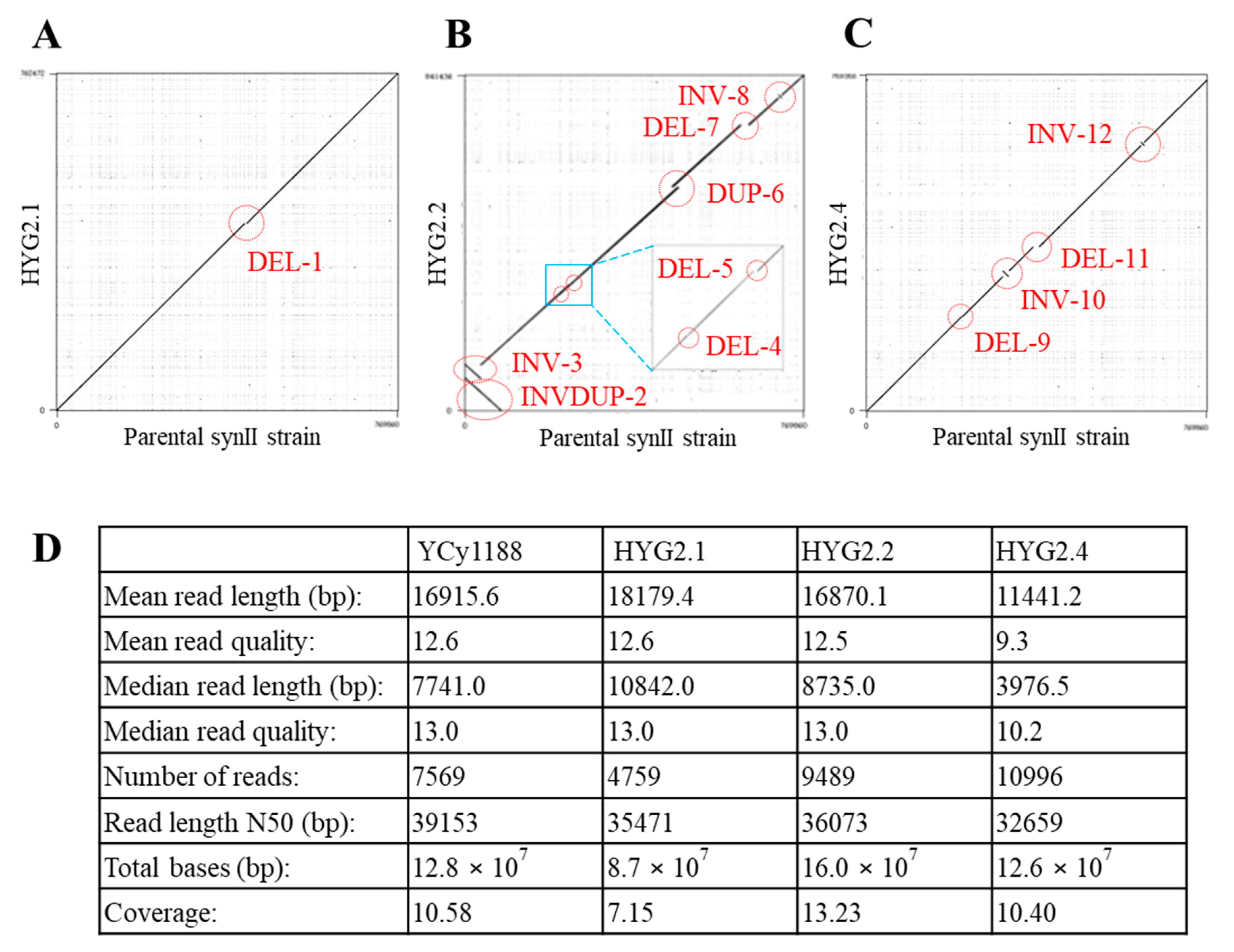
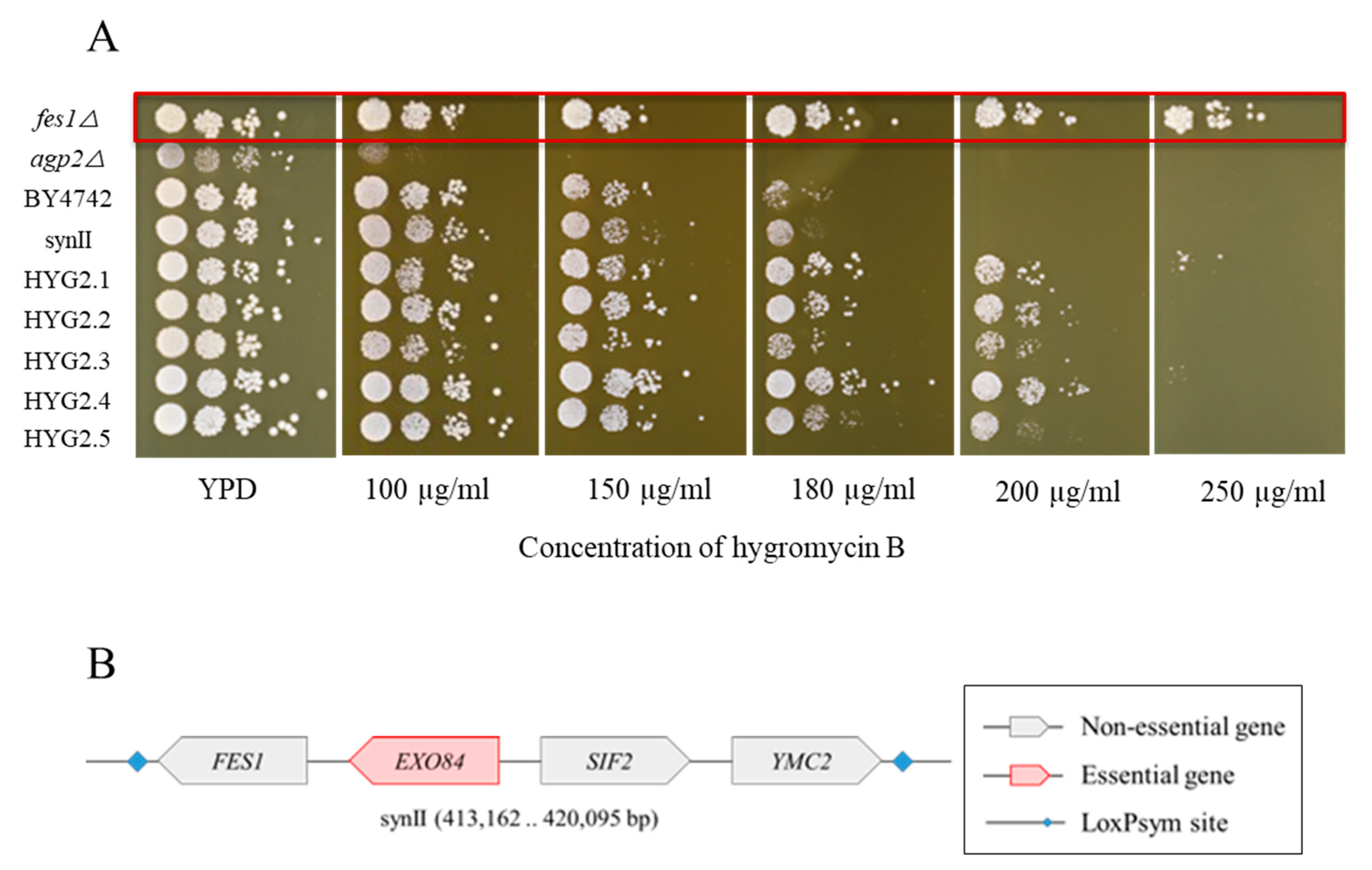
2.3. Nanopore Sequencing Analysis for Hygromycin B Resistant Strains
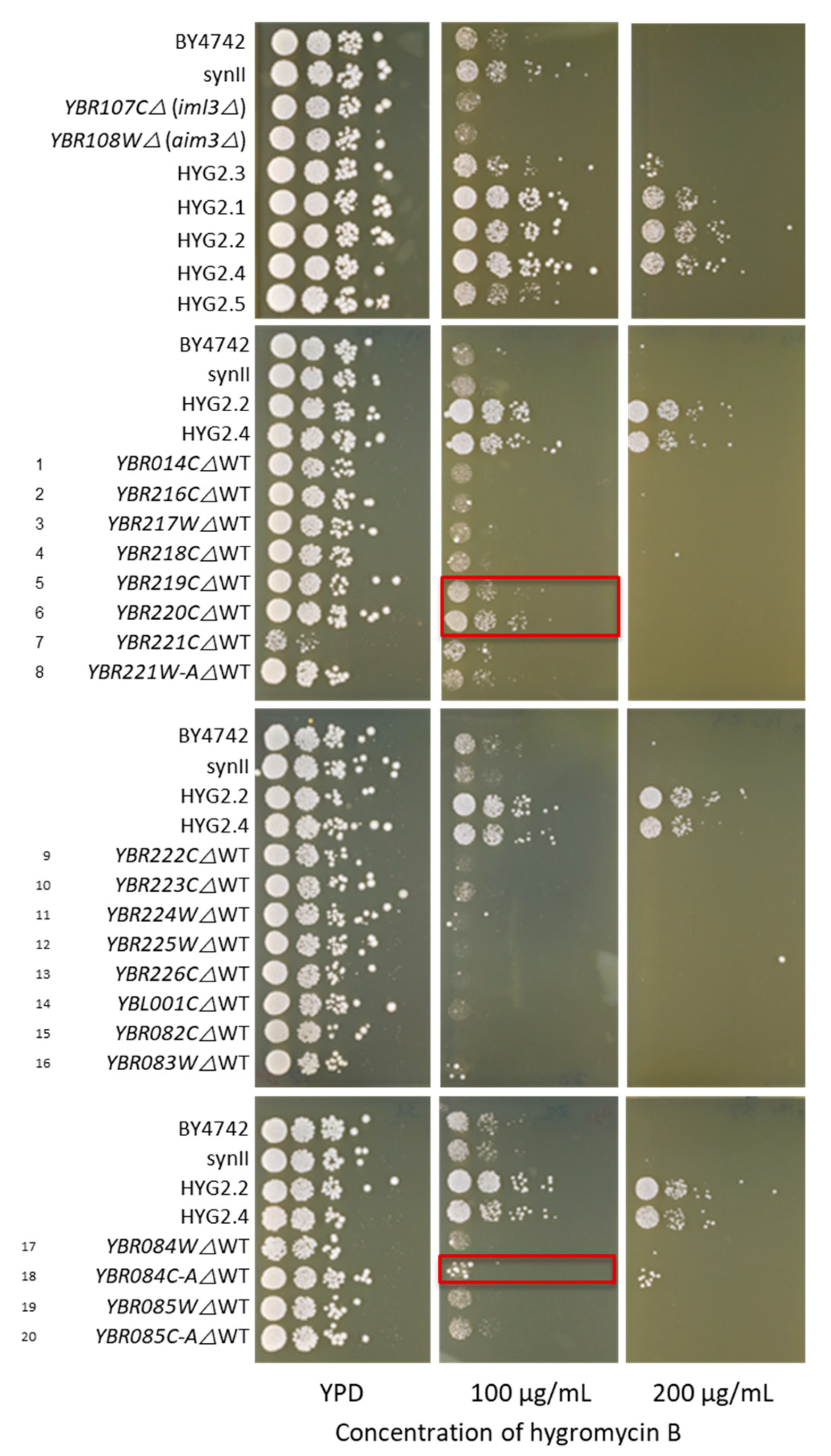
2.4. Yeast Knockout Library Assessment of Hygromycin B Resistance
3. Materials and Methods
3.1. Strains and Media
3.2. Yeast Transformations
3.3. Genomic and Plasmid DNA Isolation
3.4. Double Deletion Strains
3.5. SCRaMbLE Workflow
3.6. Post-SCRaMbLE Selection for Hygromycin B Resistance
3.7. PCRTag Analysis of HYG2.1 to HYG2.5 Strains
3.8. Nanopore Sequencing
4. Conclusions and Perspectives
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gibson, D.G.; Benders, G.A.; Andrews-Pfannkoch, C.; Denisova, E.A.; Baden-Tillson, H.; Zaveri, J.; Stockwell, T.B.; Brownley, A.; Thomas, D.W.; Algire, M.A.; et al. Complete chemical synthesis, assembly, and cloning of a Mycoplasma genitalium genome. Science 2008, 319, 1215–1220. [Google Scholar] [CrossRef] [PubMed]
- Richardson, S.M.; Mitchell, L.A.; Stracquadanio, G.; Yang, K.; Dymond, J.S.; DiCarlo, J.E.; Lee, D.; Huang, C.L.V.; Chandrasegaran, S.; Cai, Y.; et al. Design of a synthetic yeast genome. Science 2017, 355, 1040–1044. [Google Scholar] [CrossRef] [PubMed]
- Dymond, J.S.; Richardson, S.M.; Coombes, C.E.; Babatz, T.; Muller, H.; Annaluru, N.; Blake, W.J.; Schwerzmann, J.W.; Dai, J.; Lindstrom, D.L.; et al. Synthetic chromosome arms function in yeast and generate phenotypic diversity by design. Nature 2011, 477, 471. [Google Scholar] [CrossRef]
- Shen, Y.; Stracquadanio, G.; Wang, Y.; Yang, K.; Mitchell, L.A.; Xue, Y.; Cai, Y.; Chen, T.; Dymond, J.S.; Kang, K.; et al. SCRaMbLE generates designed combinatorial stochastic diversity in synthetic chromosomes. Genome Res. 2016, 26, 36–49. [Google Scholar] [CrossRef]
- Blount, B.A.; Gowers, G.O.F.; Ho, J.C.H.; Ledesma-Amaro, R.; Jovicevic, D.; McKiernan, R.M.; Xie, Z.X.; Li, B.Z.; Yuan, Y.J.; Ellis, T. Rapid host strain improvement by in vivo rearrangement of a synthetic yeast chromosome. Nat. Commun. 2018, 9, 1932. [Google Scholar] [CrossRef]
- Liu, W.; Luo, Z.; Wang, Y.; Pham, N.T.; Tuck, L.; Pérez-Pi, I.; Liu, L.; Shen, Y.; French, C.; Auer, M.; et al. Rapid pathway prototyping and engineering using in vitro and in vivo synthetic genome SCRaMbLE-in methods. Nat. Commun. 2018, 9, 1936. [Google Scholar] [CrossRef] [PubMed]
- Jia, B.; Wu, Y.; Li, B.-Z.; Mitchell, L.A.; Liu, H.; Pan, S.; Wang, J.; Zhang, H.-R.; Jia, N.; Li, B.; et al. Precise control of SCRaMbLE in synthetic haploid and diploid yeast. Nat. Commun. 2018, 9, 1933. [Google Scholar] [CrossRef]
- Gowers, G.O.F.; Chee, S.M.; Bell, D.; Suckling, L.; Kern, M.; Tew, D.; McClymont, D.W.; Ellis, T. Improved betulinic acid biosynthesis using synthetic yeast chromosome recombination and semi-automated rapid LC-MS screening. Nat. Commun. 2020, 11, 868. [Google Scholar] [CrossRef]
- Luo, Z.; Wang, L.; Wang, Y.; Zhang, W.; Guo, Y.; Shen, Y.; Jiang, L.; Wu, Q.; Zhang, C.; Cai, Y.; et al. Identifying and characterizing SCRaMbLEd synthetic yeast using ReSCuES. Nat. Commun. 2018, 9, 1930. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Li, Y.; Chen, X.; Ding, M.; Wu, Y.; Yuan, Y.-J. SCRaMbLE generates evolved yeasts with increased alkali tolerance. Microb. Cell Factories 2019, 18, 52. [Google Scholar] [CrossRef]
- Bongomin, F.; Gago, S.; Oladele, R.O.; Denning, D.W. Global and multi-national prevalence of fungal diseases-estimate precision. J. Fungi. 2017, 3, 57. [Google Scholar] [CrossRef]
- Mann, R.L.; Bromer, W.W. The isolation of a second antibiotic from Streptomyces hygroscopicus. J. Am. Chem. Soc. 1958, 80, 2714–2716. [Google Scholar] [CrossRef]
- Borovinskaya, M.A.; Shoji, S.; Fredrick, K.; Cate, J.H.D. Structural basis for hygromycin B inhibition of protein biosynthesis. RNA 2008, 14, 1590–1599. [Google Scholar] [CrossRef] [PubMed]
- Zierhut, G.; Piepersberg, W.; Bock, A. Comparative analysis of the effect of aminoglycosides on bacterial protein synthesis in vitro. Eur. J. Biochem. 1979, 98, 577–583. [Google Scholar] [CrossRef] [PubMed]
- Brodersen, D.E.; Clemons, W.M., Jr.; Carter, A.P.; Morgan-Warren, R.J.; Wimberly, B.T.; Ramakrishnan, V. The structural basis for the action of the antibiotics tetracycline, pactamycin, and hygromycin B on the 30S ribosomal subunit. Cell 2000, 103, 1143–1154. [Google Scholar] [CrossRef]
- Moazed, D.; Noller, H.F. Interaction of antibiotics with functional sites in 16S ribosomal RNA. Nature 1987, 327, 389–394. [Google Scholar] [CrossRef] [PubMed]
- González, A.; Jiménez, A.; Vázquez, D.; Davies, J.E.; Schindler, D. Studies on the mode of action of hygromycin B, an inhibitor of translocation in eukaryotes. Biochim. Biophys. Acta (Bba) Nucleic Acids Protein Synth. 1978, 521, 459–469. [Google Scholar] [CrossRef]
- Leboul, J.; Davies, J. Enzymatic modification of hygromycin B in Streptomyces hygroscopicus. J. Antibiot. 1982, 35, 527–528. [Google Scholar] [CrossRef]
- Pardo, J.M.; Malpartida, F.; Rico, M.; Jimenez, A. Biochemical basis of resistance to hygromycin B in Streptomyces hygroscopicus--the producing organism. J. Gen. Microbiol. 1985, 131, 1289–1298. [Google Scholar] [CrossRef] [PubMed]
- Zalacain, M.; Pardo, J.M.; Jimenez, A. Purification and characterization of a hygromycin B phosphotransferase from Streptomyces hygroscopicus. Eur. J. Biochem. 1987, 162, 419–422. [Google Scholar] [CrossRef]
- Rao, R.N.; Allen, N.E.; Hobbs, J.N., Jr.; Alborn, W.E., Jr.; Kirst, H.A.; Paschal, J.W. Genetic and enzymatic basis of hygromycin B resistance in Escherichia coli. Antimicrob. Agents Chemother. 1983, 24, 689–695. [Google Scholar] [CrossRef]
- Pfister, P.; Risch, M.; Brodersen, D.E.; Böttger, E.C. Role of 16S rRNA helix 44 in ribosomal resistance to hygromycin B. Antimicrob. Agents Chemother. 2003, 47, 1496–1502. [Google Scholar] [CrossRef][Green Version]
- De Stasio, E.A.; Dahlberg, A.E. Effects of mutagenesis of a conserved base-paired site near the decoding region of Escherichia coli 16 S ribosomal RNA. J. Mol. Biol. 1990, 212, 127–133. [Google Scholar] [CrossRef]
- Spangler, E.A.; Blackburn, E.H. The nucleotide sequence of the 17S ribosomal RNA gene of Tetrahymena thermophila and the identification of point mutations resulting in resistance to the antibiotics paromomycin and hygromycin. J. Biol. Chem. 1985, 260, 6334–6340. [Google Scholar] [CrossRef]
- Gritz, L.; Davies, J. Plasmid-encoded hygromycin B resistance: The sequence of hygromycin B phosphotransferase gene and its expression in Escherichia coli and Saccharomyces cerevisiae. Gene 1983, 25, 179–188. [Google Scholar] [CrossRef]
- Kaster, K.R.; Burgett, S.G.; Ingolia, T.D. Hygromycin B resistance as dominant selectable marker in yeast. Curr. Genet. 1984, 8, 353–358. [Google Scholar] [CrossRef] [PubMed]
- Aouida, M.; Khodami-Pour, A.; Ramotar, D. Novel role for the Saccharomyces cerevisiae oligopeptide transporter Opt2 in drug detoxification. Biochem. Cell Biol. Biochim. Biol. Cell. 2009, 87, 653–661. [Google Scholar] [CrossRef] [PubMed]
- Barreto, L.; Canadell, D.; Petrezsélyová, S.; Navarrete, C.; Marešová, L.; Peréz-Valle, J.; Herrera, R.; Olier, I.; Giraldo, J.; Sychrová, H.; et al. A genomewide screen for tolerance to cationic drugs reveals genes important for potassium homeostasis in Saccharomyces cerevisiae. Eukaryot. Cell 2011, 10, 1241–1250. [Google Scholar] [CrossRef] [PubMed]
- Lussier, M.; White, A.M.; Sheraton, J.; di Paolo, T.; Treadwell, J.; Southard, S.B.; Horenstein, C.I.; Chen-Weiner, J.; Ram, A.F.; Kapteyn, J.C.; et al. Large scale identification of genes involved in cell surface biosynthesis and architecture in Saccharomyces cerevisiae. Genetics 1997, 147, 435–450. [Google Scholar] [CrossRef]
- Chawla, B.; Jhingran, A.; Panigrahi, A.; Stuart, K.D.; Madhubala, R. Paromomycin affects translation and vesicle-mediated trafficking as revealed by proteomics of paromomycin -susceptible -resistant Leishmania donovani. PLoS ONE 2011, 6, e26660. [Google Scholar] [CrossRef]
- Lindstrom, D.L.; Gottschling, D.E. The mother enrichment program: A genetic system for facile replicative life span analysis in Saccharomyces cerevisiae. Genetics 2009, 183, 413–422. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.K.; Poddar, A.; Hajra, S.; Sanyal, K.; Sinha, P. The IML3/MCM19 gene of Saccharomyces cerevisiae is required for a kinetochore-related process during chromosome segregation. Mol. Genet. Genom. 2001, 265, 249–257. [Google Scholar] [CrossRef]
- Michelot, A.; Grassart, A.; Okreglak, V.; Costanzo, M.; Boone, C.; Drubin, D.G. Actin Filament Elongation in Arp2/3-Derived Networks Is Controlled by Three Distinct Mechanisms. Dev. Cell 2013, 24, 182–195. [Google Scholar] [CrossRef] [PubMed]
- Giaever, G.; Chu, A.M.; Ni, L.; Connelly, C.; Riles, L.; Véronneau, S.; Dow, S.; Lucau-Danila, A.; Anderson, K.; André, B.; et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature 2002, 418, 387–391. [Google Scholar] [CrossRef] [PubMed]
- Gowda, N.K.C.; Kandasamy, G.; Froehlich, M.S.; Dohmen, R.J.; Andréasson, C. Hsp70 nucleotide exchange factor Fes1 is essential for ubiquitin-dependent degradation of misfolded cytosolic proteins. Proc. Natl. Acad. Sci. USA 2013, 110, 5975–5980. [Google Scholar] [CrossRef]
- Gowda, N.K.C.; Kaimal, J.M.; Masser, A.E.; Kang, W.; Friedländer, M.R.; Andréasson, C. Cytosolic splice isoform of Hsp70 nucleotide exchange factor Fes1 is required for the degradation of misfolded proteins in yeast. Mol. Biol. Cell 2016, 27, 1210–1219. [Google Scholar] [CrossRef]
- Aouida, M.; Page, N.; Leduc, A.; Peter, M.; Ramotar, D. A genome-wide screen in Saccharomyces cerevisiae reveals altered transport as a mechanism of resistance to the anticancer drug bleomycin. Cancer Res 2004, 64, 1102–1109. [Google Scholar] [CrossRef]
- Winston, F.; Dollard, C.; Ricupero-Hovasse, S.L. Construction of a set of convenient saccharomyces cerevisiae strains that are isogenic to S288C. Yeast 1995, 11, 53–55. [Google Scholar] [CrossRef]
- Baker Brachmann, C.; Davies, A.; Cost, G.J.; Caputo, E.; Li, J.; Hieter, P.; Boeke, J.D. Designer deletion strains derived from Saccharomyces cerevisiae S288C: A useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 1998, 14, 115–132. [Google Scholar] [CrossRef]
- Guthrie, C.; Fink, G.R. Guide to yeast genetics and molecular biology. Methods Enzym. 1991, 194, 1–863. [Google Scholar]
- Gietz, R.D.; Schiestl, R.H.; Willems, A.R.; Woods, R.A. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast 1995, 11, 355–360. [Google Scholar] [CrossRef]
- Hoffman, C.S.; Winston, F. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene 1987, 57, 267–272. [Google Scholar] [CrossRef]
- Li, H. Minimap2: Pairwise alignment for nucleotide sequences. Bioinformatics 2018, 34, 3094–3100. [Google Scholar] [CrossRef]
- Sedlazeck, F.J.; Rescheneder, P.; Smolka, M.; Fang, H.; Nattestad, M.; von Haeseler, A.; Schatz, M.C. Accurate detection of complex structural variations using single-molecule sequencing. Nat. Methods 2018, 15, 461–468. [Google Scholar] [CrossRef] [PubMed]
- Koren, S.; Walenz, B.P.; Berlin, K.; Miller, J.R.; Bergman, N.H.; Phillippy, A.M. Canu: Scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017, 27, 722–736. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Xu, H.; Li, H.; Chen, H.; Zhou, S.; Tian, F.; Li, B.-Z.; Bo, X.; Wu, Y.; Yuan, Y.-J. SCRaMbLEing of a synthetic yeast chromosome with clustered essential genes reveals synthetic lethal interactions. ACS Synth. Biol. 2020, 9, 1181–1189. [Google Scholar] [CrossRef] [PubMed]
- Dai, J.; Boeke, J.D.; Luo, Z.; Jiang, S.; Cai, Y. Sc3.0: Revamping and minimizing the yeast genome. Genome Biol. 2020, 21, 205. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ong, J.Y.; Swidah, R.; Monti, M.; Schindler, D.; Dai, J.; Cai, Y. SCRaMbLE: A Study of Its Robustness and Challenges through Enhancement of Hygromycin B Resistance in a Semi-Synthetic Yeast. Bioengineering 2021, 8, 42. https://doi.org/10.3390/bioengineering8030042
Ong JY, Swidah R, Monti M, Schindler D, Dai J, Cai Y. SCRaMbLE: A Study of Its Robustness and Challenges through Enhancement of Hygromycin B Resistance in a Semi-Synthetic Yeast. Bioengineering. 2021; 8(3):42. https://doi.org/10.3390/bioengineering8030042
Chicago/Turabian StyleOng, Jun Yang, Reem Swidah, Marco Monti, Daniel Schindler, Junbiao Dai, and Yizhi Cai. 2021. "SCRaMbLE: A Study of Its Robustness and Challenges through Enhancement of Hygromycin B Resistance in a Semi-Synthetic Yeast" Bioengineering 8, no. 3: 42. https://doi.org/10.3390/bioengineering8030042
APA StyleOng, J. Y., Swidah, R., Monti, M., Schindler, D., Dai, J., & Cai, Y. (2021). SCRaMbLE: A Study of Its Robustness and Challenges through Enhancement of Hygromycin B Resistance in a Semi-Synthetic Yeast. Bioengineering, 8(3), 42. https://doi.org/10.3390/bioengineering8030042








