Polo-Like Kinase 4 (PLK4) Is Overexpressed in Central Nervous System Neuroblastoma (CNS-NB)
Abstract
1. Introduction
2. Materials and Methods
2.1. Quantitative Real-Time PCR (qRT-PCR)
2.2. Gene Expression Meta-Analysis
3. Results
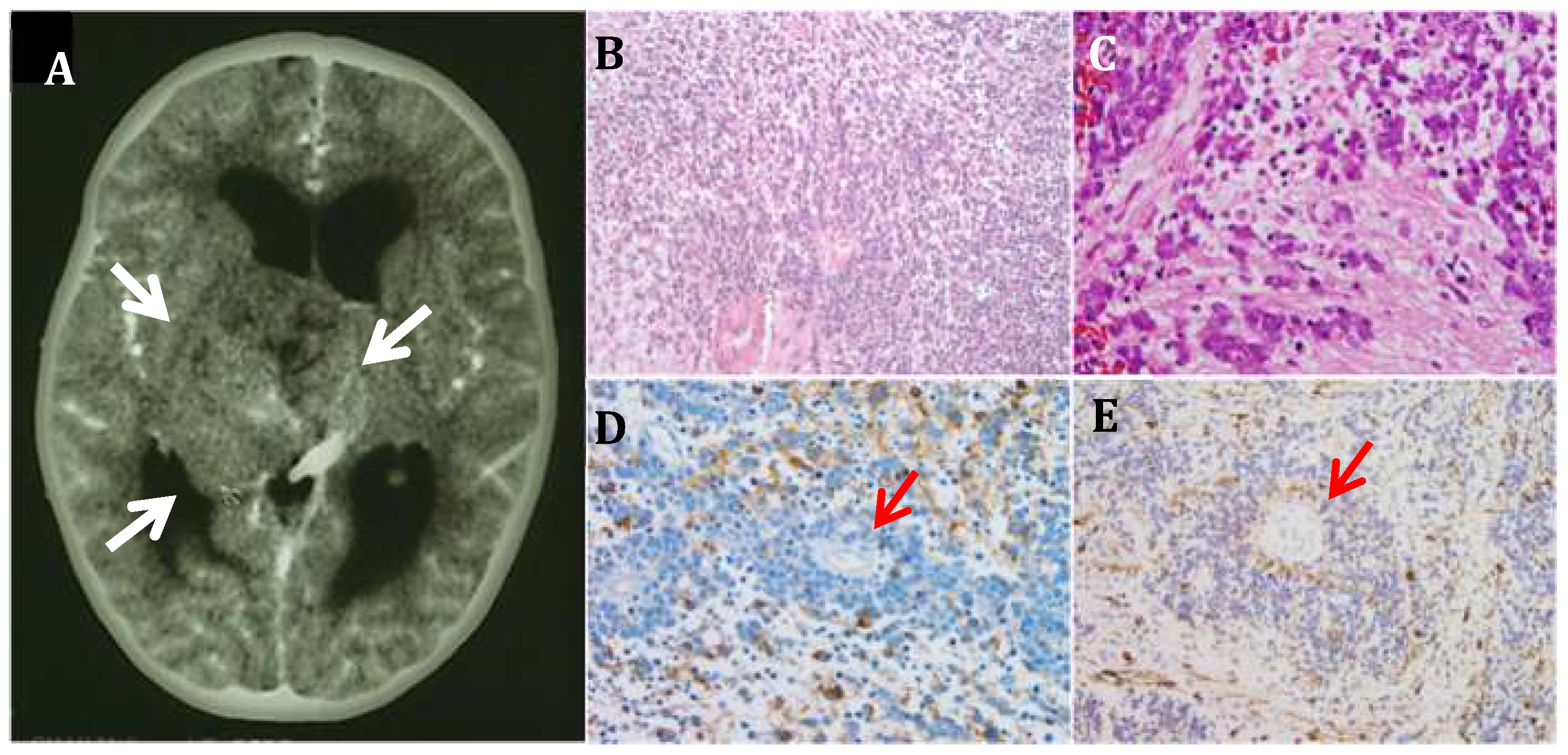
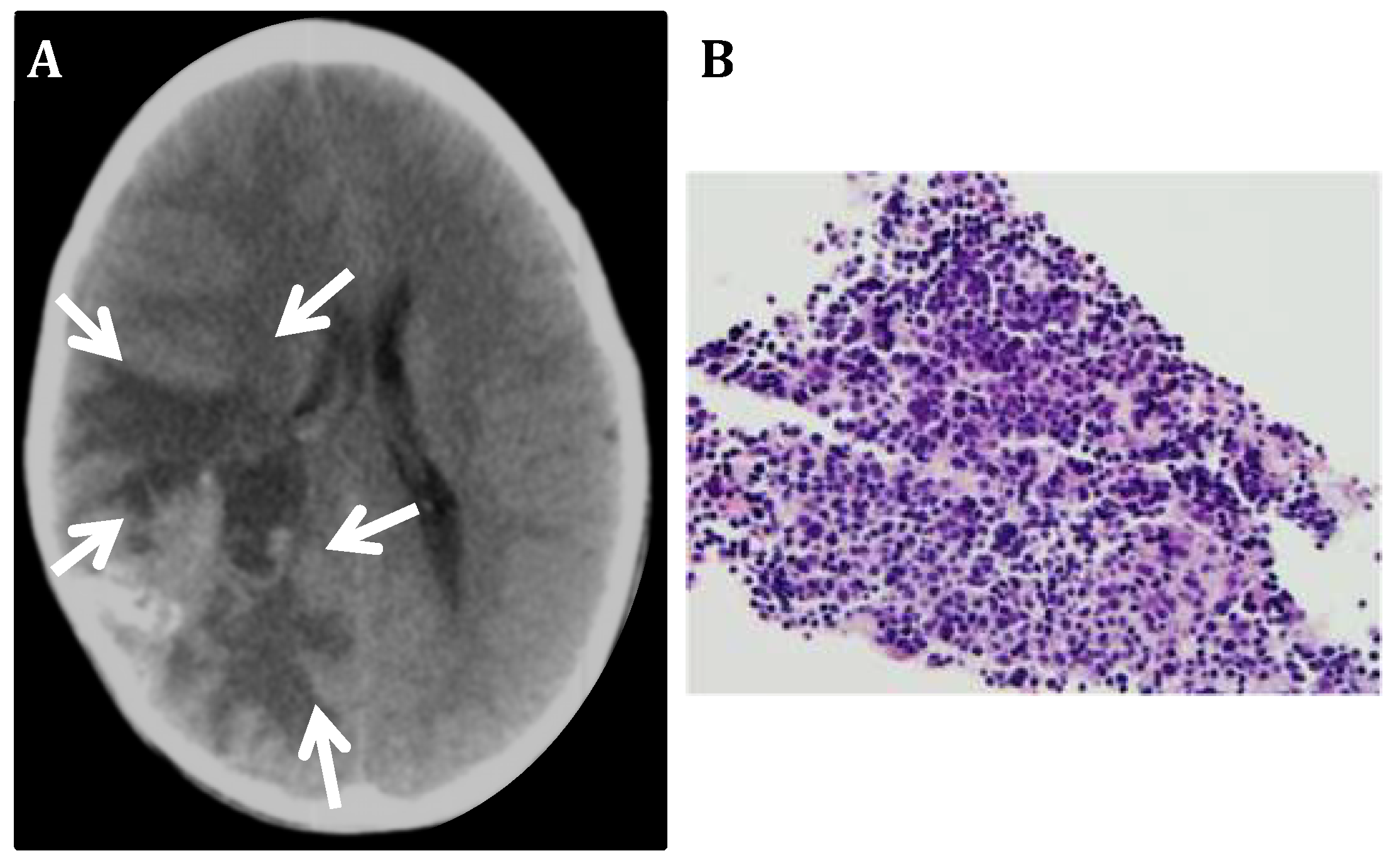
3.1. CNS Neuroblastoma
3.2. PLK4 Expression in CNS-NB Samples Determined by qRT-PCR
3.3. Gene Expression Meta-Analysis
4. Discussion and Literature Review
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Ramaswamy, V.; Taylor, M.D. Medulloblastoma: From myth to molecular. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2017, 35, 2355–2363. [Google Scholar] [CrossRef] [PubMed]
- Sredni, S.T.; Tomita, T. Rhabdoid tumor predisposition syndrome. Pediatr. Dev. Pathol. 2015, 18, 49–58. [Google Scholar] [CrossRef] [PubMed]
- Tariq, M.U.; Ahmad, Z.; Minhas, M.K.; Memon, A.; Mushtaq, N.; Hawkins, C. Embryonal tumor with multilayered rosettes, c19mc-altered: Report of an extremely rare malignant pediatric central nervous system neoplasm. SAGE Open Med. Case Rep. 2017, 5, 2050313X17745208. [Google Scholar] [CrossRef] [PubMed]
- Bianchi, F.; Tamburrini, G.; Gessi, M.; Frassanito, P.; Massimi, L.; Caldarelli, M. Central nervous system (cns) neuroblastoma. A case-based update. Child. Nerv. Syst. 2018, 34, 817–823. [Google Scholar] [CrossRef] [PubMed]
- Maris, J.M.; Hogarty, M.D.; Bagatell, R.; Cohn, S.L. Neuroblastoma. Lancet (London, England) 2007, 369, 2106–2120. [Google Scholar] [CrossRef]
- Tian, X.; Zhou, D.; Chen, L.; Tian, Y.; Zhong, B.; Cao, Y.; Dong, Q.; Zhou, M.; Yan, J.; Wang, Y.; et al. Polo-like kinase 4 mediates epithelial-mesenchymal transition in neuroblastoma via pi3k/akt signaling pathway. Cell Death Dis. 2018, 9, 54. [Google Scholar] [CrossRef] [PubMed]
- Sredni, S.T.; Suzuki, M.; Yang, J.P.; Topczewski, J.; Bailey, A.W.; Gokirmak, T.; Gross, J.N.; de Andrade, A.; Kondo, A.; Piper, D.R.; et al. A functional screening of the kinome identifies the polo-like kinase 4 as a potential therapeutic target for malignant rhabdoid tumors, and possibly, other embryonal tumors of the brain. Pediatr. Blood Cancer 2017, 64, e26551. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.K.; Davies, K.P.; Allen, J.; Zhu, L.; Pestell, R.G.; Zagzag, D.; Kalpana, G.V. Cell cycle arrest and repression of cyclin d1 transcription by ini1/hsnf5. Mol. Cell. Biol. 2002, 22, 5975–5988. [Google Scholar] [CrossRef] [PubMed]
- Albanese, P.; Belin, M.F.; Delattre, O. The tumour suppressor hsnf5/ini1 controls the differentiation potential of malignant rhabdoid cells. Eur. J. Cancer (Oxford, England: 1990) 2006, 42, 2326–2334. [Google Scholar] [CrossRef] [PubMed]
- Sredni, S.T.; Bailey, A.W.; Suri, A.; Hashizume, R.; He, X.; Louis, N.; Gokirmak, T.; Piper, D.R.; Watterson, D.M.; Tomita, T. Inhibition of polo-like kinase 4 (plk4): A new therapeutic option for rhabdoid tumors and pediatric medulloblastoma. Oncotarget 2017, 8, 111190–111212. [Google Scholar] [CrossRef] [PubMed]
- Sredni, S.T.; Tomita, T. The polo-like kinase 4 gene (plk4) is overexpressed in pediatric medulloblastoma. Child. Nerv. Syst. 2017, 33, 1031. [Google Scholar] [CrossRef] [PubMed]
- Sampson, P.B.; Liu, Y.; Forrest, B.; Cumming, G.; Li, S.W.; Patel, N.K.; Edwards, L.; Laufer, R.; Feher, M.; Ban, F.; et al. The discovery of polo-like kinase 4 inhibitors: Identification of (1r,2s).2-(3-((e).4-(((cis).2,6-dimethylmorpholino)methyl)styryl). 1h.Indazol-6-yl)-5’-methoxyspiro[cyclopropane-1,3’-indolin]-2’-one (cfi-400945) as a potent, orally active antitumor agent. J. Med. Chem. 2015, 58, 147–169. [Google Scholar] [CrossRef] [PubMed]
- Mason, J.M.; Lin, D.C.; Wei, X.; Che, Y.; Yao, Y.; Kiarash, R.; Cescon, D.W.; Fletcher, G.C.; Awrey, D.E.; Bray, M.R.; et al. Functional characterization of cfi-400945, a polo-like kinase 4 inhibitor, as a potential anticancer agent. Cancer Cell 2014, 26, 163–176. [Google Scholar] [CrossRef] [PubMed]
- Yu, B.; Yu, Z.; Qi, P.P.; Yu, D.Q.; Liu, H.M. Discovery of orally active anticancer candidate cfi-400945 derived from biologically promising spirooxindoles: Success and challenges. Eur. J. Med. Chem. 2015, 95, 35–40. [Google Scholar] [CrossRef] [PubMed]
- Sillibourne, J.E.; Bornens, M. Polo-like kinase 4: The odd one out of the family. Cell Division 2010, 5, 25. [Google Scholar] [CrossRef] [PubMed]
- Rosario, C.O.; Kazazian, K.; Zih, F.S.; Brashavitskaya, O.; Haffani, Y.; Xu, R.S.; George, A.; Dennis, J.W.; Swallow, C.J. A novel role for plk4 in regulating cell spreading and motility. Oncogene 2015, 34, 3441–3451. [Google Scholar] [CrossRef] [PubMed]
- Bettencourt-Dias, M.; Rodrigues-Martins, A.; Carpenter, L.; Riparbelli, M.; Lehmann, L.; Gatt, M.K.; Carmo, N.; Balloux, F.; Callaini, G.; Glover, D.M. Sak/plk4 is required for centriole duplication and flagella development. Curr. Biol. CB 2005, 15, 2199–2207. [Google Scholar] [CrossRef] [PubMed]
- Levine, M.S.; Bakker, B.; Boeckx, B.; Moyett, J.; Lu, J.; Vitre, B.; Spierings, D.C.; Lansdorp, P.M.; Cleveland, D.W.; Lambrechts, D.; et al. Centrosome amplification is sufficient to promote spontaneous tumorigenesis in mammals. Dev. Cell 2017, 40, 313–322. [Google Scholar] [CrossRef] [PubMed]
- Shinmura, K.; Kurabe, N.; Goto, M.; Yamada, H.; Natsume, H.; Konno, H.; Sugimura, H. Plk4 overexpression and its effect on centrosome regulation and chromosome stability in human gastric cancer. Mol. Biol. Rep. 2014, 41, 6635–6644. [Google Scholar] [CrossRef] [PubMed]
- Macmillan, J.C.; Hudson, J.W.; Bull, S.; Dennis, J.W.; Swallow, C.J. Comparative expression of the mitotic regulators sak and plk in colorectal cancer. Ann. Surg. Oncol. 2001, 8, 729–740. [Google Scholar] [CrossRef] [PubMed]
- Marina, M.; Saavedra, H.I. Nek2 and plk4: Prognostic markers, drivers of breast tumorigenesis and drug resistance. Front. Biosci. (Landmark Edition) 2014, 19, 352–365. [Google Scholar] [CrossRef]
- Kawakami, M.; Mustachio, L.M.; Zheng, L.; Chen, Y.; Rodriguez-Canales, J.; Mino, B.; Kurie, J.M.; Roszik, J.; Villalobos, P.A.; Thu, K.L.; et al. Polo-like kinase 4 inhibition produces polyploidy and apoptotic death of lung cancers. Proc. Natl. Acad. Sci. USA 2018, 115, 1913–1918. [Google Scholar] [CrossRef] [PubMed]
- Denu, R.A.; Shabbir, M.; Nihal, M.; Singh, C.K.; Longley, B.J.; Burkard, M.E.; Ahmad, N. Centriole overduplication is the predominant mechanism leading to centrosome amplification in melanoma. Mol. Cancer Res. MCR 2018, 16, 517–527. [Google Scholar] [CrossRef] [PubMed]
- Goroshchuk, O.; Kolosenko, I.; Vidarsdottir, L.; Azimi, A.; Palm-Apergi, C. Polo-like kinases and acute leukemia. Oncogene 2018. [Google Scholar] [CrossRef] [PubMed]
- Lohse, I.; Mason, J.; Cao, P.M.; Pintilie, M.; Bray, M.; Hedley, D.W. Activity of the novel polo-like kinase 4 inhibitor cfi-400945 in pancreatic cancer patient-derived xenografts. Oncotarget 2017, 8, 3064–3071. [Google Scholar] [CrossRef] [PubMed]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative rt-pcr data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, Research0034. [Google Scholar] [CrossRef] [PubMed]
- Haller, F.; Kulle, B.; Schwager, S.; Gunawan, B.; von Heydebreck, A.; Sultmann, H.; Fuzesi, L. Equivalence test in quantitative reverse transcription polymerase chain reaction: Confirmation of reference genes suitable for normalization. Anal. Biochem. 2004, 335, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Valente, V.; Teixeira, S.A.; Neder, L.; Okamoto, O.K.; Oba-Shinjo, S.M.; Marie, S.K.; Scrideli, C.A.; Paco-Larson, M.L.; Carlotti, C.G., Jr. Selection of suitable housekeeping genes for expression analysis in glioblastoma using quantitative rt-pcr. Ann. Neurosci. 2014, 21, 62–63. [Google Scholar] [CrossRef] [PubMed]
- Goodenberger, M.L.; Jenkins, R.B. Genetics of adult glioma. Cancer Genet. 2012, 205, 613–621. [Google Scholar] [CrossRef] [PubMed]
- Packer, R.J.; Pfister, S.; Bouffet, E.; Avery, R.; Bandopadhayay, P.; Bornhorst, M.; Bowers, D.C.; Ellison, D.; Fangusaro, J.; Foreman, N.; et al. Pediatric low-grade gliomas: Implications of the biologic era. Neuro-Oncol. 2017, 19, 750–761. [Google Scholar] [CrossRef] [PubMed]
- Georgantzi, K.; Sköldenberg, E.G.; Stridsberg, M.; Kogner, P.; Jakobson, Å.; Janson, E.T.; Christofferson, R.H.B. Chromogranin a and neuron-specific enolase in neuroblastoma: Correlation to stage and prognostic factors. Pediatr. Hematol. Oncol. 2018, 35, 156–165. [Google Scholar] [CrossRef] [PubMed]
- Popko, B.; Pearl, D.K.; Walker, D.M.; Comas, T.C.; Baerwald, K.D.; Burger, P.C.; Scheithauer, B.W.; Yates, A.J. Molecular markers that identify human astrocytomas and oligodendrogliomas. J. Neuropathol. Exp. Neurol. 2002, 61, 329–338. [Google Scholar] [CrossRef] [PubMed]
- Hol, E.M.; Pekny, M. Glial fibrillary acidic protein (gfap) and the astrocyte intermediate filament system in diseases of the central nervous system. Curr. Opin. Cell Biol. 2015, 32, 121–130. [Google Scholar] [CrossRef] [PubMed]
- Consortium, G.T.; Aguet, F.; Brown, A.A.; Castel, S.E.; Davis, J.R.; He, Y.; Jo, B.; Mohammadi, P.; Park, Y.; Parsana, P.; et al. Genetic effects on gene expression across human tissues. Nature 2017, 550, 204. [Google Scholar]
- Siddiqui, S.; White, M.W.; Schroeder, A.M.; DeLuca, N.V.; Leszczynski, A.L.; Raimondi, S.L. Aberrant dnmt3b7 expression correlates to tissue type, stage, and survival across cancers. PLoS ONE 2018, 13, e0201522. [Google Scholar] [CrossRef] [PubMed]
- Loir, P. Models for transcript quantification for rna-seq. arXiv 2011, arXiv:1104.3889. [Google Scholar]
- Kleihues, P.; Burger, P.C.; Scheithauer, B.W. The new who classification of brain tumours. Brain Pathol. (Zurich, Switzerland) 1993, 3, 255–268. [Google Scholar] [CrossRef] [PubMed]
- Kleihues, P.; Louis, D.N.; Scheithauer, B.W.; Rorke, L.B.; Reifenberger, G.; Burger, P.C.; Cavenee, W.K. The who classification of tumors of the nervous system. J. Neuropathol. Exp. Neurol. 2002, 61, 215–225. [Google Scholar] [CrossRef] [PubMed]
- Horten, B.C.; Rubinstein, L.J. Primary cerebral neuroblastoma. A clinicopathological study of 35 cases. Brain A J. Neurol. 1976, 99, 735–756. [Google Scholar] [CrossRef]
- Etus, V.; Kurtkaya, O.; Sav, A.; Ilbay, K.; Ceylan, S. Primary cerebral neuroblastoma: A case report and review. Tohoku J. Exp. Med. 2002, 197, 55–65. [Google Scholar] [CrossRef] [PubMed]
- Latchaw, R.E.; L’Heureux, P.R.; Young, G.; Priest, J.R. Neuroblastoma presenting as central nervous system disease. AJNR. Am. J. Neuroradiol. 1982, 3, 623–630. [Google Scholar] [PubMed]
- Zulch, K.J. Principles of the new world health organization (who) classification of brain tumors. Neuroradiology 1980, 19, 59–66. [Google Scholar] [CrossRef] [PubMed]
- Louis, D.N.; Ohgaki, H.; Wiestler, O.D.; Cavenee, W.K.; Burger, P.C.; Jouvet, A.; Scheithauer, B.W.; Kleihues, P. The 2007 who classification of tumours of the central nervous system. Acta Neuropathol. 2007, 114, 97–109. [Google Scholar] [CrossRef] [PubMed]
- Louis, D.N.; Perry, A.; Reifenberger, G.; von Deimling, A.; Figarella-Branger, D.; Cavenee, W.K.; Ohgaki, H.; Wiestler, O.D.; Kleihues, P.; Ellison, D.W. The 2016 world health organization classification of tumors of the central nervous system: A summary. Acta Neuropathol. 2016, 131, 803–820. [Google Scholar] [CrossRef] [PubMed]
- Sturm, D.; Orr, B.A.; Toprak, U.H.; Hovestadt, V.; Jones, D.T.W.; Capper, D.; Sill, M.; Buchhalter, I.; Northcott, P.A.; Leis, I.; et al. New brain tumor entities emerge from molecular classification of cns-pnets. Cell 2016, 164, 1060–1072. [Google Scholar] [CrossRef] [PubMed]
- Alexandrescu, S.; Paulson, V.; Dubuc, A.; Ligon, A.; Lidov, H.G. Phox2b is a reliable immunomarker in distinguishing peripheral neuroblastic tumours from cns embryonal tumours. Histopathology 2018. [Google Scholar] [CrossRef] [PubMed]
- Cardani, S.; Di Lascio, S.; Belperio, D.; Di Biase, E.; Ceccherini, I.; Benfante, R.; Fornasari, D. Desogestrel down-regulates phox2b and its target genes in progesterone responsive neuroblastoma cells. Exp. Cell Res. 2018, 370, 671–679. [Google Scholar] [CrossRef] [PubMed]
- Zimmerman, R.A.; Bilaniuk, L.T. Ct of primary and secondary craniocerebral neuroblastoma. Am. J. Roentgenol. 1980, 135, 1239–1242. [Google Scholar] [CrossRef] [PubMed]
- Matthay, K.K.; Brisse, H.; Couanet, D.; Couturier, J.; Benard, J.; Mosseri, V.; Edeline, V.; Lumbroso, J.; Valteau-Couanet, D.; Michon, J. Central nervous system metastases in neuroblastoma: Radiologic, clinical, and biologic features in 23 patients. Cancer 2003, 98, 155–165. [Google Scholar] [CrossRef] [PubMed]
- Petrov, I.; Suntsova, M.; Ilnitskaya, E.; Roumiantsev, S.; Sorokin, M.; Garazha, A.; Spirin, P.; Lebedev, T.; Gaifullin, N.; Larin, S.; et al. Gene expression and molecular pathway activation signatures of mycn-amplified neuroblastomas. Oncotarget 2017, 8, 83768–83780. [Google Scholar] [CrossRef] [PubMed]
- Mishra, A.; Beniwal, M.; Nandeesh, B.N.; Srinivas, D.; Somanna, S. Primary pediatric intracranial neuroblastoma: A report of two cases. J Pediatr. Neurosci. 2018, 13, 366–370. [Google Scholar] [PubMed]
- Kramer, K.; Kushner, B.; Heller, G.; Cheung, N.K. Neuroblastoma metastatic to the central nervous system. The memorial sloan-kettering cancer center experience and a literature review. Cancer 2001, 91, 1510–1519. [Google Scholar] [CrossRef]
- Kholodenko, I.V.; Kalinovsky, D.V.; Doronin, I.I.; Deyev, S.M.; Kholodenko, R.V. Neuroblastoma origin and therapeutic targets for immunotherapy. J. Immunol. Res. 2018, 2018, 7394268. [Google Scholar] [CrossRef] [PubMed]
- Matthay, K.K.; Maris, J.M.; Schleiermacher, G.; Nakagawara, A.; Mackall, C.L.; Diller, L.; Weiss, W.A. Neuroblastoma. Nat. Rev. Dis. Primers 2016, 2, 16078. [Google Scholar] [CrossRef] [PubMed]
- Kushner, B.H.; Ostrovnaya, I.; Cheung, I.Y.; Kuk, D.; Kramer, K.; Modak, S.; Yataghene, K.; Cheung, N.-K.V. Prolonged progression-free survival after consolidating second or later remissions of neuroblastoma with anti-gd2 immunotherapy and isotretinoin: A prospective phase ii study. OncoImmunology 2015, 4, e1016704. [Google Scholar] [CrossRef] [PubMed]
- Kramer, K.; Kushner, B.H.; Modak, S.; Pandit-Taskar, N.; Smith-Jones, P.; Zanzonico, P.; Humm, J.L.; Xu, H.; Wolden, S.L.; Souweidane, M.M.; et al. Compartmental intrathecal radioimmunotherapy: Results for treatment for metastatic cns neuroblastoma. J. Neuro-Oncol. 2010, 97, 409–418. [Google Scholar] [CrossRef] [PubMed]
- Ko, M.A.; Rosario, C.O.; Hudson, J.W.; Kulkarni, S.; Pollett, A.; Dennis, J.W.; Swallow, C.J. Plk4 haploinsufficiency causes mitotic infidelity and carcinogenesis. Nat. Genet. 2005, 37, 883–888. [Google Scholar] [CrossRef] [PubMed]
- Holland, A.J.; Lan, W.; Niessen, S.; Hoover, H.; Cleveland, D.W. Polo-like kinase 4 kinase activity limits centrosome overduplication by autoregulating its own stability. J. Cell Biol. 2010, 188, 191–198. [Google Scholar] [CrossRef] [PubMed]
- Fode, C.; Binkert, C.; Dennis, J.W. Constitutive expression of murine sak-a suppresses cell growth and induces multinucleation. Mol. Cell. Biol. 1996, 16, 4665–4672. [Google Scholar] [CrossRef] [PubMed]
- Sillibourne, J.E.; Tack, F.; Vloemans, N.; Boeckx, A.; Thambirajah, S.; Bonnet, P.; Ramaekers, F.C.; Bornens, M.; Grand-Perret, T. Autophosphorylation of polo-like kinase 4 and its role in centriole duplication. Mol. Biol. Cell 2010, 21, 547–561. [Google Scholar] [CrossRef] [PubMed]
- Sampson, P.B.; Liu, Y.; Patel, N.K.; Feher, M.; Forrest, B.; Li, S.W.; Edwards, L.; Laufer, R.; Lang, Y.; Ban, F.; et al. The discovery of polo-like kinase 4 inhibitors: Design and optimization of spiro[cyclopropane-1,3’[3h]indol]-2’(1’h).Ones as orally bioavailable antitumor agents. J. Med. Chem. 2015, 58, 130–146. [Google Scholar] [CrossRef] [PubMed]
- Dzhindzhev, N.S.; Tzolovsky, G.; Lipinszki, Z.; Abdelaziz, M.; Debski, J.; Dadlez, M.; Glover, D.M. Two-step phosphorylation of ana2 by plk4 is required for the sequential loading of ana2 and sas6 to initiate procentriole formation. Open Biol. 2017, 7, 170247. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; O’Rourke, B.P.; Soni, R.K.; Jallepalli, P.V.; Hendrickson, R.C.; Tsou, M.B. Promotion and suppression of centriole duplication are catalytically coupled through plk4 to ensure centriole homeostasis. Cell Rep. 2016, 16, 1195–1203. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.; Seo, M.Y.; Chang, J.; Hwang, D.S.; Rhee, K. Plk4 phosphorylation of cp110 is required for efficient centriole assembly. Cell Cycle 2017, 16, 1225–1234. [Google Scholar] [CrossRef] [PubMed]
- Bonni, S.; Ganuelas, M.L.; Petrinac, S.; Hudson, J.W. Human plk4 phosphorylates cdc25c. Cell Cycle 2008, 7, 545–547. [Google Scholar] [CrossRef] [PubMed]
- Puklowski, A.; Homsi, Y.; Keller, D.; May, M.; Chauhan, S.; Kossatz, U.; Grunwald, V.; Kubicka, S.; Pich, A.; Manns, M.P.; et al. The scf-fbxw5 e3-ubiquitin ligase is regulated by plk4 and targets hssas-6 to control centrosome duplication. Nat. Cell Biol. 2011, 13, 1004–1009. [Google Scholar] [CrossRef] [PubMed]
- Bury, L.; Coelho, P.A.; Simeone, A.; Ferries, S.; Eyers, C.E.; Eyers, P.A.; Zernicka-Goetz, M.; Glover, D.M. Plk4 and aurora a cooperate in the initiation of acentriolar spindle assembly in mammalian oocytes. J. Cell Biol. 2017, 216, 3571–3590. [Google Scholar] [CrossRef] [PubMed]
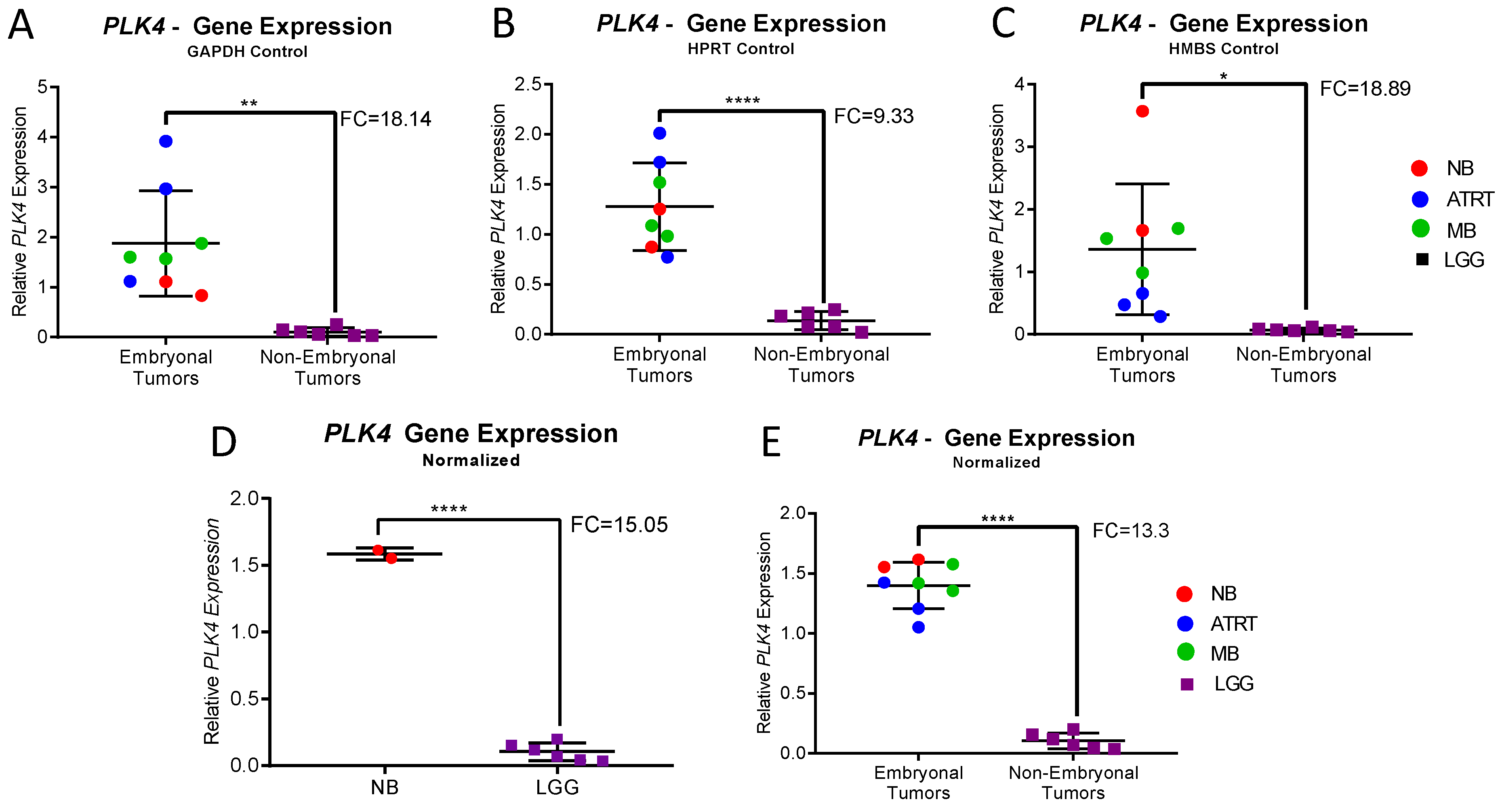

| Normalized Expression | CNS-NB | LGG | Fold Change | p-Value | Embryonal Tumors | Non-Embryonal Tumors | Fold Change | p-Value |
| PLK4/GAPDH | 0.97 | 0.1 | 9.4 | 0.0016 | 1.88 | 0.1 | 18.14 | 0.006 |
| PLK4/HPRT 1 | 1.062 | 0.14 | 7.78 | <0.0001 | 1.28 | 0.14 | 9.33 | 0.0031 |
| PLK4/HMBS | 2.62 | 0.07 | 36.41 | 0.0116 | 1.36 | 0.07 | 18.89 | 0.0116 |
| Normalized Expression | CNS-NB | LGG | Fold Change | p-Value | Embryonal Tumors | Non-Embryonal Tumors | Fold Change | p-Value |
| PLK4 | 1.58 | 0.1 | 15.05 | <0.0001 | 1.4 | 0.1 | 13.3 | <0.0001 |
| Organ # | Organ Name | Sample Size | PLK4 | CHGA | MBP | GFAP |
|---|---|---|---|---|---|---|
| Neuroblastoma | 153 | 14.0 | 658.1 | 2.9 | 0.3 | |
| Low Grade Glioma | 508 | 2.2 | 45.4 | 212.4 | 8535.2 | |
| 1 | Adipose—Subcutaneous | 442 | 1.4 | 0.1 | 6.3 | 1.9 |
| 2 | Adipose—Visceral (Omentum) | 355 | 0.8 | 0.1 | 6.3 | 1.2 |
| 3 | Adrenal Gland | 190 | 0.9 | 7.5 | 1.5 | 0.8 |
| 4 | Artery—Aorta | 299 | 0.5 | 0.2 | 6.8 | 2.9 |
| 5 | Artery—Coronary | 173 | 0.8 | 0.2 | 6.5 | 2.1 |
| 6 | Artery—Tibial | 441 | 0.7 | 0.2 | 6.7 | 1.4 |
| 7 | Bladder | 11 | 1.1 | 0.6 | 7.5 | 0.4 |
| 8 | Brain—Amygdala | 100 | 0.6 | 29.2 | 905.8 | 1669.7 |
| 9 | Brain—Anterior cingulate cortex (BA24) | 121 | 0.6 | 87.7 | 302.3 | 1027.0 |
| 10 | Brain—Caudate (Basal ganglia) | 160 | 0.6 | 26.9 | 422.6 | 1577.2 |
| 11 | Brain Cerebellar Hemisphere | 136 | 0.2 | 4.8 | 208.6 | 600.3 |
| 12 | Brain—Cerebellum | 173 | 0.2 | 4.4 | 177.7 | 696.6 |
| 13 | Brain—Cortex | 158 | 0.6 | 219.6 | 267.8 | 1200.6 |
| 14 | Brain—Frontal Cortex (BA9) | 129 | 0.8 | 335.6 | 332.5 | 961.1 |
| 15 | Brain—Hippocampus | 123 | 0.4 | 40.5 | 1472.2 | 2225.0 |
| 16 | Brain—Hypothalamus | 121 | 0.7 | 79.3 | 890.0 | 3809.2 |
| 17 | Brain—Nucleus accumbens (basal ganglia) | 147 | 0.9 | 32.6 | 335.9 | 913.3 |
| 18 | Brain—Putamen (basal ganglia) | 124 | 0.5 | 23.0 | 884.8 | 985.9 |
| 19 | Brain—Spinal cord (cervical c-1) | 91 | 0.8 | 5.7 | 9405.2 | 12,714.4 |
| 20 | Brain—Substantia nigra | 88 | 0.5 | 32.2 | 2607.8 | 4370.6 |
| 21 | Breast—Mammary Tissue | 290 | 1.2 | 0.3 | 6.5 | 2.8 |
| 22 | Cervix—Ectocervix | 6 | 1.4 | 0.4 | 8.9 | 0.2 |
| 23 | Cervix—Endocervix | 5 | 1.2 | 1.6 | 11.5 | 0.5 |
| 24 | Colon—Sigmoid | 233 | 0.5 | 5.8 | 5.5 | 0.9 |
| 25 | Colon—Transverse | 274 | 1.7 | 38.4 | 5.9 | 0.5 |
| 26 | Esophagus—Gastroesophageal Junction | 244 | 0.5 | 1.9 | 6.1 | 0.8 |
| 27 | Esophagus—Mucosa | 407 | 4.5 | 0.2 | 7.1 | 0.3 |
| 28 | Esophabus—Musclaris | 370 | 0.5 | 2.2 | 5.4 | 0.6 |
| 29 | Fallopian Tube | 7 | 1.1 | 1.8 | 7.3 | 0.4 |
| 30 | Heart—Atrial Appendage | 297 | 0.2 | 0.1 | 2.8 | 2.0 |
| 31 | Heart—Left Ventricle | 303 | 0.1 | 0.1 | 2.5 | 1.4 |
| 32 | Kidney—Cortex | 45 | 0.5 | 0.4 | 5.1 | 0.6 |
| 33 | Liver | 175 | 0.2 | 0.1 | 3.6 | 0.3 |
| 34 | Lung | 427 | 1.2 | 0.3 | 9.9 | 0.9 |
| 35 | Minor Salivary Gland | 97 | 0.1 | 0.2 | 7.0 | 1.1 |
| 36 | Muscle—Skeletal | 564 | 0.1 | 0.1 | 9.8 | 0.6 |
| 37 | Nerve—Tibial | 414 | 1.2 | 0.3 | 418.9 | 13.3 |
| 38 | Ovary | 133 | 1.4 | 0.3 | 4.9 | 0.6 |
| 39 | Pancreas | 248 | 0.3 | 42.3 | 3.4 | 0.5 |
| 40 | Pituitary | 183 | 0.4 | 781.6 | 7.6 | 16.3 |
| 41 | Prostate | 152 | 0.9 | 7.7 | 6.2 | 0.8 |
| 42 | Skin—Not Sun Exposed (Suprapubic) | 387 | 2.7 | 0.4 | 9.9 | 0.9 |
| 45 | Skin—Sun Exposed (Lower Leg) | 473 | 2.7 | 0.4 | 9.5 | 1.0 |
| 43 | Small Intestine—Terminal Ileum | 137 | 2.0 | 86.5 | 8.1 | 0.4 |
| 44 | Spleen | 162 | 2.1 | 0.2 | 11.3 | 0.5 |
| 46 | Stomach | 262 | 0.7 | 226.5 | 5.3 | 0.4 |
| 47 | Testis | 259 | 23.7 | 158.6 | 3.3 | 1.5 |
| 48 | Thyroid | 446 | 1.0 | 0.3 | 9.4 | 1.4 |
| 49 | Uterus | 111 | 1.1 | 0.3 | 7.8 | 0.4 |
| 50 | Vagina | 115 | 2.3 | 0.6 | 7.9 | 0.6 |
| 51 | Whole Blood | 407 | 0.3 | 0.2 | 18.3 | 1.0 |
| Neuroblastoma | Low Grade Glioma | Fold Change | p-Value | |
|---|---|---|---|---|
| PLK4 | 14.92 | 3.49 | 4.28 | p < 0.0001 |
| CHGA | 879.97 | 75.63 | 11.63 | p < 0.0001 |
| MBP | 5.26 | 411.70 | −78.32 | p < 0.0001 |
| GFAP | 1.68 | 11,046.77 | −6578.74 | p < 0.0001 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bailey, A.W.; Suri, A.; Chou, P.M.; Pundy, T.; Gadd, S.; Raimondi, S.L.; Tomita, T.; Sredni, S.T. Polo-Like Kinase 4 (PLK4) Is Overexpressed in Central Nervous System Neuroblastoma (CNS-NB). Bioengineering 2018, 5, 96. https://doi.org/10.3390/bioengineering5040096
Bailey AW, Suri A, Chou PM, Pundy T, Gadd S, Raimondi SL, Tomita T, Sredni ST. Polo-Like Kinase 4 (PLK4) Is Overexpressed in Central Nervous System Neuroblastoma (CNS-NB). Bioengineering. 2018; 5(4):96. https://doi.org/10.3390/bioengineering5040096
Chicago/Turabian StyleBailey, Anders W., Amreena Suri, Pauline M. Chou, Tatiana Pundy, Samantha Gadd, Stacey L. Raimondi, Tadanori Tomita, and Simone Treiger Sredni. 2018. "Polo-Like Kinase 4 (PLK4) Is Overexpressed in Central Nervous System Neuroblastoma (CNS-NB)" Bioengineering 5, no. 4: 96. https://doi.org/10.3390/bioengineering5040096
APA StyleBailey, A. W., Suri, A., Chou, P. M., Pundy, T., Gadd, S., Raimondi, S. L., Tomita, T., & Sredni, S. T. (2018). Polo-Like Kinase 4 (PLK4) Is Overexpressed in Central Nervous System Neuroblastoma (CNS-NB). Bioengineering, 5(4), 96. https://doi.org/10.3390/bioengineering5040096






