Hydrogel Models with Stiffness Gradients for Interrogating Pancreatic Cancer Cell Fate
Abstract
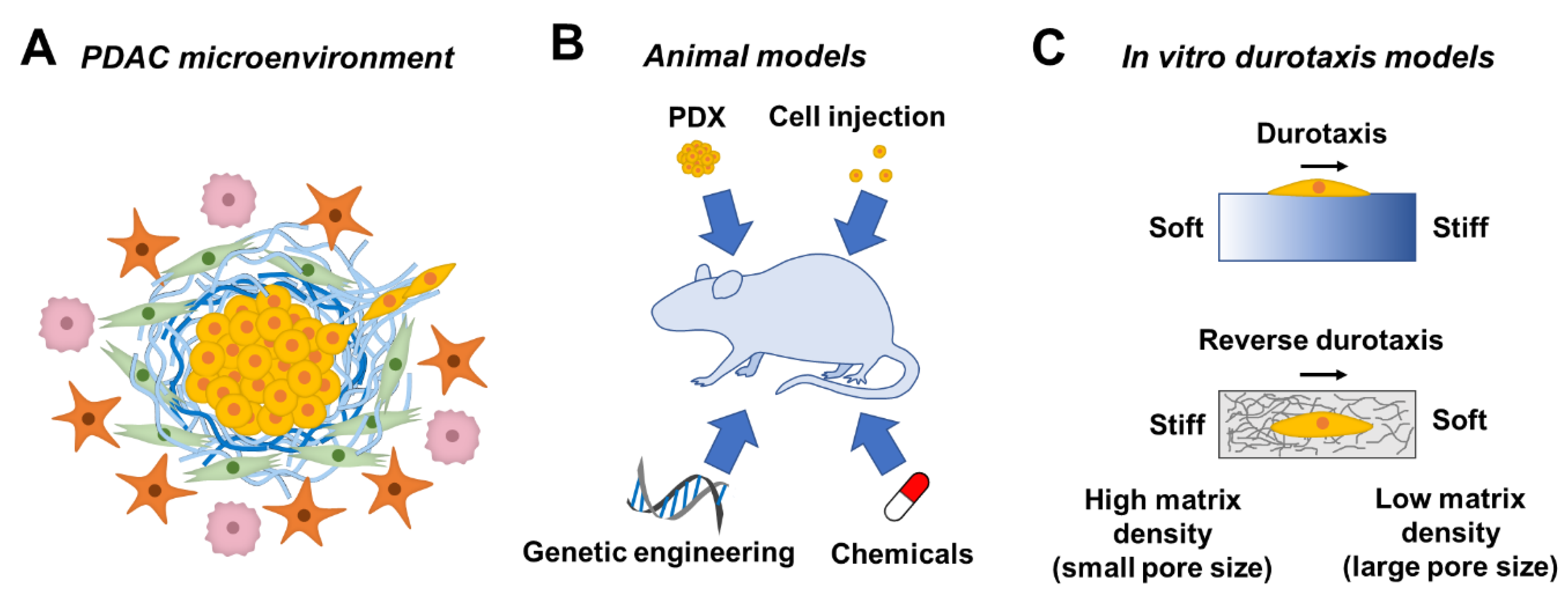
1. Tumor Microenvironment in Pancreatic Ductal Adenocarcinoma
2. Animal Models for Studying PDAC Progression
3. Mimicking a Stiffening Matrix Using Dynamic Hydrogels
| Mechanism | Material | Ref. |
|---|---|---|
| Physical crosslinking | ||
| Supramolecular interactions | Azobenzene-HA + β-cyclodextrin-HA | [82] |
| Supramolecular interactions | o-Nitrobenzyl-methacrylate-HA + dithiothreitol | [83] |
| Supramolecular interactions | β-cyclodextrin-acrylamide + adamantane-acrylamide | [84] |
| Supramolecular interactions | Thiolated poly(vinyl alcohol) + PEG-allylether + β-cyclodextrin-allylether | [85] |
| Ionic crosslinking | Alginate | [86] |
| Temperature-responsive polymers | Polyisocyanide + poly(N-isopropylacrylamide) | [87] |
| Chemical crosslinking | ||
| UV-based photocrosslinking | PEG-norbornene + thiol-bearing peptide | [69] |
| UV-based photocrosslinking | Methacrylated HA + dithiothreitol | [75] |
| UV-based photocrosslinking | PEG-anthracene | [70] |
| Visible light-based photocrosslinking | PEG-acrylamidylpyrene | [71] |
| Visible light-based photocrosslinking | PEG-norbornene + thiol/tyrosine-bearing peptide | [72] |
| Enzymatic crosslinking | Gelatin-norbornene-HPA + thiolated HA (or PEG4SH) | [79] |
| Enzymatic crosslinking | PEG-norbornene + thiol/tyrosine-bearing peptide | [80] |
| Click chemistry | PEG-norbornene + thiol-bearing peptide (or PEG4SH) | [81] |
4. Stiffness Gradient in TME-Mimetic Hydrogels
4.1. Durotaxis in Cancer Cells
| Material | Method | Gradient Range/Strength | Studied Cell Response | Ref. |
|---|---|---|---|---|
| Natural polymers | ||||
| Collagen | Compression on the material | 1057–2305 kPa ~ 31.2 kPa/mm | Durotaxis | [104] |
| Collagen | Juxtaposition of soft and stiff gel | - - | Migration | [105] |
| Collagen | Juxtaposition of soft and stiff gel | 50–217 Pa - | Migration | [106] |
| Matrigel | Patterned underlying substrate | - 0.27–0.37 N/cm * | Durotaxis | [107] |
| Fibrin | Strain-stiffening | - - | Orientation | [108] |
| Modified natural polymers | ||||
| Methacrylated gelatin | Photopolymerization | 4–13 kPa 0.68 kPa/mm | Morphology, differentiation, and durotaxis | [109] |
| Methacrylated gelatin | Photopolymerization | 23.7–1536.7 Pa (G’) - | Morphology and migration | [110] |
| Styrenated gelatin | Photopolymerization | 2.2–83 kPa 40–1600 kPa/mm | Durotaxis | [102] |
| Methacrylated HA | Photopolymerization | 0.5–1.5 kPa - | Spreading and differentiation | [111] |
| Synthetic polymers | ||||
| PA | Photopolymerization | 1–80 kPa 0–40 kPa/mm | Morphology and durotaxis | [94] |
| PA | Photopolymerization | 1–12 kPa 1/10/ ≥ 100 kPa/mm | Durotaxis | [95] |
| PA | Patterned underlying substrate | 1–3.5 kPa - | Durotaxis | [112] |
| PA | Patterned underlying substrate | 3–20 kPa 49.4–190.6 kPa/mm | Orientation and migration | [91] |
| PA | Gradient of diffusion rate | 1–40 kPa 0.5–8.2 kPa/mm | Differentiation | [103] |
| PEGDM | Photopolymerization | 2.05–6.11 kPa - | Phenotype maintenance and ECM deposition | [113] |
| PEGNB | Photopolymerization | 100–360 Pa (G’) | Migration | [96] |
| PDMS | Thermal gradient | 0.19–3.1 MPa 241 kPa/mm | Differentiation | [114] |
| PVA | Freeze–thaw cycle | 1–24 kPa - | Adhesion, proliferation, and differentiation | [115] |
| PAH + PAA | Gradient of crosslinker | 0.5–110 MPa - | Adhesion and proliferation | [116] |
4.2. Creating Stiffness Gradient in Natural Matrices
4.3. Creating Stiffness Gradient in Hybrid and Synthetic Hydrogels

5. Conclusions and Future Directions
Funding
Conflicts of Interest
Abbreviations
| PDAC | Pancreatic ductal adenocarcinoma |
| ECM | Extracellular matrix |
| HA | Hyaluronic acid |
| TAM | Tumor-associated |
| CAF | Cancer-associated fibroblast |
| TME | Tumor microenvironment |
| αSMA | Alpha smooth muscle actin |
| PSC | Pancreatic stellate cell |
| TGF β | Transforming growth factor β |
| IL | Interleukin |
| MMP | Matrix metalloproteinase |
| LIF | Leukemia inhibitory factor |
| GAS | Growth arrest-specific protein |
| FGF | Fibroblast growth factor |
| GDF | Growth differentiation factor |
| HGF | Hepatocyte growth factor |
| EMT | Epithelial–mesenchymal transition |
| MAPK | Mitogen-activated protein kinase |
| PI3K | Phosphatidylinositol 3-kinase |
| AKT | Protein kinase B |
| LOX | Lysyl oxidase |
| HAS | Hyaluronan synthase |
| EPR | Enhanced permeability and retention |
| PanIN | Pancreatic intraepithelial neoplasia |
| SCID | Severe combined immunodeficient |
| PDX | Patient-derived xenograft |
| PA | Polyacrylamide |
| PEG | Poly(ethylene glycol) |
| GelMA | Gelatin-methacryloyl |
| GelNB | Gelatin-norbornene |
| HPA | Hydroxyphenyl acetic acid |
| hiPSC | Human induced pluripotent stem cells |
| PNIPAM | Poly(N-isopropylacrylamide) |
| PEGDM | Polyethylene glycol dimethacrylate |
| PEGNB | Polyethylene glycol norbornene |
| PDMS | Polydimethylsiloxane |
| PVA | Polyvinyl alcohol |
| PAH | Poly (allylamine) hydrochloride |
| PAA | Poly (acrylic acid) |
| EDC | 1-ethyl-3-(3-dimethylaminopropyl) carbo diimide |
| AFM | Atomic force microscope |
| YAP | Yes-associated protein |
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2020. CA A Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Kamisawa, T.; Wood, L.D.; Itoi, T.; Takaori, K. Pancreatic cancer. Nat. Rev. Dis. Primers 2016, 2, 16022. [Google Scholar] [CrossRef]
- Provenzano, P.P.; Cuevas, C.; Chang, A.E.; Goel, V.K.; Von Hoff, D.D.; Hingorani, S.R. Enzymatic Targeting of the Stroma Ablates Physical Barriers to Treatment of Pancreatic Ductal Adenocarcinoma. Cancer Cell 2012, 21, 418–429. [Google Scholar] [CrossRef]
- Whatcott, C.J.; Diep, C.H.; Jiang, P.; Watanabe, A.; LoBello, J.; Sima, C.; Hostetter, G.; Shepard, H.M.; Von Hoff, D.D.; Han, H. Desmoplasia in Primary Tumors and Metastatic Lesions of Pancreatic Cancer. Clin. Cancer Res. 2015, 21, 3561–3568. [Google Scholar] [CrossRef]
- Chauhan, V.P.; Boucher, Y.; Ferrone, C.R.; Roberge, S.; Martin, J.D.; Stylianopoulos, T.; Bardeesy, N.; Depinho, R.A.; Padera, T.P.; Munn, L.L.; et al. Compression of Pancreatic Tumor Blood Vessels by Hyaluronan Is Caused by Solid Stress and Not Interstitial Fluid Pressure. Cancer Cell 2014, 26, 14–15. [Google Scholar] [CrossRef] [PubMed]
- Rhim, A.D.; Oberstein, P.E.; Thomas, D.H.; Mirek, E.T.; Palermo, C.F.; Sastra, S.A.; Dekleva, E.N.; Saunders, T.; Becerra, C.P. Stromal Elements Act to Restrain, Rather than Support, Pancreatic Ductal Adenocarcinoma. Cancer Cell 2014, 25, 735–747. [Google Scholar] [CrossRef] [PubMed]
- Jeong, S.-Y.; Lee, J.-H.; Shin, Y.; Chung, S.; Kuh, H.-J. Co-Culture of Tumor Spheroids and Fibroblasts in a Collagen Matrix-Incorporated Microfluidic Chip Mimics Reciprocal Activation in Solid Tumor Microenvironment. PLoS ONE 2016, 11, e0159013. [Google Scholar] [CrossRef] [PubMed]
- Saini, H.; Eliato, K.R.; Veldhuizen, J.; Zare, A.; Allam, M.; Silva, C.; Kratz, A.; Truong, D.; Mouneimne, G.; LaBaer, J. The role of tumor-stroma interactions on desmoplasia and tumorigenicity within a microengineered 3D platform. Biomaterials 2020, 247, 119975. [Google Scholar] [CrossRef] [PubMed]
- Neri, S.; Hashimoto, H.; Kii, H.; Watanabe, H.; Masutomi, K.; Kuwata, T.; Date, H.; Tsuboi, M.; Goto, K.; Ochiai, A.; et al. Cancer cell invasion driven by extracellular matrix remodeling is dependent on the properties of cancer-associated fibroblasts. J. Cancer Res. Clin. Oncol. 2016, 142, 437–446. [Google Scholar] [CrossRef]
- Sahai, E.; Astsaturov, I.; Cukierman, E.; DeNardo, D.G.; Egeblad, M.; Evans, R.M.; Fearon, D.; Greten, F.R.; Hingorani, S.R.; Hunter, T.; et al. A framework for advancing our understanding of cancer-associated fibroblasts. Nat. Rev. Cancer 2020, 20, 174–186. [Google Scholar] [CrossRef]
- Shan, T.; Chen, S.; Chen, X.; Lin, W.R.; Li, W.; Ma, J.; Wu, T.; Ji, H.; Li, Y.; Cui, X.; et al. Prometastatic mechanisms of CAF-mediated EMT regulation in pancreatic cancer cells. Int. J. Oncol. 2017, 50, 121–128. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, M.F.B.; Mortensen, M.B.; Detlefsen, S. Key players in pancreatic cancer-stroma interaction: Cancer-associated fibroblasts, endothelial and inflammatory cells. World J. Gastroenterol. 2016, 22, 2678–2700. [Google Scholar] [CrossRef] [PubMed]
- Lachowski, D.; Cortes, E.; Pink, D. Substrate Rigidity Controls Activation and Durotaxis in Pancreatic Stellate Cells. Sci. Rep. 2017, 7, 2506. [Google Scholar] [CrossRef]
- Sapudom, J.; Müller, C.D.; Nguyen, K.-T.; Martin, S. Matrix Remodeling and Hyaluronan Production by Myofibroblasts and Cancer-Associated Fibroblasts in 3D Collagen Matrices. Gels 2020, 6, 33. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Zhou, L.; Li, D.; Andl, T.; Zhang, Y. Cancer-Associated Fibroblasts Build and Secure the Tumor Microenvironment. Front. Cell Dev. Biol. 2019, 7, 60. [Google Scholar] [CrossRef]
- Erdogan, B.; Ao, M.; White, L.M.; Means, A.L.; Brewer, B.M.; Yang, L.; Washington, M.K.; Shi, C.; Franco, O.E.; Weaver, A.M.; et al. Cancer-associated fibroblasts promote directional cancer cell migration by aligning fibronectin. J. Cell Biol. 2017, 216, 3799–3816. [Google Scholar] [CrossRef] [PubMed]
- Gaggioli, C.; Hooper, S.; Hidalgo-Carcedo, C.; Grosse, R.; Marshall, J.F.; Harrington, K.; Sahai, E. Fibroblast-led collective invasion of carcinoma cells with differing roles for RhoGTPases in leading and following cells. Nat. Cell Biol. 2007, 9, 1392–1400. [Google Scholar] [CrossRef] [PubMed]
- Zeltz, C.; Primac, I.; Erusappan, P.; Alam, J.; Noel, A.; Gullberg, D. Cancer-associated fibroblasts in desmoplastic tumors: Emerging role of integrins. Semin. Cancer Biol. 2020, 62, 166–181. [Google Scholar] [CrossRef] [PubMed]
- Labernadie, A.; Kato, T.; Brugués, A.; Serra-Picamal, X.; Derzsi, S.; Arwert, E.; Weston, A.; González-Tarragó, V.; Elosegui-Artola, A.; Albertazzi, L.; et al. A mechanically active heterotypic E-cadherin/N-cadherin adhesion enables fibroblasts to drive cancer cell invasion. Nat. Cell Biol. 2017, 19, 224–237. [Google Scholar] [CrossRef]
- Stylianou, A.; Gkretsi, V.; Stylianopoulos, T. Transforming growth factor-β modulates pancreatic cancer associated fibroblasts cell shape, stiffness and invasion. Biochim. Biophys. Acta (BBA) Gen. Subj. 2018, 1862, 1537–1546. [Google Scholar] [CrossRef]
- Massagué, J. TGFβ in Cancer. Cell 2008, 134, 215–230. [Google Scholar] [CrossRef]
- Yang, L.; Pang, Y.; Moses, H.L. TGF-β and immune cells: An important regulatory axis in the tumor microenvironment and progression. Trends Immunol. 2010, 31, 220–227. [Google Scholar] [CrossRef]
- Pickup, M.; Novitskiy, S.; Moses, H.L. The roles of TGFβ in the tumour microenvironment. Nat. Rev. Cancer 2013, 13, 788–799. [Google Scholar] [CrossRef]
- Straussman, R.; Morikawa, T.; Shee, K.; Barzily-Rokni, M.; Qian, Z.R.; Du, J.; Davis, A.; Mongare, M.M.; Gould, J.; Frederick, D.T.; et al. Tumour micro-environment elicits innate resistance to RAF inhibitors through HGF secretion. Nature 2012, 487, 500–504. [Google Scholar] [CrossRef] [PubMed]
- Flint, T.R.; Janowitz, T.; Connell, C.M.; Roberts, E.W.; Denton, A.E.; Coll, A.P.; Jodrell, D.I.; Fearon, D.T. Tumor-Induced IL-6 Reprograms Host Metabolism to Suppress Anti-tumor Immunity. Cell Metab. 2016, 24, 672–684. [Google Scholar] [CrossRef] [PubMed]
- Neuwirt, H.; Bouchal, J.; Kharaishvili, G.; Ploner, C.; Jöhrer, K.; Pitterl, F.; Weber, A.; Klocker, H.; Eder, I.E. Cancer-associated fibroblasts promote prostate tumor growth and progression through upregulation of cholesterol and steroid biosynthesis. Cell Commun. Signal. 2020, 18, 11. [Google Scholar] [CrossRef]
- Walker, C.; Mojares, E.; Del Río Hernández, A. Role of Extracellular Matrix in Development and Cancer Progression. Int. J. Mol. Sci. 2018, 19, 3028. [Google Scholar] [CrossRef] [PubMed]
- Mouw, J.K.; Ou, G.; Weaver, V.M. Extracellular matrix assembly: A multiscale deconstruction. Nat. Rev. Mol. Cell Biol. 2014, 15, 771–785. [Google Scholar] [CrossRef] [PubMed]
- van der Slot, A.J.; van Dura, E.A.; de Wit, E.C.; DeGroot, J.; Huizinga, T.W.; Bank, R.A.; Zuurmond, A.M. Elevated formation of pyridinoline cross-links by profibrotic cytokines is associated with enhanced lysyl hydroxylase 2b levels. Biochim. Biophys. Acta (BBA) Mol. Basis Dis. 2005, 1741, 95–102. [Google Scholar] [CrossRef]
- Amorim, S.; Da Costa, D.S.; Freitas, D.; Reis, C.A.; Reis, R.L.; Pashkuleva, I.; Pires, R.A. Molecular weight of surface immobilized hyaluronic acid influences CD44-mediated binding of gastric cancer cells. Sci. Rep. 2018, 8, 16058. [Google Scholar] [CrossRef]
- Sapudom, J.; Nguyen, K.-T.; Martin, S.; Wippold, T.; Möller, S.; Schnabelrauch, M.; Anderegg, U.; Pompe, T. Biomimetic tissue models reveal the role of hyaluronan in melanoma proliferation and invasion. Biomater. Sci. 2020, 8, 1405–1417. [Google Scholar] [CrossRef]
- Sapudom, J.; Ullm, F.; Martin, S.; Kalbitzer, L.; Naab, J.; Möller, S.; Schnabelrauch, M.; Anderegg, U.; Schmidt, S.; Pompe, T. Molecular weight specific impact of soluble and immobilized hyaluronan on CD44 expressing melanoma cells in 3D collagen matrices. Acta Biomater. 2017, 50, 259–270. [Google Scholar] [CrossRef] [PubMed]
- Price, Z.K.; Lokman, N.A.; Ricciardelli, C. Differing Roles of Hyaluronan Molecular Weight on Cancer Cell Behavior and Chemotherapy Resistance. Cancers 2018, 10, 482. [Google Scholar] [CrossRef]
- Gurski, L.A.; Xu, X.; Labrada, L.N.; Nguyen, N.T.; Xiao, L.; Van Golen, K.L.; Jia, X. Hyaluronan (HA) Interacting Proteins RHAMM and Hyaluronidase Impact Prostate Cancer Cell Behavior and Invadopodia Formation in 3D HA-Based Hydrogels. PLoS ONE 2012, 7, e50075. [Google Scholar] [CrossRef]
- Götte, M.; Yip, G.W. Heparanase, Hyaluronan, and CD44 in Cancers: A Breast Carcinoma Perspective. Cancer Res. 2006, 66, 10233–10237. [Google Scholar] [CrossRef] [PubMed]
- Spill, F.; Reynolds, D.S.; Kamm, R.D.; Zaman, M.H. Impact of the physical microenvironment on tumor progression and metastasis. Curr. Opin. Biotechnol. 2016, 40, 41–48. [Google Scholar] [CrossRef] [PubMed]
- Rice, A.J.; Cortes, E.; Lachowski, D.; Cheung, B.C.H.; A Karim, S.; Morton, J.P.; Hernández, A.D.R. Matrix stiffness induces epithelial–mesenchymal transition and promotes chemoresistance in pancreatic cancer cells. Oncogenesis 2017, 6, e352. [Google Scholar] [CrossRef]
- Cox, T.R.; Erler, J.T. Remodeling and homeostasis of the extracellular matrix: Implications for fibrotic diseases and cancer. Dis. Models Mech. 2011, 4, 165–178. [Google Scholar] [CrossRef] [PubMed]
- Lee, G.; Han, S.-B.; Lee, J.-H.; Kim, H.-W.; Kim, D.-H. Cancer Mechanobiology: Microenvironmental Sensing and Metastasis. ACS Biomater. Sci. Eng. 2019, 5, 3735–3752. [Google Scholar] [CrossRef]
- Ahmadzadeh, H.; Webster, M.R.; Behera, R.; Valencia, A.M.J.; Wirtz, D.; Weeraratna, A.T.; Shenoy, V.B. Modeling the two-way feedback between contractility and matrix realignment reveals a nonlinear mode of cancer cell invasion. Proc. Natl. Acad. Sci. USA 2017, 114, E1617–E1626. [Google Scholar] [CrossRef]
- Han, Y.L.; Ronceray, P.; Xu, G.; Malandrino, A.; Kamm, R.D.; Lenz, M.; Broedersz, C.P.; Guo, M. Cell contraction induces long-ranged stress stiffening in the extracellular matrix. Proc. Natl. Acad. Sci. USA 2018, 115, 4075–4080. [Google Scholar] [CrossRef] [PubMed]
- Chaudhuri, O.; Koshy, S.T.; Da Cunha, C.B.; Shin, J.-W.; Verbeke, C.S.; Allison, K.H.; Mooney, D.J. Extracellular matrix stiffness and composition jointly regulate the induction of malignant phenotypes in mammary epithelium. Nat. Mater. 2014, 13, 970–978. [Google Scholar] [CrossRef]
- Gilkes, D.M.; Semenza, G.L.; Wirtz, D. Hypoxia and the extracellular matrix: Drivers of tumour metastasis. Nat. Rev. Cancer 2014, 14, 430–439. [Google Scholar] [CrossRef] [PubMed]
- Maeda, H.; Wu, J.; Sawa, T.; Matsumura, Y.; Hori, K. Tumor vascular permeability and the EPR effect in macromolecular therapeutics: A review. J. Control. Release 2000, 65, 271–284. [Google Scholar] [CrossRef]
- Prabhakar, U.; Maeda, H.; Jain, R.K.; Sevick-Muraca, E.M.; Zamboni, W.; Farokhzad, O.C.; Barry, S.T.; Gabizon, A.; Grodzinski, P.; Blakey, D.C. Challenges and Key Considerations of the Enhanced Permeability and Retention Effect for Nanomedicine Drug Delivery in Oncology. Cancer Res. 2013, 73, 2412–2417. [Google Scholar] [CrossRef] [PubMed]
- Di Maggio, F.; Arumugam, P.; Delvecchio, F.R.; Batista, S.; Lechertier, T.; Hodivala-Dilke, K.; Kocher, H.M. Pancreatic stellate cells regulate blood vessel density in the stroma of pancreatic ductal adenocarcinoma. Pancreatology 2016, 16, 995–1004. [Google Scholar] [CrossRef]
- Nguyen, D.-H.T.; Lee, E.; Alimperti, S.; Norgard, R.J.; Wong, A.; Lee, J.J.-K.; Eyckmans, J.; Stanger, B.Z.; Chen, C.S. A biomimetic pancreatic cancer on-chip reveals endothelial ablation via ALK7 signaling. Sci. Adv. 2019, 5, eaav6789. [Google Scholar] [CrossRef]
- Saiyin, H.; Ardito-Abraham, C.M.; Wu, Y.; Wei, Y.; Fang, Y.; Han, X.; Li, J.; Zhou, P.; Yi, Q.; Maitra, A.; et al. Identification of novel vascular projections with cellular trafficking abilities on the microvasculature of pancreatic ductal adenocarcinoma. J. Pathol. 2015, 236, 142–154. [Google Scholar] [CrossRef] [PubMed]
- Ayuso, J.M.; Gong, M.M.; Skala, M.C.; Harari, P.M.; Beebe, D.J. Human Tumor-Lymphatic Microfluidic Model Reveals Differential Conditioning of Lymphatic Vessels by Breast Cancer Cells. Adv. Healthc. Mater. 2020, 9, 1900925. [Google Scholar] [CrossRef] [PubMed]
- Moon, J.J.; Saik, J.E.; Poché, R.A.; Leslie-Barbick, J.E.; Lee, S.-H.; Smith, A.A.; Dickinson, M.E.; West, J.L. Biomimetic hydrogels with pro-angiogenic properties. Biomaterials 2010, 31, 3840–3847. [Google Scholar] [CrossRef]
- Roudsari, L.C.; Jeffs, S.E.; Witt, A.S.; Gill, B.J.; West, J.L. A 3D Poly(ethylene glycol)-based Tumor Angiogenesis Model to Study the Influence of Vascular Cells on Lung Tumor Cell Behavior. Sci. Rep. 2016, 6, 32726. [Google Scholar] [CrossRef]
- Rao, M.S. Animal models of exocrine pancreatic carcinogenesis. Cancer Metastasis Rev. 1987, 6, 665–676. [Google Scholar] [CrossRef]
- Hayashi, Y.; Hasegawa, T. Experimental Pancreatic Tumor in Rats after Intravenous Injection of 4-Hydroxyaminoquinoline 1-oxide. GANN Jpn. J. Cancer Res. 1971, 62, 329–330_1. [Google Scholar]
- Reddy, J.K.; Rao, M.S. Malignant Tumors in Rats Fed Nafenopin, a Hepatic Peroxisome Proliferator2. JNCI J. Natl. Cancer Inst. 1977, 59, 1645–1650. [Google Scholar] [CrossRef] [PubMed]
- Reddy, J.K.; Qureshi, S.A. Tumorigenicity of the hypolipidaemic peroxisome proliferator ethyl-α-p-chlorophenoxyisobutyrate (clofibrate) in rats. Br. J. Cancer 1979, 40, 476–482. [Google Scholar] [CrossRef]
- Hingorani, S.R.; Petricoin, E.F.; Maitra, A.; Rajapakse, V.; King, C.; Jacobetz, M.A.; Ross, S.; Conrads, T.P.; Veenstra, T.D.; Hitt, B.A.; et al. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell 2003, 4, 437–450. [Google Scholar] [CrossRef]
- Aguirre, A.J.; Bardeesy, N.; Sinha, M.; Lopez, L.; Tuveson, D.A.; Horner, J.; Redston, M.S.; Depinho, R.A. Activated Kras and Ink4a/Arf deficiency cooperate to produce metastatic pancreatic ductal adenocarcinoma. Genes Dev. 2003, 17, 3112–3126. [Google Scholar] [CrossRef] [PubMed]
- Kong, K.; Guo, M.; Liu, Y.; Zheng, J. Progress in Animal Models of Pancreatic Ductal Adenocarcinoma. J. Cancer 2020, 11, 1555–1567. [Google Scholar] [CrossRef]
- Yu, X.; Li, H.; Fu, D.; Jin, C.; Li, J. Characterization of the role of the photosensitizer, deuteporfin, in the detection of lymphatic metastases in a pancreatic cancer xenograft model. Oncol. Lett. 2015, 10, 1430–1436. [Google Scholar] [CrossRef]
- Saur, D.; Seidler, B.; Schneider, G.; Algül, H.; Beck, R.; Senekowitsch–Schmidtke, R.; Schwaiger, M.; Schmid, R.M. CXCR4 Expression Increases Liver and Lung Metastasis in a Mouse Model of Pancreatic Cancer. Gastroenterology 2005, 129, 1237–1250. [Google Scholar] [CrossRef] [PubMed]
- Vezeridis, M.P.; Tzanakakis, G.N.; Meitner, P.A.; Doremus, C.M.; Tibbetts, L.M.; Calabresi, P. In vivo selection of a highly metastatic cell line from a human pancreatic carcinoma in the nude mouse. Cancer 1992, 69, 2060–2063. [Google Scholar] [CrossRef]
- Hidalgo, M.; Amant, F.; Biankin, A.V.; Budinská, E.; Byrne, A.T.; Caldas, C.; Clarke, R.B.; De Jong, S.; Jonkers, J.; Mælandsmo, G.M.; et al. Patient-Derived Xenograft Models: An Emerging Platform for Translational Cancer Research. Cancer Discov. 2014, 4, 998–1013. [Google Scholar] [CrossRef]
- Erstad, D.J.; Sojoodi, M.; Taylor, M.S.; Ghoshal, S.; Razavi, A.A.; Graham-O’Regan, K.A.; Bardeesy, N.; Ferrone, C.R.; Lanuti, M.; Caravan, P.; et al. Orthotopic and heterotopic murine models of pancreatic cancer and their different responses to FOLFIRINOX chemotherapy. Dis. Models Mech. 2018, 11, dmm034793. [Google Scholar] [CrossRef]
- Awasthi, N.; Kronenberger, D.; Stefaniak, A.; Hassan, S.; Von Holzen, U.; Schwarz, M.A.; Schwarz, R.E. Dual inhibition of the PI3K and MAPK pathways enhances nab-paclitaxel/gemcitabine chemotherapy response in preclinical models of pancreatic cancer. Cancer Lett. 2019, 459, 41–49. [Google Scholar] [CrossRef]
- Kim, M.P.; Truty, M.J.; Choi, W.; Kang, Y.; Chopin-Lally, X.; Gallick, G.E.; Wang, H.; McConkey, D.J.; Logsdon, C.; Abbruzzesse, J.; et al. Molecular Profiling of Direct Xenograft Tumors Established from Human Pancreatic Adenocarcinoma After Neoadjuvant Therapy. Ann. Surg. Oncol. 2012, 19, 395–403. [Google Scholar] [CrossRef] [PubMed]
- Von Hoff, D.D.; Ramanathan, R.K.; Borad, M.J.; Laheru, D.A.; Smith, L.S.; Wood, T.E.; Korn, R.L.; Desai, N.; Trieu, V.; Iglesias, J.L.; et al. Gemcitabine Plus nab-Paclitaxel Is an Active Regimen in Patients With Advanced Pancreatic Cancer: A Phase I/II Trial. J. Clin. Oncol. 2011, 29, 4548–4554. [Google Scholar] [CrossRef] [PubMed]
- Von Hoff, D.D.; Ervin, T.; Arena, F.P.; Chiorean, E.G.; Infante, J.; Moore, M.; Seay, T.; Tjulandin, S.A.; Ma, W.W.; Saleh, M.N.; et al. Increased Survival in Pancreatic Cancer with nab-Paclitaxel plus Gemcitabine. N. Engl. J. Med. 2013, 369, 1691–1703. [Google Scholar] [CrossRef]
- Denisin, A.K.; Pruitt, B.L. Tuning the Range of Polyacrylamide Gel Stiffness for Mechanobiology Applications. ACS Appl. Mater. Interfaces 2016, 8, 21893–21902. [Google Scholar] [CrossRef] [PubMed]
- Mabry, K.M.; Lawrence, R.L.; Anseth, K.S. Dynamic stiffening of poly(ethylene glycol)-based hydrogels to direct valvular interstitial cell phenotype in a three-dimensional environment. Biomaterials 2015, 49, 47–56. [Google Scholar] [CrossRef]
- Günay, K.A.; Ceccato, T.L.; Silver, J.S.; Bannister, K.L.; Bednarski, O.J.; Leinwand, L.A.; Anseth, K.S. PEG–Anthracene Hydrogels as an On-Demand Stiffening Matrix To Study Mechanobiology. Angew. Chem. Int. Ed. 2019, 58, 9912–9916. [Google Scholar] [CrossRef]
- Kalayci, K.; Frisch, H.; Barner-Kowollik, C.; Truong, V.X. Wavelength-Dependent Stiffening of Hydrogel Matrices via Redshifted [2+2] Photocycloadditions. Adv. Funct. Mater. 2020, 30, 1908171. [Google Scholar] [CrossRef]
- Liu, H.-Y.; Nguyen, H.D.; Lin, C.-C. Dynamic PEG–Peptide Hydrogels via Visible Light and FMN-Induced Tyrosine Dimerization. Adv. Healthc. Mater. 2018, 7, 1800954. [Google Scholar] [CrossRef] [PubMed]
- Khetan, S.; Katz, J.S.; Burdick, J.A. Sequential crosslinking to control cellular spreading in 3-dimensional hydrogels. Soft Matter 2009, 5, 1601–1606. [Google Scholar] [CrossRef]
- Khetan, S.; Guvendiren, M.; Legant, W.R.; Cohen, D.M.; Chen, C.S.; Burdick, J.A. Degradation-mediated cellular traction directs stem cell fate in covalently crosslinked three-dimensional hydrogels. Nat. Mater. 2013, 12, 458–465. [Google Scholar] [CrossRef] [PubMed]
- Caliari, S.R.; Perepelyuk, M.; Cosgrove, B.D.; Tsai, S.J.; Lee, G.Y.; Mauck, R.L.; Wells, R.G.; Burdick, J.A. Stiffening hydrogels for investigating the dynamics of hepatic stellate cell mechanotransduction during myofibroblast activation. Sci. Rep. 2016, 6, 21387. [Google Scholar] [CrossRef] [PubMed]
- Mũnoz, Z.; Shih, H.; Lin, C.-C. Gelatin hydrogels formed by orthogonal thiol–norbornene photochemistry for cell encapsulation. Biomater. Sci. 2014, 2, 1063–1072. [Google Scholar] [CrossRef]
- Shih, H.; Greene, T.; Korc, M.; Lin, C.-C. Modular and Adaptable Tumor Niche Prepared from Visible Light Initiated Thiol-Norbornene Photopolymerization. Biomacromolecules 2016, 17, 3872–3882. [Google Scholar] [CrossRef]
- Koshy, S.T.; Desai, R.M.; Joly, P.; Li, J.; Bagrodia, R.K.; Lewin, S.A.; Joshi, N.S.; Mooney, D.J. Click-Crosslinked Injectable Gelatin Hydrogels. Adv. Healthc. Mater. 2016, 5, 541–547. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.-Y.; Korc, M.; Lin, C.-C. Biomimetic and enzyme-responsive dynamic hydrogels for studying cell-matrix interactions in pancreatic ductal adenocarcinoma. Biomaterials 2018, 160, 24–36. [Google Scholar] [CrossRef]
- Liu, H.-Y.; Greene, T.; Lin, T.-Y.; Dawes, C.S.; Korc, M.; Lin, C.-C. Enzyme-mediated stiffening hydrogels for probing activation of pancreatic stellate cells. Acta Biomater. 2017, 48, 258–269. [Google Scholar] [CrossRef]
- Arkenberg, M.R.; Dimmitt, N.H.; Johnson, H.C.; Koehler, K.R.; Lin, C.-C. Dynamic Click Hydrogels for Xeno-Free Culture of Induced Pluripotent Stem Cells. Adv. Biosyst. 2020, 4, 2000129. [Google Scholar] [CrossRef]
- Rosales, A.M.; Rodell, C.B.; Chen, M.H.; Morrow, M.G.; Anseth, K.S.; Burdick, J.A. Reversible Control of Network Properties in Azobenzene-Containing Hyaluronic Acid-Based Hydrogels. Bioconjugate Chem. 2018, 29, 905–913. [Google Scholar] [CrossRef] [PubMed]
- Rosales, A.M.; Vega, S.L.; DelRio, F.W.; Burdick, J.A.; Anseth, K.S. Hydrogels with Reversible Mechanics to Probe Dynamic Cell Microenvironments. Angew. Chem. Int. Ed. 2017, 56, 12132–12136. [Google Scholar] [CrossRef] [PubMed]
- Hörning, M.; Nakahata, M.; Linke, P.; Yamamoto, A.; Veschgini, M.; Kaufmann, S.; Takashima, Y.; Harada, A.; Tanaka, M. Dynamic Mechano-Regulation of Myoblast Cells on Supramolecular Hydrogels Cross-Linked by Reversible Host-Guest Interactions. Sci. Rep. 2017, 7, 7660. [Google Scholar] [CrossRef] [PubMed]
- Shih, H.; Lin, C.-C. Tuning stiffness of cell-laden hydrogel via host–guest interactions. J. Mater. Chem. B 2016, 4, 4969–4974. [Google Scholar] [CrossRef]
- Stowers, R.S.; Allen, S.C.; Suggs, L.J. Dynamic phototuning of 3D hydrogel stiffness. Proc. Natl. Acad. Sci. USA 2015, 112, 1953–1958. [Google Scholar] [CrossRef]
- de Almeida, P.; Jaspers, M.; Vaessen, S.; Tagit, O.; Portale, G.; Rowan, A.E.; Kouwer, P.H. Cytoskeletal stiffening in synthetic hydrogels. Nat. Commun. 2019, 10, 609. [Google Scholar] [CrossRef]
- Cooperstein, M.A.; Canavan, H.E. Assessment of cytotoxicity of (N-isopropyl acrylamide) and Poly(N-isopropyl acrylamide)-coated surfaces. Biointerphases 2013, 8, 19. [Google Scholar] [CrossRef]
- Zhu, M.; Tao, H.; Samani, M.; Luo, M.; Wang, X.; Hopyan, S.; Sun, Y. Spatial mapping of tissue properties in vivo reveals a 3D stiffness gradient in the mouse limb bud. Proc. Natl. Acad. Sci. USA 2020, 117, 4781–4791. [Google Scholar] [CrossRef] [PubMed]
- Berry, M.F.; Engler, A.J.; Woo, Y.J.; Pirolli, T.J.; Bish, L.T.; Jayasankar, V.; Morine, K.J.; Gardner, T.J.; Discher, D.E.; Sweeney, H.L. Mesenchymal stem cell injection after myocardial infarction improves myocardial compliance. Am. J. Physiol. -Heart Circ. Physiol. 2006, 290, H2196–H2203. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Lin, F.; Huang, J.; Xiong, C. Anisotropic stiffness gradient-regulated mechanical guidance drives directional migration of cancer cells. Acta Biomater. 2020, 106, 181–192. [Google Scholar] [CrossRef]
- DuChez, B.J.; Doyle, A.D.; Dimitriadis, E.K.; Yamada, K.M. Durotaxis by Human Cancer Cells. Biophys. J. 2019, 116, 670–683. [Google Scholar] [CrossRef]
- Lo, C.-M.; Wang, H.-B.; Dembo, M.; Wang, Y.-L. Cell Movement Is Guided by the Rigidity of the Substrate. Biophys. J. 2000, 79, 144–152. [Google Scholar] [CrossRef]
- Isenberg, B.C.; DiMilla, P.A.; Walker, M.; Kim, S.; Wong, J.Y. Vascular Smooth Muscle Cell Durotaxis Depends on Substrate Stiffness Gradient Strength. Biophys. J. 2009, 97, 1313–1322. [Google Scholar] [CrossRef]
- Vincent, L.G.; Choi, Y.S.; Alonso-Latorre, B.; del Álamo, J.C.; Engler, A.J. Mesenchymal stem cell durotaxis depends on substrate stiffness gradient strength. Biotechnol. J. 2013, 8, 472–484. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.P.; Schwartz, M.P.; Lee, J.Y.; Fairbanks, B.D.; Anseth, K.S. A peptide functionalized poly(ethylene glycol) (PEG) hydrogel for investigating the influence of biochemical and biophysical matrix properties on tumor cell migration. Biomater. Sci. 2014, 2, 1024–1034. [Google Scholar] [CrossRef] [PubMed]
- Storm, C.; Pastore, J.J.; MacKintosh, F.C.; Lubensky, T.C.; Janmey, P.A. Nonlinear elasticity in biological gels. Nature 2005, 435, 191–194. [Google Scholar] [CrossRef] [PubMed]
- van Helvert, S.; Friedl, P. Strain Stiffening of Fibrillar Collagen during Individual and Collective Cell Migration Identified by AFM Nanoindentation. ACS Appl. Mater. Interfaces 2016, 8, 21946–21955. [Google Scholar] [CrossRef] [PubMed]
- Engler, A.J.; Sen, S.; Sweeney, H.L.; Discher, D.E. Matrix elasticity directs stem cell lineage specification. Cell 2006, 126, 677–689. [Google Scholar] [CrossRef]
- Ulrich, T.A.; de Juan Pardo, E.M.; Kumar, S. The Mechanical Rigidity of the Extracellular Matrix Regulates the Structure, Motility, and Proliferation of Glioma Cells. Cancer Res. 2009, 69, 4167–4174. [Google Scholar] [CrossRef]
- Bauer, J.; Emon, M.A.B.; Staudacher, J.J.; Thomas, A.L.; Zessner-Spitzenberg, J.; Mancinelli, G.; Krett, N.; Saif, M.T.; Jung, B. Increased stiffness of the tumor microenvironment in colon cancer stimulates cancer associated fibroblast-mediated prometastatic activin A signaling. Sci. Rep. 2020, 10, 50. [Google Scholar] [CrossRef]
- Moriyama, K.; Kidoaki, S. Cellular Durotaxis Revisited: Initial-Position-Dependent Determination of the Threshold Stiffness Gradient to Induce Durotaxis. Langmuir 2019, 35, 7478–7486. [Google Scholar] [CrossRef] [PubMed]
- Hadden, W.J.; Young, J.L.; Holle, A.W.; McFetridge, M.L.; Kim, D.Y.; Wijesinghe, P.; Taylor-Weiner, H.; Wen, J.H.; Lee, A.R.; Bieback, K.; et al. Stem cell migration and mechanotransduction on linear stiffness gradient hydrogels. Proc. Natl. Acad. Sci. USA 2017, 114, 5647–5652. [Google Scholar] [CrossRef]
- Hadjipanayi, E.; Mudera, V.; Brown, R.A. Guiding cell migration in 3D: A collagen matrix with graded directional stiffness. Cell Motil. 2009, 66, 121–128. [Google Scholar] [CrossRef] [PubMed]
- Bordeleau, F.; Tang, L.N.; Reinhart-King, C.A. Topographical guidance of 3D tumor cell migration at an interface of collagen densities. Phys. Biol. 2013, 10, 065004. [Google Scholar] [CrossRef] [PubMed]
- Sapudom, J.; Rubner, S.; Martin, S.; Pompe, T. Mimicking Tissue Boundaries by Sharp Multiparameter Matrix Interfaces. Adv. Healthc. Mater. 2016, 5, 1861–1867. [Google Scholar] [CrossRef] [PubMed]
- Joaquin, D.; Grigola, M.; Kwon, G.; Blasius, C.; Han, Y.; Perlitz, D.; Jiang, J.; Ziegler, Y.; Nardulli, A.; Hsia, K.J. Cell migration and organization in three-dimensional in vitro culture driven by stiffness gradient. Biotechnol. Bioeng. 2016, 113, 2496–2506. [Google Scholar] [CrossRef]
- Kotlarchyk, M.A.; Shreim, S.G.; Alvarez-Elizondo, M.B.; Estrada, L.C.; Singh, R.; Valdevit, L.; Kniazeva, E.; Gratton, E.; Putnam, A.J.; Botvinick, E.L. Concentration Independent Modulation of Local Micromechanics in a Fibrin Gel. PLoS ONE 2011, 6, e20201. [Google Scholar] [CrossRef]
- Kim, C.; Young, J.L.; Holle, A.W.; Jeong, K.; Major, L.G.; Jeong, J.H.; Aman, Z.M.; Han, D.-W.; Hwang, Y.; Spatz, J.P.; et al. Stem Cell Mechanosensation on Gelatin Methacryloyl (GelMA) Stiffness Gradient Hydrogels. Ann. Biomed. Eng. 2020, 48, 893–902. [Google Scholar] [CrossRef] [PubMed]
- Lavrentieva, A.; Fleischhammer, T.; Enders, A.; Pirmahboub, H.; Bahnemann, J.; Pepelanova, I. Fabrication of Stiffness Gradients of GelMA Hydrogels Using a 3D Printed Micromixer. Macromol. Biosci. 2020, 20, 2000107. [Google Scholar] [CrossRef] [PubMed]
- Rape, A.D.; Zibinsky, M.; Murthy, N.; Kumar, S. A synthetic hydrogel for the high-throughput study of cell–ECM interactions. Nat. Commun. 2015, 6, 8129. [Google Scholar] [CrossRef]
- Kuo, C.-H.R.; Xian, J.; Brenton, J.D.; Franze, K.; Sivaniah, E. Complex Stiffness Gradient Substrates for Studying Mechanotactic Cell Migration. Adv. Mater. 2012, 24, 6059–6064. [Google Scholar] [CrossRef]
- Smith Callahan, L.A.; Ganios, A.M.; Childers, E.P.; Weiner, S.D.; Becker, M.L. Primary human chondrocyte extracellular matrix formation and phenotype maintenance using RGD-derivatized PEGDM hydrogels possessing a continuous Young’s modulus gradient. Acta Biomater. 2013, 9, 6095–6104. [Google Scholar] [CrossRef]
- Wang, P.-Y.; Tsai, W.-B.; Voelcker, N.H. Screening of rat mesenchymal stem cell behaviour on polydimethylsiloxane stiffness gradients. Acta Biomater. 2012, 8, 519–530. [Google Scholar] [CrossRef]
- Kim, T.H.; An, D.B.; Oh, S.H.; Kang, M.K.; Song, H.H.; Lee, J.H. Creating stiffness gradient polyvinyl alcohol hydrogel using a simple gradual freezing–thawing method to investigate stem cell differentiation behaviors. Biomaterials 2015, 40, 51–60. [Google Scholar] [CrossRef] [PubMed]
- Hopp, I.; Michelmore, A.; Smith, L.E.; Robinson, D.E.; Bachhuka, A.; Mierczynska, A.; Vasilev, K. The influence of substrate stiffness gradients on primary human dermal fibroblasts. Biomaterials 2013, 34, 5070–5077. [Google Scholar] [CrossRef]
- Puls, T.J.; Tan, X.; Whittington, C.F.; Voytik-Harbin, S.L. 3D collagen fibrillar microstructure guides pancreatic cancer cell phenotype and serves as a critical design parameter for phenotypic models of EMT. PLoS ONE 2017, 12, e0188870. [Google Scholar] [CrossRef] [PubMed]
- Seo, B.R.; Chen, X.; Ling, L.; Song, Y.H.; Shimpi, A.A.; Choi, S.; Gonzalez, J.; Sapudom, J.; Wang, K.; Eguiluz, R.C.A.; et al. Collagen microarchitecture mechanically controls myofibroblast differentiation. Proc. Natl. Acad. Sci. USA 2020, 117, 11387–11398. [Google Scholar] [CrossRef]
- Licup, A.J.; Münster, S.; Sharma, A.; Sheinman, M.; Jawerth, L.M.; Fabry, B.; Weitz, D.A.; Mackintosh, F.C. Stress controls the mechanics of collagen networks. Proc. Natl. Acad. Sci. USA 2015, 112, 9573–9578. [Google Scholar] [CrossRef] [PubMed]
- Sapudom, J.; Rubner, S.; Martin, S.; Kurth, T.; Riedel, S.; Mierke, C.T.; Pompe, T. The phenotype of cancer cell invasion controlled by fibril diameter and pore size of 3D collagen networks. Biomaterials 2015, 52, 367–375. [Google Scholar] [CrossRef] [PubMed]
- Sapudom, J.; Kalbitzer, L.; Wu, X.; Martin, S.; Kroy, K.; Pompe, T. Fibril bending stiffness of 3D collagen matrices instructs spreading and clustering of invasive and non-invasive breast cancer cells. Biomaterials 2019, 193, 47–57. [Google Scholar] [CrossRef]
- Taufalele, P.V.; VanderBurgh, J.A.; Muñoz, A.; Zanotelli, M.R.; Reinhart-King, C.A. Fiber alignment drives changes in architectural and mechanical features in collagen matrices. PLoS ONE 2019, 14, e0216537. [Google Scholar] [CrossRef]
- Zhu, D.; Trinh, P.; Li, J.; Grant, G.A.; Yang, F. Gradient hydrogels for screening stiffness effects on patient-derived glioblastoma xenograft cellfates in 3D. J. Biomed. Mater. Res. Part A 2020. [Google Scholar] [CrossRef] [PubMed]
- Pedron, S.; Polishetty, H.; Pritchard, A.M.; Mahadik, B.P.; Woese, C.R.; Sarkaria, J.N.; Harley, B.A.C. Spatially graded hydrogels for preclinical testing of glioblastoma anticancer therapeutics. MRS Commun. 2017, 7, 442–449. [Google Scholar] [CrossRef]
- Sunyer, R.; Jin, A.J.; Nossal, R.; Sackett, D.L. Fabrication of Hydrogels with Steep Stiffness Gradients for Studying Cell Mechanical Response. PLoS ONE 2012, 7, e46107. [Google Scholar] [CrossRef] [PubMed]
- Nemir, S.; Hayenga, H.N.; West, J.L. PEGDA hydrogels with patterned elasticity: Novel tools for the study of cell response to substrate rigidity. Biotechnol. Bioeng. 2010, 105, 636–644. [Google Scholar] [CrossRef]
- Olivares, O.; Mayers, J.R.; Gouirand, V.; Torrence, M.E.; Gicquel, T.; Borge, L.; Lac, S.; Roques, J.; Lavaut, M.-N.; Berthezène, P.; et al. Collagen-derived proline promotes pancreatic ductal adenocarcinoma cell survival under nutrient limited conditions. Nat. Commun. 2017, 8, 16031. [Google Scholar] [CrossRef]
- Kamphorst, J.J.; Nofal, M.; Commisso, C.; Hackett, S.R.; Lu, W.; Grabocka, E.; Heiden, M.G.V.; Miller, G.; Drebin, J.A.; Bar-Sagi, D.; et al. Human Pancreatic Cancer Tumors Are Nutrient Poor and Tumor Cells Actively Scavenge Extracellular Protein. Cancer Res. 2015, 75, 544–553. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chang, C.-Y.; Lin, C.-C. Hydrogel Models with Stiffness Gradients for Interrogating Pancreatic Cancer Cell Fate. Bioengineering 2021, 8, 37. https://doi.org/10.3390/bioengineering8030037
Chang C-Y, Lin C-C. Hydrogel Models with Stiffness Gradients for Interrogating Pancreatic Cancer Cell Fate. Bioengineering. 2021; 8(3):37. https://doi.org/10.3390/bioengineering8030037
Chicago/Turabian StyleChang, Chun-Yi, and Chien-Chi Lin. 2021. "Hydrogel Models with Stiffness Gradients for Interrogating Pancreatic Cancer Cell Fate" Bioengineering 8, no. 3: 37. https://doi.org/10.3390/bioengineering8030037
APA StyleChang, C.-Y., & Lin, C.-C. (2021). Hydrogel Models with Stiffness Gradients for Interrogating Pancreatic Cancer Cell Fate. Bioengineering, 8(3), 37. https://doi.org/10.3390/bioengineering8030037







