Abstract
The production of high yields of soluble recombinant protein is one of the main objectives of protein biotechnology. Several factors, such as expression system, vector, host, media composition and induction conditions can influence recombinant protein yield. Identifying the most important factors for optimum protein expression may involve significant investment of time and considerable cost. To address this problem, statistical models such as Design of Experiments (DoE) have been used to optimise recombinant protein production. This review examines the application of DoE in the production of recombinant proteins in prokaryotic expression systems with specific emphasis on media composition and culture conditions. The review examines the most commonly used DoE screening and optimisation designs. It provides examples of DoE applied to optimisation of media and culture conditions.
1. Introduction
Advances in biotechnology, including the development of genetic engineering and cloning, have provided a means for the large scale expression of heterologous proteins for different applications [1]. Currently, recombinant proteins are widely used in the biological and biomedical industries as well as in research with their market share increasing rapidly [2,3]. The production of high yields of soluble and functional recombinant protein is the ultimate goal in protein biotechnology [4]. To achieve this objective, many key aspects such as the expression system, the expression vector, the host strain, the purification tag, the media composition, the induction conditions and the purification methods need to be carefully evaluated and optimised before embarking on large scale production of a recombinant protein of interest [5,6,7].
Although both eukaryotic and prokaryotic expression systems are used for overproduction of soluble recombinant protein, choosing the right system for your protein depends, amongst other things, on the growth rate and culturing conditions of host cells, the level of the target gene expression and post translational processing of the synthesized protein [8,9]. The most commonly used prokaryotic systems are based on expression in bacteria, including E. coli and Bacillus species [10,11]. There is no single method which is universally successful for protein expression that will ensure the production of a desired concentration of soluble and functional protein [12,13,14]. Varying factors that influence protein expression in a trial-and-error process to achieve optimum protein expression has been troublesome [15]. To overcome this problem, statistical approaches have been used to evaluate the variables that have the largest influence on the production of a recombinant protein of interest in terms of yield [16,17], product quality [18], purity [19,20] and solubility [21,22]. These statistical processes include the Design of Experiment (DoE) approach [23,24]. This approach advances the traditional one-factor-at-a-time (OFAT) method, which involves varying one factor while other factors are held constant. This single variable OFAT approach results in the need to run multiple experiments with a high risk of failing to identify the true optimum [25]. The DoE method provides for a significantly reduced experimental matrix [26,27,28].
There are an increasing number of published studies on the application of statistically based optimization processes in the field of protein biotechnology [18,29]. This has been matched by a corresponding increase in the application of DoE methods, such as screening and optimisation designs, to enhance protein production. This review examines the literature on the DoE methodologies commonly employed to evaluate the effect of media composition and culture conditions on recombinant protein expression. It will focus on the application of DoE to increase recombinant protein expression in prokaryotic systems, where high yields can be achieved but poor product quality remains a risk [30]. It also provides an overview of the important statistical analysis tools embedded in common DoE software. These tools facilitate the interpretation of experimental data which ultimately allows the identification of optimal factor levels for maximum yield. Finally, the review provides some thoughts on the benefits of the common DoE methods typically used in recombinant protein production in order to direct future research efforts.
2. Production of Recombinant Proteins in a Prokaryotic Expression System
2.1. Factors that Inform the Choice of Expression System
Protein purification from natural sources can require a large quantity of the source organism and may yield only small amount of target protein after several rounds of extraction and purification [4,31]. Recombinant expression of proteins has become an indispensable tool to produce proteins to satisfactory yields [32] and to meet the demands of industry and research [1,33]. With the aid of genetic engineering, a desired gene cloned into a suitable expression vector can be overexpressed as a recombinant protein of interest [34]. Recombinant proteins can be expressed in cell cultures of bacteria [35], yeasts [36], mammalian cells [37,38], plants [39] and insects [40]. However, the prokaryotic systems remains the most attractive hosts due to their low cost, high productivity and rapid production rates [30]. Prokaryotic heterologous protein expression is mainly carried out in the bacteria E. coli, although increasingly the Bacillus species are being employed [41,42,43]. Drawbacks of prokaryotic expression systems include poor protein quality, due to the inability of prokaryotic cells to carry out post-translational modifications such as glycosylation, the presence of toxic cell wall pyrogens, along with the formation of inclusion bodies resulting in aggregated and insoluble heterologous protein [44]. Some widely used bacterial expression systems that are commercially available are listed in Table 1.

Table 1.
Summary of the most widely used recombinant expression strains from E. coli and Bacillus species outlining their advantages and disadvantages.
While there are a variety of expression vectors commercially available, their choice is strongly based on the combination of replicons, promoters, selection markers, multiple cloning sites and fusion proteins [11]. An informed decision on the best expression plasmid [10,51,52,53,54] can be confusing. The most commonly used expression plasmids [22,55,56,57,58] and their key features such as promoters [59,60,61,62,63], affinity tags [64,65] and selection markers [7] have been extensively reviewed in the literature, primarily focusing on the E. coli prokaryotic expression system. Widely used Bacillus strains [66,67], vectors and promoters have also been reviewed [68,69,70].
2.2. Factors that Influence Media Composition and Culture Conditions in an Expression System
A careful selection of expression system, expression vector and host does not always guarantee the production of a large amount of target protein in soluble and active form [7]. Media composition and induction conditions have a significant influence on recombinant protein expression levels [71,72,73] and solubility [45]. For example, media containing a defined concentration of salts, peptone and yeast influences the yield of a recombinant glucosidase [47]; while media composition does not always have a major effect on protein solubility [51]. Prosthetic groups in media are known to prevent the formation of inclusion bodies [74] where required by the protein [41,75]. The most common media used in prokaryotic expression systems, along with their advantages and disadvantages, have been reviewed elsewhere [76]. Culture conditions are another set of factors that must be carefully optimised to achieve high yields of heterologous protein [14]. Factors such as cell density prior to induction, inducer concentration, induction temperature and induction duration are all known to influence yield [77,78,79,80,81].
2.3. Enhancing the Production of Recombinant Proteins in a Prokaryotic Expression System by DoE
It can be difficult to make informed decisions regarding the optimal combination of expression system, conditions and media components. Oftentimes this results in an unsatisfactory and costly trial-and-error process being employed to enhance the overall production yield [64]. To address this problem more effective, statistically supported, approaches have been developed and have gained significant traction. In this approach, a controlled model is developed defining media components, induction and expression conditions based on the recombinant protein of interest [16]. DoE, employed in this way, has provided powerful tools to screen and optimise factors affecting recombinant protein expression [82]. This is due to DoEs’ ability to identify factors affecting recombinant protein production and optimise the process with the minimum number of experiments [83]. A typical DoE workflow is depicted in diagrammatic form (see Figure 1). The desired output, or response, is to achieve a high yield of a protein of interest and involves three main stages:
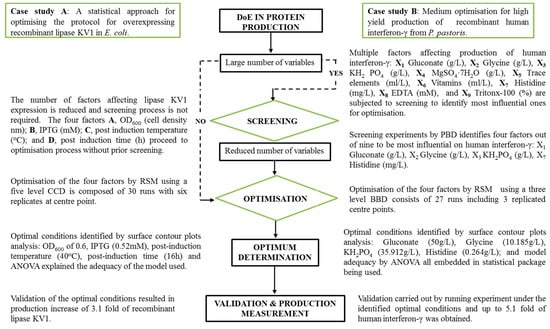
Figure 1.
A typical DoE workflow in protein production. Case study A illustrates the optimization of recombinant lipase KV1 expression in E. coli [84] where a screening process was not required since the number of factors affecting this enzyme is not large (four factors). The four factors (A, B, C, D), therefore, underwent optimisation by Central Composite Design (CCD) under Response Surface Methodology (RSM) which resulted in a yield increase in protein expression of 3.1-fold. Case study B describes the optimisation process for high yield production of recombinant human interferon-γ [85]. In this case, the number of factors involved is large (nine factors) and they were subjected to a screening process before optimisation. Four factors (X1, X2, X3, X7) out of nine were identified by Plackett-Burman Design (PBD) based screening to be the most influential and subsequently used for further optimisation. A Box-Benkhn Design (BBD) also under RSM was selected to optimize the screened factors and increased the production of human interferon-γ up to 5.1 fold. Further details of these two case studies can be found in the references provided and similar cases are found in Tables 4 and 7.
- Stage 1. The first stage of the process is to compile a list of factors that can influence protein expression. These are usually such factors as; induction temperature, induction duration, pH, media components (carbon source, nitrogen source, micronutrients).
- Stage 2. At this stage, a suitable software package such as MINITAB, JMP or Design Experts will be acquired for the statistical analysis. The second stage of DoE aims to reduce the number of factors to a smaller subset, these being the most important factors (i.e. those with the greatest impact on expression). This process is known as screening. Having a smaller set of significant factors greatly simplifies the statistical process. Sometimes, if the number of factors is small (between 2 and 4) there is no need to carry out the screening stage. When looking at a factor that influences protein expression the concept of levels is important: temperature, for example, may be examined between 20 °C and 40 °C. These two temperatures represent the lowest and highest “level” of this parameter that will influence expression. For the purposes of modelling these two levels are input into the model for this factor. Similarly, the upper and lower levels are input for all other relevant parameters. It is important to note that the levels are input into the DoE package as +1 (highest value of a parameter) and −1 (lowest value of a parameter). This “coding” is carried out to avoid the use of multiple different measurement units for parameters such as pH, temperature. The software will then suggest a minimal set of experiments to explore the significance of each factor. The design of the experimental matrix can be selected from a range of choices such as Full Factorial Design, Plackett Burman Design or indeed a custom design. The objective is to assess the “main effect” of a factor (its direct effect on a response) as well as its “interaction effects” (the effect on other factors). The suggested experiments are carried out and the results are used to inform the next stage of the process—optimisation.
- Stage 3. The final stage of the process is optimisation and is typically carried out with a set of three to four factors. An experimental RSM (Response Surface Methodology) design strategy is selected and experiments are run as for the screening stage. The optimisation process expresses the response surface as a polynomial and uses the input data to estimate its coefficients. The derivative of this polynomial is used to obtain inflection points corresponding to maxima or minima in the model. The model can be evaluated by looking at the goodness of fit between the model and experimental data. Finally, experiments using the optimum conditions predicted by the model are carried out to validate the model.
3. Design of Experiments (DoE) to Optimise Recombinant Protein Production
3.1. DoE; a Brief Overview
DoE is a statistical technique used to plan experiments and analyse data using a controlled set of tests designed to model and explore the relationship between factors and observed responses [14]. This technique allows the researcher to use the minimum number of experiments, in which the experimental parameters can be varied simultaneously, to make evidence based decisions [86]. It uses a mathematical model to analyse the process data, such as protein expression levels [87]. The model allows a researcher to understand the influence of the experimental parameters (inputs) on the response (outputs) and to identify a process optimum [88]. Furthermore, DoE software uses three-dimensional surface and contour plots, to visualise and understand the relationship between factors and responses [55,89]. In recombinant protein production, a DoE approach can significantly improve the efficiency in screening for most influential experimental parameters (e.g., media composition, culture condition etc.) and determine optimal experimental conditions [90].
The mathematical models employed in DoE define the process under study [91]. Screening designs such as Plackett Burman Design are based on a first order model [92] as shown in Equation (1).
where Y is the response, β0 is the model intercept, βi is the linear coefficient and Xi is the level of the independent variables. A statistically significant level of 5% (p-value = 0.05) is commonly used to identify the most influential factors. The significance level (or p-value) of each variable is based on its effect on the response and is calculated using Student’s T-test [85] in Equation (1).
where E(Xi) is the effect of variable Xi and S.E., the associated standard error. Factors with p-value < 0.05 are statistically significant while factors with p-value > 0.05 are not statistically significant (see Table 5 for more details). Statistically significant factors are subjected to further optimisation by Response Surface Methodology. A second-order polynomial equation in which independent variables are coded using Equation (3) is used to input factors into the model (see Section 5.4.1).
where xi is a dimensionless value of an independent variable; Xi is real value of an independent variable; Xcp is real value of an independent variable at the design centre point; and ∆Xi is step change in the real value of the variable i [93]. Replicates at the central point are required to check for the absence of bias between sets of experiments. The fit of the model is then evaluated through analysis of variance (ANOVA) which determines the significance of each term in the equation and estimates the goodness of fit in each case [94] (see Figure 5 and Table 9 for more details).
3.2. DoE Versus One-Factor-At-a-Time (OFAT)
DoE advances the traditional OFAT approach; OFAT fails to account for variables interacting with and influencing, each other and also requires significantly more experiments to converge on an optimum; all of which increases cost and time [95]. Figure 2 provides a brief comparative description between DoE and OFAT.
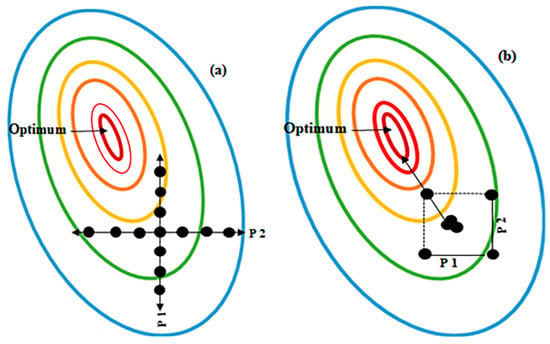
Figure 2.
Comparison between Design of Experiments (DoE) and One-Factor-at-A-Time (OFAT) by examining the effect of two parameters, P1 (Parameter 1) and P2 (Parameter 2). (a) OFAT is performed using more experiments than DoE (each black dot represents an experiment) and does not identify the true optimum (indicated as a red oval). However, with the DoE approach (b) fewer experiments are used and the likelihood of finding the optimum conditions (in red) for the process being studied is high. With DoE the combined or interaction effect of P1 and P2 on the response can be identified and measured. The ovals indicate production yields, blue indicates the lowest yields, whereas red indicates highest yields, where the optimum is found. The DoE approach also identifies a pathway to the optimum response (indicated by the arrow).
In recombinant protein expression, where various independent variables do not always act in isolation, it is likely that their interaction effects can significantly influence protein production [96]. Therefore, it is necessary to use a controlled set of tests that can examine the effects of many interacting factors to achieve optimal expression [97].
4. Defining a DoE Workflow to Optimise Recombinant Protein Production
Employing DoE to optimise the production of a recombinant protein can be divided into two main work packages, initial screening and subsequent optimisation. To evaluate all the factors that influence a production process, it is initially required to carry out a wide-ranging experimental screening. This first screening step will identify all factors that significantly influence recombinant protein production [98]. The second step in the workflow is to use a DoE optimisation design to achieve optimum production focusing only on the factors identified through the initial screening design. A variety of DoE software packages such as MINITAB (Minitab Ltd., State College, PA, USA), JMP (SAS Institute, Cary, NC, USA) and Design Experts (Science Plus Group, Groningen, the Netherlands) are commercially available and provide a variety of factorial designs depending upon the objective of the experiment. Regardless of the statistical package used, the main steps of a typical DoE workflow include planning the test, screening and optimisation (detailed schematically in Figure 3).
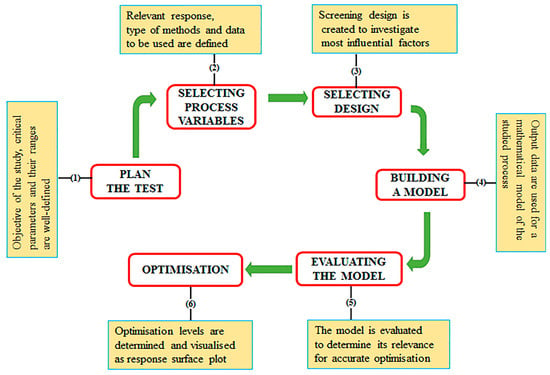
Figure 3.
A typical DoE workflow for the optimisation of recombinant protein production. The figure describes the main steps involved in the experimental design when both screening and optimisation designs are used. (1) The objectives of the study are defined including the selection of factors, levels and responses. (2) Process variables and expected responses are identified; the process variable levels (for a 2 level study) are set as high (+1), low (−1), (on occasion a 0 point is included). (3) The experimental screening design is selected based on the objectives of the study and the number of factors involved. (4) A mathematical model is built with certain conditions to meet the desired objectives (e.g., measurement of all the desired responses, process stability and accurate approximation by polynomial models). (5) The response data are analysed and visualised using plots for ease of data interpretation. At this stage, a reduced number of factors (i.e., the most influential) are retained for the subsequent optimisation phase. (6) Further optimisation can be carried out (via an optimisation DoE design).
5. A Suggested DoE Workflow for Recombinant Protein Production
5.1. Planning the Test; Selection of Factors and Associated Levels Influencing Recombinant Protein Production
The DoE workflow in protein production, like in any other DoE process optimisation, starts with the planning the test [99]. This involves defining the objective of the study, identifying factors involved and associated levels (i.e., high, central and low). Thus, preliminary experiments are recommended when knowledge of effects of factors on the experiment is not sufficient to set levels. The factors are input parameters that can be modified in the experiment and are referred to as the controllable factors. The levels of factors are fixed based on their working limits [82]. The most popular experimental designs are two level designs although more levels can be used depending upon the type of design and objective of the study. Table 2 depicts a two level experimental design.

Table 2.
An example of a two level experimental design having nine factors that are known to influence recombinant protein expression. In this case the nine factors relate to two experimental components; media composition and induction conditions. When planning the screening phase the selected factors (yeast extract, tryptone, glycerol, NaCl, Inoculum size, IPTG concentration, induction temperature, incubation time and pH, labelled X1 to X9 respectively) and associated levels (high, defined as +1 and low defined as −1 are selected to cover the intended experimental space (i.e., to cover the productive range). The levels are defined as the range between the known working limits.
In general, for recombinant protein expression subjected to DoE, the most commonly selected factors relate to media composition and include components such as yeast extract [100], K2HPO4, MgSO4, starch, glucose, peptone, NaCl, sucrose, glycerine [101]. For induction conditions, common factors selected are incubation time, incubation temperature, pH, agitation, inoculum age and size [102,103]; induction period, induction temperature, culture inoculation concentration [48,104]; Optical Density (OD), Isopropyl β-D-1-thiogalactopyranoside (IPTG) concentration [21].
5.2. Screening Designs to Identify Factors that Significantly Affect Recombinant Protein Expression
Screening designs are used to devise a matrix using factors and levels as formulated in the planning stage. [105]. By employing the statistical tools embedded in the DoE software, screening designs establish the relationships between variables and responses. The interaction effects between variables on a given response are also investigated [106]. In protein biotechnology, screening designs are mainly utilised to identify media composition and culture condition factors that significantly influence protein production [107]. Various researchers have explored the effects of both media components [94,107,108,109,110] and culture conditions [111,112] on protein expression. There are many different types of screening designs and their choice depends upon the nature of experiment and the objective of the study. The classical screening designs include Full Factorial Designs, Fractional Factorial Designs and Plackett-Burman Designs. Current DoE software, such as JMP from the SAS Institute, provides additional screening designs such as Definitive Screening Designs and Custom Designs. The most common screening designs are compared in Table 3.

Table 3.
A comparison of DoE screening designs commonly used in optimizing recombinant protein production. The table lists the types of screening designs; the effect explained by the model along with number of factors and associated number of runs (a rune refers to an experiment). It should be noted that extra runs (such as those related to central points) can be added when required. Custom design is more flexible and allows the designer to select the number of experimental runs.
5.2.1. Full Factorial Design
When little is known about the effects of the factors on a response, a full factorial design is recommended. This design includes all combinations of all factor levels and provides a predictive model that includes the main effects and all possible interactions [113]. This design consists of two, or more, levels with experimental runs that encompass all possible combinations of these levels, across all factors. In a full factorial design where k represents number of factors; 2k represents the number of experiments required to carry out a two level design with k factors. Similar to other screening designs, Full Factorial Design can include centre points, randomisation and blocking variables to improve the efficiency of the design [14]. This approach was significant in screening for the most influential factors affecting recombinant protein production for a variety of proteins [114,115] (see Table 4).

Table 4.
A selection of the widely used screening designs and their application in identifying the influential factors on the production of recombinant proteins.
5.2.2. Fractional Factorial Design (FFD)
FFD is a recommended screening design when a large number of factors are involved. This design consists of reducing the initially large number of potential factors to a subset of the most effective ones and is represented using the following notation:
where 2 represents number of levels, k the number of factors, p the extra columns required and R the resolution of the method. The method resolution describes the degree to which the estimated main effects are aligned with the estimated interactions associated with levels [22,116,117].
5.2.3. Plackett-Burman Designs (PBD)
PBD design is often used as an alternative to fractional and full factorial designs because of its potential to reduce the gaps found in fractional designs and to strengthen the estimation of the main effects, which may have been disregarded when full factorial designs are used [118,119,120,121,122].
5.2.4. Definitive Screening Design (DSD) and Custom Design (CD)
DSD and CD are a class of screening designs that have potential applications in recombinant protein expression for assessing the impact of a large number of factors on a given response. DSD has recently been reported to be particularly advantageous as it allows the estimation of the main effects of certain components alone but also the interactions between components as well as the factors with non-linear effects such as quadratic effects (an interaction term where a factor interacts with itself); all executed with the minimum number of experimental runs [123]. CD enables tailoring a design, whilst simultaneously minimising resource usage: it is highly flexible and more cost-effective than other screening designs. It allows for the best use of the experimental budget and tackles a wide range of challenges with the capability to model effects including centre points and replicates. However, in most cases this design allows for the estimation of main effects only. Table 4 summarises the most common screening designs, along with their roles in identifying most influential independent factors, in recombinant protein production.
The rationale of screening designs lies in identifying the variables that are statistically significant in influencing protein production among a large number of potentially important variables [128,129]. Table 5 illustrates how screening analysis identifies statistically significant factors based on their effect and probability values.

Table 5.
Identification of the statistically significant factors during a screening process using a Fractional Factorial Design. The table depicts the effect, positive or negative and p-value for seven factors examined (labelled X1 to X7 respectively). The effect of each factor, positive (+) or negative (−) is identified during the analysis stage using the statistical formula imbedded in DoE software used (JMP in this example). Interaction effects are also identified (e.g., X5*X1 and X3*X7; where * indicates an interaction). The p-value of each factor is also shown, at the significance level of 0.05. In this example, the highlighted factors, (X3, X6, X1), were identified as the most influential based on their high effects (−1.11273, 0.2252, 0.17492) and p-values < 0.05 (0.001, 0.0143, 0.0296). Thus, only factors X3, X6 and X1 are statistically significant at the level of 0.05, with X3 having a negative effect while X6 and X1 have positive effects. Other factors, X2, X4, X5, X7 and interactions X5*X1, X3*X7 are not statistically significant.
The screening process identifies most influential factors on the process under investigation (i.e., X1 and X6 in the example shown in Table 5) and thus paves the way for effective optimisation by reducing the number of factors to be optimised in the third work package of the DoE workflow [130].
5.3. Optimisation Designs to Maximise Recombinant Protein Production in Prokaryotic Systems
As a collection of statistical design and numerical optimisation techniques [131], optimisation uses the reduced number of variables identified in the previous screening process and focuses on finding the variable levels that result in an optimal yield [132,133]. Figure 4, describes the benefit of carrying out an optimisation process after a screening process has identified a small number of key variables.
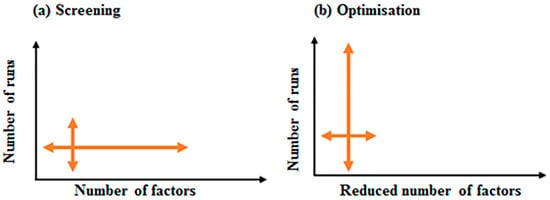
Figure 4.
A comparative illustration of screening and optimisation designs. (a) In screening designs a large number of factors, with reduced number of runs, are used to screen for important factors affecting the process. (b) In optimisation designs, a reduced number of factors, with large number of runs, are utilised to find the optimum conditions for high yield of recombinant protein.
Response Surface Methodology (RSM) is the most popular optimisation method [134]. It consists of mathematical and statistical techniques used to build empirical models capable of exploring the process space and studying the relationship between the response and process variables to find the optimal response [99,133,135]. In general, for a given number of factors, RSM requires more runs than screening designs, thus, the number of factors to consider should initially be reduced through an appropriate screening process. Central composite designs (CCD) and Box-Behnken designs (BBD) are the two of the major Response Surface Designs commonly used in recombinant protein optimization [136].
5.3.1. Central Composite Design (CCD)
CCDs are favoured in process optimisation due to determine the coefficients of a second-degree polynomial which fit a full quadratic during response surface analysis [127]. CCD has been widely used in optimising protein production process specifically addressing the aim of increasing productivity and solubility [137]. There are different types of central composite designs such as uniform precision, orthogonal/block and so forth. However, a common standard characteristic includes the number of runs per design [138], which depends on the number factors (see Table 6). Central composite uniform precision designs are used to provide protection against bias in the regression coefficients while central composite orthogonal designs can be used to avoid correlations between coefficients of variables [139].

Table 6.
Common CCD components and the possible total number of runs. Factorial, axial and central points are the main components of a typical CCD and the total number of runs is dictated by the number of factors being tested. As the number of factors increases, the number of component points increase and so the total number of runs. In some cases, CCDs do not contain axial points, especially when the variance of model prediction is not suspected [140].
5.3.2. Box Behnken Design (BBD)
BBDs are also a class of response surface designs; however, they differ from CCD in their design structure. For example, a CCD with 4 factors requires 31 runs (experiments), whereas a BBD only has 27 runs for the same number of factors. For 5 factors, CCD has 52 runs while BBD has 46 runs. Reduced runs can result in significant time and cost savings in an optimisation process. In optimisation experiments BBD is widely used as a good design to fit the quadratic model with fewer experiments [141]. Several studies show that BBDs have contributed to production increases for recombinant proteins (see Table 7).

Table 7.
RSM methods used to optimise the production of recombinant proteins along with their effect on yield and citing reference.
5.3.3. Summary and Choice of Optimisation Methods
Both CCD and BBD optimisation methods are widely used, the choice depends on the number of factors and objectives of the study (see Figure 1). The standard characteristic is that all response surface designs feature a second-order polynomial model to describe the process where interaction terms introduce curvature into the response function and a first-order equation is inadequate to fit the model [159,160]. CCD is the most preferred RSM [16,161] due to the fact that this design contains full factorial or fractional factorial modes, with the potential to add central points to evaluate the experimental error and axial points to check the variance of the model [14,140]. The number of runs (N) in CCD is calculated using Equation (4).
where k is the number of factors and Cp the number of centre points [162]. Table 8 is an example of a two level CCD with two centre point replicates along with responses such as actual, predicted and residues (see Table 8).

Table 8.
Central Composite Design of four independent factors (labelled X1, X2, X3, X4 respectively) studied at two levels (+1 and −1) including two central point replicates (0 and 0). The table also shows different types of common responses found in optimisation process; (1) Actual data refers to experimental results; (2) predicted data are generated by software based on the design and actual results. The residuals are the difference between actual and predicted data.
5.4. Analysis and Interpretation of Optimisation Data
Regardless of the DoE design employed, the goal is to provide a methodology for conducting controlled experiments with the aim of identifying the vital process inputs and investigating interactions between them [163]. At a screening level, after the experimental data are entered, the DoE software generates a variety of graphs that are used to interpret the results obtained. These may be scatter plots, histograms, bar charts and Pareto charts that allow the researcher to identify the distribution of the data and statistical significance of the variables tested [85]. Different screening analysis methods have been used in the field of protein production [77,92,112,164]. Figure 5 illustrates a typical DoE data analysis and interpretation route from data visualisation, through experiment validation to conclusion.
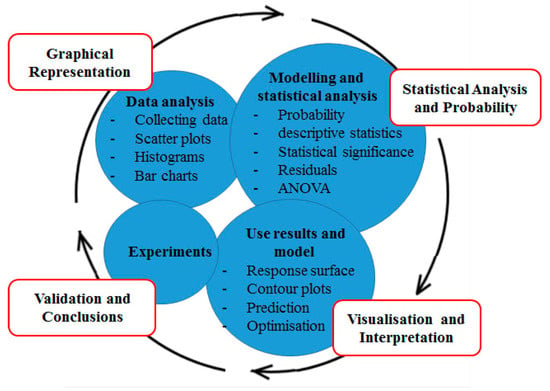
Figure 5.
A typical DoE analysis route from initial Experiments to validation and conclusions. The rationale for data analysis is to evaluate the effects of variables on response. Graphical Representation shows how the data are distributed. The Statistical Analysis and Probability stage identifies variables that are statistically significant. This will identify variables that are important to bring forward to the subsequent optimisation step based on their statistical significance. The Visualization and Interpretation stage will focus on representational analysis that identifies optimal levels.
Evaluation of Experimental Design and Predictive Model Validation
For RSM analysis, the goals are to (i) develop a predictive model that describes how the process inputs influence the process output and (ii) determine the optimal settings of the inputs [165,166]. Following the completion of the optimisation experiments, the results are used to fit a second-order polynomial equation (Equation (5)) [85].
where Yi is the predicted response, β0, βi, βii and βij are regression coefficients for the intercept, first-order model coefficients, quadratic coefficient and linear model coefficient for the interaction respectively [167,168]. The fit of the model is then evaluated through analysis of variance (ANOVA, Table 9) which compares the variation due to the change in the combination of variable levels with the variation due to the random errors [14,169].

Table 9.
An example of Analysis of Variance (ANOVA) for Response Surface Methodology fitted to a second-order polynomial equation. The table depicts R-squared (R2), Adjusted R-squared (Adj-R2), Predicted R-squared (Pred-R2), degree of freedom (DF), adjusted sum of square (Adj SS), adjusted mean square (Adj MS), F-value and p-value of the model.
The coefficient value of R2 defines how well the model fits the data. The closer the R2 is to 1, the better it describes the experimental data [21]. The Adjusted R2 is used to check the adequacy of the model by measuring the amount of variation about the mean derived from the model; the closer the value is to 1, the better it describes the model [130]. For example, in Table 9, the R2 = 0.9971 indicates the significance of regression of the fitting equation and therefore, adequacy of discrimination, indicating that only 0.29% of the total variation could not be explained by the fitting equation [142]. When R2 = 99.71%, Adj-R2 = 99.63%, Pred-R2 = 99.48% are in good agreement with each other (as in Table 9), this provides confidence in the accuracy of the model [156].
Additionally, the p-value and signal-to-noise ratio are used to estimate the quality of the model. For a significant model, a p-value < 0.05 is desirable [170]. Appropriate precision measures the signal-to-noise ratio; where a ratio greater than 4 indicates an adequate model [171] and is commonly used in protein production optimisation [172,173]. Furthermore, the p-value lack of fit and the plot of observed values versus predicted values are used to estimate the quality of the model. With a good model, the p-value lack of fit should be >0.05 [168] as shown in Table 9. Finally, all data should fall on the straight line on the observed versus predicted plots [145] as shown in Figure 6.
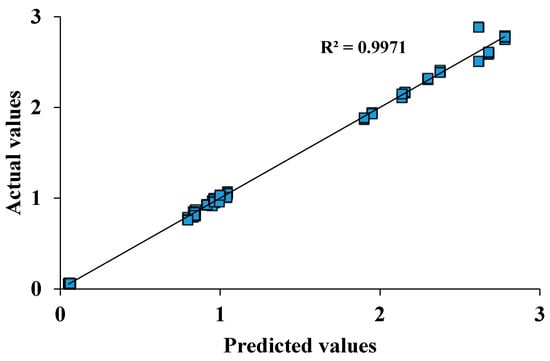
Figure 6.
A linear plot estimating accuracy of a regression model by comparing actual versus predicted data sets. The plot determines the correlation between the model’s predictions and actual data and thereby indicates how well the model fits the data. The closer the value of R2 is to 1, the better the fit of the line to the data and the goodness of the model.
5.5. Optimum Determination
Once the predictive model has been validated, it can be used to determine the optimised parameters. The statistical tools embedded in DoE software are used to generate 3D-graphs, called surface contour plots that visually describe the relationship between variables and response [174,175]. The 3-D surface and contour graphs are generated as a combination of two test variables with the others maintained at their respective zero levels [176] see Figure 7. Surface, contour and residual plots, along with ANOVA, are the main optimisation analysis tools commonly used to determine optimum levels for high yields of recombinant protein [20,177,178,179].
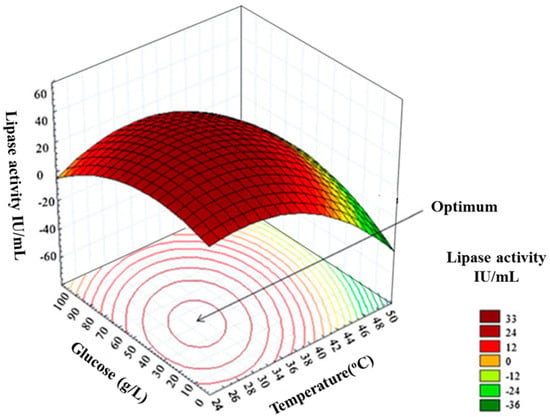
Figure 7.
An example of response surface and contour plot adapted from Nelofer et al., 2012 [163]. The figure depicts the two-factor interaction (in this case the two factors explored are glucose and culturing temperature) where one factor influences the response of another factor. It also shows the visualisation of optimum levels. The colour scale indicates the level of lipase activity (IU/mL) where red indicates the region of optimal yield, yellow indicates medium yield, and green indicates low yield. In this case, the optimal enzyme activity (33 IU/mL) was achieved at a culture temperature between 30 °C and 34 °C; and a glucose concentration between 40 g/mL–50 g/mL. Image used with permission.
6. Conclusions; Getting It ‘Just Right’
DoE offers many choices for screening and optimisation designs which advance traditional optimisation methodologies, such as one-factor-at-a-time. The statistical approach offered by DoE has proven to be applicable in protein biotechnology effectively investigating media composition and culture condition factors in recombinant protein production. DoE’s ability to identify the most influential factors in recombinant protein expression through screening designs and identify the factor/levels that give the maximum yield has considerably enhanced the production of soluble, active recombinant protein. With the recent development of more flexible screening and optimisation designs and enhancements in computational processing DoE will continue to find applications in biotechnology; in recombinant protein production and beyond.
Author Contributions
All authors contributed substantially to this body of work, in the form of; Conceptualization, A.U., G.K.K., G.T.H. and B.J.R.; Data Curation, A.U.; Writing-Original Draft Preparation, A.U., G.K.K., G.T.H. and B.J.R.; Writing-Review & Editing, A.U., G.K.K., G.T.H. and B.J.R.; Supervision, G.K.K., G.T.H. and B.J.R.; Project Administration, B.J.R.; Funding Acquisition, B.J.R., G.T.H. and A.U.
Funding
This research was funded by the Dublin Institute of Technology Fiosraigh Postgraduate Scheme [AU].
Conflicts of Interest
The authors declare no conflict of interest.
References
- Adrio, J.L.; Demain, A.L. Recombinant organisms for production of industrial products. Bioeng. Bugs 2010, 1, 116–131. [Google Scholar] [CrossRef] [PubMed]
- Mitragotri, S.; Burke, P.A.; Langer, R. Overcoming the challenges in administering biopharmaceuticals: Formulation and delivery strategies. Nat. Rev. Drug Discov. 2014, 13, 655–672. [Google Scholar] [CrossRef] [PubMed]
- Manning, M.C.; Chou, D.K.; Murphy, B.M.; Payne, R.W.; Katayama, D.S. Stability of protein pharmaceuticals: An update. Pharm. Res. 2010, 27, 544–575. [Google Scholar] [CrossRef] [PubMed]
- Ferrer-miralles, N.; Villaverde, A. Bacterial Cell Factories for Recombinant Protein Production; expanding the catalogue. Microb. Cell Fac. 2013, 12, 133. [Google Scholar] [CrossRef] [PubMed]
- Papaneophytou, C.; Kontopidis, G.A. Optimization of TNF-α overexpression in Escherichia coli using response surface methodology: Purification of the protein and oligomerization studies. Protein Expr. Purif. 2012, 86, 35–44. [Google Scholar] [CrossRef] [PubMed]
- Marini, G.; Luchese, M.D.; Argondizzo, A.P.C.; de Góes, A.M.; Galler, R.; Alves, T.L.M. Experimental design approach in recombinant protein expression: Determining medium composition and induction conditions for expression of pneumolysin from Streptococcus pneumoniae in Escherichia coli and preliminary purification process. BMC Biotechnol. 2014, 14, 1. [Google Scholar] [CrossRef] [PubMed]
- Rosano, G.L.; Ceccarelli, E.A. Recombinant protein expression in Escherichia coli: Advances and challenges. Front. Microbiol. 2014, 5, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Fakruddin, M.; Mohammad Mazumdar, R.; Bin Mannan, K.S.; Chowdhury, A.; Hossain, M.N. Critical Factors Affecting the Success of Cloning, Expression, and Mass Production of Enzymes by Recombinant E. coli. ISRN Biotechnol. 2013, 3, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Lambertz, C.; Garvey, M.; Klinger, J.; Heesel, D.; Klose, H.; Fischer, R. Challenges and advances in the heterologous expression of cellulolytic enzymes: A review. Biotechnol. Biofuels 2014, 7, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.H.; Keum, K.C.; Lee, S.Y. Production of recombinant proteins by high cell density culture of Escherichia coli. Chem. Eng. Sci. 2006, 61, 876–885. [Google Scholar] [CrossRef]
- Berlec, A.; Štrukelj, B. Current state and recent advances in biopharmaceutical production in Escherichia coli, yeasts and mammalian cells. J. Ind. Microbiol. Biotechnol. 2013, 40, 257–274. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Zhang, L.; Zhang, Y.; Zhang, T.; Feng, Y.; Lu, X. Highly efficient production of soluble proteins from insoluble inclusion bodies by a Two-Step-Denaturing and refolding method. PLoS ONE 2011, 6, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Sørensen, H.P.; Mortensen, K.K. Soluble expression of recombinant proteins in the cytoplasm of Escherichia coli. Microb. Cell Fact. 2005, 8, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Papaneophytou, C.P.; Kontopidis, G. Statistical approaches to maximize recombinant protein expression in Escherichia coli: A general review. Protein Expr. Purif. 2014, 94, 22–32. [Google Scholar] [CrossRef] [PubMed]
- Makino, T.; Skretas, G.; Georgiou, G. Strain engineering for improved expression of recombinant proteins in bacteria. Microb. Cell. Fact. 2011, 10, 32. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.; Liu, M.; Wang, B.; Jiang, K.; Wang, L. Statistical optimization of medium composition and culture condition for the production of recombinant anti-lipopolysaccharide factor of Eriocheir sinensis in Escherichia coli. Chin. J. Oceanol. Limnol. 2011, 29, 1249–1259. [Google Scholar] [CrossRef]
- Xu, Y.; Li, Y.; Xu, S.; Liu, Y.; Wang, X.; Tang, J. Improvement of xylanase production by Aspergillus niger XY-1 using response surface methodology for optimizing the medium composition. J. Zhejiang Univ. Sci. B 2008, 9, 558–566. [Google Scholar] [CrossRef] [PubMed]
- Shafiee, F.; Rabbani, M.; Jahanian-Najafabadi, A. Optimization of the Expression of DT386-BR2 Fusion Protein in Escherichia coli using Response Surface Methodology. Adv. Biomed. Res. 2017, 6, 22. [Google Scholar] [CrossRef] [PubMed]
- Akbarzadeh, A.; Dehnavi, E.; Aghaeepoor, M.; Amani, J. Optimization of recombinant expression of synthetic bacterial phytase in Pichia pastoris using response surface methodology. Jundishapur J. Microbiol. 2015, 8, e27553. [Google Scholar] [CrossRef] [PubMed]
- Maharjan, S.; Singh, B.; Bok, J.D.; Kim, J.I.; Jiang, T.; Cho, C.S. Exploring codon optimization and response surface methodology to express biologically active transmembrane RANKL in E. Coli. PLoS ONE 2014, 9, e96259. [Google Scholar] [CrossRef] [PubMed]
- Larentis, A.L.; Nicolau, J.F.M.Q.; Esteves, G.D.S.; Vareschini, D.T.; de Almeida, F.V.R.; dos Reis, M.G. Evaluation of pre-induction temperature, cell growth at induction and IPTG concentration on the expression of a leptospiral protein in E. coli using shaking flasks and microbioreactor. BMC Res. Notes 2014, 7, 671. [Google Scholar] [CrossRef] [PubMed]
- Beigi, L.; Karbalaei-Heidari, H.R.; Kharrati-Kopaei, M. Optimization of an extracellular zinc-metalloprotease (SVP2) expression in Escherichia coli BL21 (DE3) using response surface methodology. Protein Expr. Purif. 2012, 84, 161–166. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Ruan, H.; Zhang, H.; Ni, H.; He, G. Enhanced production of elastase by Bacillus licheniformis ZJUEL31410: Optimization of cultivation conditions using response surface methodology. J. Zhejiang Univ. Sci. B 2007, 8, 845–852. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.W.; He, Y.L.; Niu, T.G.; Zhong, J.J. Optimization of fermentation conditions for production of anti-TMV extracellular ribonuclease by Bacillus cereus using response surface methodology. Bioprocess Biosyst. Eng. 2010, 33, 657–663. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Sun, Y.B.; He, H.W.; Feng, J.T.; Zhang, X.; Han, L.R. Optimization of medium compositions to improve a novel glycoprotein production by Streptomyces kanasenisi ZX01. AMB Express 2017, 7, 6. [Google Scholar] [CrossRef] [PubMed]
- Altekar, M.; Homon, C.A.; Kashem, M.; Mason, S.W.; Nelson, R.M.; Patnaude, L.A. Assay Optimization: A Statistical Design of Experiments Approach. Clin. Lab. Med. 2007, 27, 139–154. [Google Scholar] [CrossRef] [PubMed]
- Samaei-Nouroozi, A.; Rezaei, S.; Khoshnevis, N.; Doosti, M.; Hajihoseini, R.; Khoshayand, M.R. Medium-based optimization of an organic solvent-tolerant extracellular lipase from the isolated halophilic Alkalibacillus Salilacus. Extremophiles 2015, 19, 933–947. [Google Scholar] [CrossRef] [PubMed]
- Gangadharan, D.; Sivaramakrishnan, S.; Nampoothiri, K.M.; Sukumaran, R.K.; Pandey, A. Response surface methodology for the optimization of alpha amylase production by Bacillus Amyloliquefaciens. Bioresour. Technol. 2008, 99, 4597–4602. [Google Scholar] [CrossRef] [PubMed]
- Setiawan, D.; Fadhlillah, M.; Pertiwi, W.; Gaffar, S.; Subroto, T. Optimization of The Expression of Recombinant Universal Infleunza vaccine Candidate in Escherichia coli Using Response Surface Methodology. IOSR J. Pharm. 2018, 8, 53–60. [Google Scholar]
- Demain, A.L.; Vaishnav, P. Production of Recombinant Proteins by Microbes and Higher Organisms. Biotechnol. Adv. 2009, 27, 297–306. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Babnigg, G.; Jedrzejczak, R.; Eschenfeldt, W.H.; Li, H.; Maltseva, N. High-throughput protein purification and quality assessment for crystallization. Methods 2011, 55, 12–28. [Google Scholar] [CrossRef] [PubMed]
- Mondal, S.; Shet, D. High yield expression of proteins in E. coli for NMR studies. Adv. Biosci. 2013, 4, 751–767. [Google Scholar] [CrossRef]
- Li, S.; Yang, X.; Yang, S.; Zhu, M.; Wang, X. Technology Prospecting on Enzymes: Application, Marketing and Engineering Abstract: Enzymes are protein molecules functioning as specialized catalysts for chemical reactions. They have contributed greatly to the traditional and modern chemical industr. Comput. Struct. Biotechnol. J. 2012, 2, e201209017. [Google Scholar] [CrossRef] [PubMed]
- Dubey, A.A.; Singh, M.I.; Jain, V. Rapid and Robust PCR-Based All-Recombinant Cloning Methodology. PLoS ONE 2016, 11, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Barnes, H.; Arlotto, M.P.; Waterman, M.R. Expression and enzymatic activity of recombinant cytochrome P450 17 alpha-hydroxylase in Escherichia coli. Proc. Natl. Acad. Sci. USA 1991, 88, 5597–5601. [Google Scholar] [CrossRef] [PubMed]
- Han, M.; Yu, X. Enhanced expression of heterologous proteins in yeast cells via the modification of N-glycosylation sites. Bioengineered 2015, 6, 115–118. [Google Scholar] [CrossRef] [PubMed]
- Araki, Y.; Hamafuji, T.; Noguchi, C.; Shimizu, N. Efficient recombinant production in mammalian cells using a novel IR/MAR gene amplification method. PLoS ONE 2012, 7, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Durocher, Y. High-level and high-throughput recombinant protein production by transient transfection of suspension-growing human 293-EBNA1 cells. Nucleic Acids Res. 2002, 30, 1–11. [Google Scholar] [CrossRef]
- Zimran, A.; Brill-almon, E.; Chertkoff, R.; Petakov, M.; Blanco-favela, F.; Muñoz, T.; Solorio-Meza, S.E.; Amato, D.; Duran, G.; Giona, F.; et al. Pivotal trial with plant cell-expressed recombinant glucocerebrosidase, taliglucerase alfa, a novel enzyme replacement therapy for Gaucher disease. Blood 2011, 118, 5767–5773. [Google Scholar] [CrossRef] [PubMed]
- Bleckmann, M.; Schmelz, S.; Schinkowski, C.; Scrima, A.; van den Heuvel, J. Fast plasmid based protein expression analysis in insect cells using an automated SplitGFP screen. Biotechnol. Bioeng. 2016, 113, 1975–1983. [Google Scholar] [CrossRef] [PubMed]
- Khow, O.; Suntrarachun, S. Strategies for production of active eukaryotic proteins in bacterial expression system. Asian Pac. J. Trop. Biomed. 2012, 2, 159–162. [Google Scholar] [CrossRef]
- Tanimura, K.; Takashima, S.; Matsumoto, T.; Tanaka, T.; Kondo, A. 2,3-Butanediol production from cellobiose using exogenous beta-glucosidase-expressing Bacillus Subtilis. Appl. Microbiol. Biotechnol. 2016, 100, 5781–5789. [Google Scholar] [CrossRef] [PubMed]
- Fontani, S.; Niccolai, A.; Kapat, A.; Olivieri, R. Studies on the maximization of recombinant Helicobacter pylori neutrophil-activating protein production in Escherichia coli: Application of Taguchi robust design and response surface methodology for process optimization. World J. Microbiol. Biotechnol. 2003, 19, 711–717. [Google Scholar] [CrossRef]
- Adrio, J.L.; Demain, A.L. Microbial enzymes: Tools for biotechnological processes. Biomolecules 2014, 4, 117–139. [Google Scholar] [CrossRef] [PubMed]
- Gopal, G.J.; Kumar, A. Strategies for the production of recombinant protein in Escherichia coli. Protein J. 2013, 32, 419–425. [Google Scholar] [CrossRef] [PubMed]
- Gurung, P.; Lukens, J.R.; Kanneganti, T. Overview of the puification of recombinant proteins. HHS Public Access 2016, 3, 193–201. [Google Scholar]
- El-Helow, E.R.; Abdel-Fattah, Y.R.; Ghanem, K.M.; Mohamad, E. Application of the response surface methodology for optimizing the activity of an aprE-driven gene expression system in Bacillus Subtilis. Appl. Microbiol. Biotechnol. 2000, 54, 515–520. [Google Scholar] [CrossRef] [PubMed]
- Li, R.F.; Wang, B.; Liu, S.; Chen, S.H.; Yu, G.H.; Yang, S.Y. Optimization of the Expression Conditions of CGA-N46 in Bacillus subtilis DB1342(p-3N46) by Response Surface Methodology. Interdiscip. Sci. Comput. Life Sci. 2016, 8, 277–283. [Google Scholar] [CrossRef] [PubMed]
- Mohammadi, H.S.; Mostafavi, S.S.; Soleimanib, S.; Bozorgian, S.; Pooraskari, M.; Kianmehr, A. Response surface methodology to optimize partition and purification of two recombinant oxidoreductase enzymes, glucose dehydrogenase and D-galactose dehydrogenase in aqueous two-phase systems. Protein Expr. Purif. 2015, 108, 41–47. [Google Scholar] [CrossRef] [PubMed]
- Murashima, K.; Chen, C.; Kosugi, A.; Tamaru, Y.; Doi, R.H.; Wong, S. Heterologous Production of Clostridium cellulovorans engB, Using Protease-Deficient Bacillus subtilis, and Preparation of Active Recombinant Cellulosomes. J. Bacteriol. 2002, 184, 76–81. [Google Scholar] [CrossRef] [PubMed]
- Berrow, N.S.; Büssow, K.; Coutard, B.; Diprose, J.; Ekberg, M.; Folkers, G.E. Recombinant protein expression and solubility screening in Escherichia coli: A comparative study. Acta Crystallogr. Sect. D Biol. Crystallogr. 2006, 62, 1218–1226. [Google Scholar] [CrossRef] [PubMed]
- de Marco, A. Strategies for successful recombinant expression of disulfide bond-dependent proteins in Escherichia coli. Microb. Cell Fact. 2009, 8, 1–26. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, C.; Guimarães, P.M.R.; Domingues, L. Recombinant microbial systems for improved β-galactosidase production and biotechnological applications. Biotechnol. Adv. 2011, 29, 600–609. [Google Scholar] [CrossRef] [PubMed]
- Peti, W.; Page, R. Strategies to maximize heterologous protein expression in Escherichia coli with minimal cost. Protein Expr. Purif. 2007, 51, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Baweja, M.; Nain, L.; Kawarabayasi, Y.; Shukla, P. Current technological improvements in enzymes toward their biotechnological applications. Front. Microbiol. 2016, 7, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Jarmander, J.; Janoschek, L.; Lundh, S.; Larsson, G.; Gustavsson, M. Process optimization for increased yield of surface-expressed protein in Escherichia coli. Bioprocess Biosyst. Eng. 2014, 37, 1685–1693. [Google Scholar] [CrossRef] [PubMed]
- Mahalik, S.; Sharma, A.K.; Mukherjee, K.J. Genome engineering for improved recombinant protein expression in Escherichia coli. Microb. Cell Fact. 2014, 13, 177. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Fu, Q.; Li, W.; Wang, B.; Yin, X.; Liu, S. Identification and characterization of a novel β-glucosidase via metagenomic analysis of Bursaphelenchus xylophilus and its microbial flora. Sci. Rep. 2017, 7, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Jana, S.; Deb, J.K. Strategies for efficient production of heterologous proteins in Escherichia coli. Appl. Microbiol. Biotechnol. 2005, 67, 289–298. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.; Pozzo, T.; Megyeri, M.; Lindahl, S.; Sundin, A.; Turner, C. Aglycone specificity of Thermotoga neapolitana β-glucosidase 1A modified by mutagenesis, leading to increased catalytic efficiency in quercetin-3-glucoside hydrolysis. BMC Biochem. 2011, 12, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Khushoo, A.; Pal, Y.; Singh, B.N.; Mukherjee, K.J. Extracellular expression and single step purification of recombinant Escherichia coli l-asparaginase II. Protein Expr. Purif. 2004, 38, 29–36. [Google Scholar] [CrossRef] [PubMed]
- Volontè, F.; Marinelli, F.; Gastaldo, L.; Sacchi, S.; Pilone, M.S.; Pollegioni, L. Optimization of glutaryl-7-aminocephalosporanic acid acylase expression in E. Coli. Protein Expr. Purif. 2008, 61, 131–137. [Google Scholar] [CrossRef] [PubMed]
- Zelena, K.; Eisele, N.; Berger, R.G. Escherichia coli as a production host for novel enzymes from basidiomycota. Biotechnol. Adv. 2014, 32, 1382–1395. [Google Scholar] [CrossRef] [PubMed]
- Costa, S.; Almeida, A.; Castro, A.; Domingues, L. Fusion tags for protein solubility, purification, and immunogenicity in Escherichia coli: The novel Fh8 system. Front. Microbiol. 2014, 5, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Dickson, J.M.J.; Lee, W.; Shepherd, P.R.; Buchanan, C.M. Enzyme activity effects of N-terminal His-tag attached to catalytic sub-unit of phosphoinositide-3-kinase. Biosci. Rep. 2013, 33, 857–863. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Fu, G.; Zhang, D. Cloning, Characterization, and production of a novel lysozyme by different expression hosts. J. Microbiol. Biotechnol. 2014, 24, 1405–1412. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Nie, Y.; Zhang, Y.; Wang, Y.; Xu, Y. Improved inducible expression of Bacillus naganoensis pullulanase from recombinant Bacillus subtilis by enhancer regulation. Protein Expr. Purif. 2018, 148, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Nikoloff, J.M.; Zhang, D. Improving protein production on the level of regulation of both expression and secretion pathways in Bacillus Subtilis. J. Microbiol. Biotechnol. 2015, 25, 963–977. [Google Scholar] [CrossRef] [PubMed]
- Song, W.; Nie, Y.; Mu, X.Q.; Xu, Y. Enhancement of extracellular expression of Bacillus naganoensis pullulanase from recombinant Bacillus subtilis: Effects of promoter and host. Protein Expr. Purif. 2016, 124, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Thuy, L.A.T.; Schumann, W. A novel cold-inducible expression system for Bacillus Subtilis. Protein Expr. Purif. 2007, 53, 264–269. [Google Scholar] [CrossRef] [PubMed]
- Gomes, L.C.; Mergulh, F.J. Effects of antibiotic concentration and nutrient medium composition on Escherichia coli biofilm formation and green fluorescent protein expression. FEMS Microbiol. Lett. 2018, 364, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Jia, B.; Jeon, C.O. High-throughput recombinant protein expression in Escherichia coli: Current status and future perspectives. Open Biol. 2016, 6, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Chhetri, G.; Kalita, P.; Tripathi, T. An efficient protocol to enhance recombinant protein expression using ethanol in Escherichia coli. MethodsX 2015, 2, 385–391. [Google Scholar] [CrossRef] [PubMed]
- Ryan, B.J.; O’Fágáin, C. Effects of single mutations on the stability of horseradish peroxidase to hydrogen peroxide. Biochimie 2007, 89, 1029–1032. [Google Scholar] [CrossRef] [PubMed]
- Deller, M.C.; Kong, L.; Rupp, B. Protein stability: A crystallographer’s perspective. Acta Crystallogr. Sect. Struct. Biol. Commun. 2016, 72, 72–95. [Google Scholar] [CrossRef] [PubMed]
- Vincentelli, R.; Cimino, A.; Geerlof, A.; Kubo, A.; Satou, Y.; Cambillau, C. High-throughput protein expression screening and purification in Escherichia coli. Methods 2011, 55, 65–72. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.M.; Rhee, C.H.; Kang, C.K.; Kim, J.H. Sequential and Simultaneous Statistical Optimization by Dynamic Design of Experiment for Peptide Overexpression in Recombinant Escherichia coli. Appl. Biochem. Biotechnol. 2006, 135, 59–80. [Google Scholar] [CrossRef]
- Lebendiker, M.; Danieli, T. Production of prone-to-aggregate proteins. FEBS Lett. 2014, 588, 236–246. [Google Scholar] [CrossRef] [PubMed]
- Malik, A.; Alsenaidy, A.M.; Elrobh, M.; Khan, W.; Alanazi, M.S.; Bazzi, M.D. Optimization of expression and purification of HSPA6 protein from Camelus dromedarius in E. coli. Saudi J. Biol. Sci. 2016, 23, 410–419. [Google Scholar] [CrossRef] [PubMed]
- Ou, J.; Wang, L.; Ding, X.; Du, J.; Zhang, Y.; Chen, H. Stationary phase protein overproduction is a fundamental capability of Escherichia coli. Biochem. Biophys. Res. Commun. 2004, 314, 174–180. [Google Scholar] [CrossRef] [PubMed]
- Papaneophytou, C.P.; Rinotas, V.; Douni, E.; Kontopidis, G. A statistical approach for optimization of RANKL overexpression in Escherichia coli: Purification and characterization of the protein. Protein Expr. Purif. 2013, 90, 9–19. [Google Scholar] [CrossRef] [PubMed]
- Mandenius, C.; Brundin, A. Review: Biocatalysts and Bioreactor Design Optimization, Bioprocess Methodology, Using Design-of-experiments. Biotechnol. Progr. 2008, 24, 1191–1203. [Google Scholar] [CrossRef] [PubMed]
- Hajihassan, Z.; Biroonro, N. Enhanced expression of recombinant activin a in Escherichia coli by optimization of induction parameters. J. Sci. Islam Repub. Iran 2018, 29, 105–111. [Google Scholar]
- Batumalaie, K.; Khalili, E.; Mahat, N.A.; Huyop, F.Z.; Wahab, R.A. A statistical approach for optimizing the protocol for overexpressing lipase KV1 in Escherichia coli: Purification and characterization. Biotechnol. Biotechnol. Equip. 2018, 32, 69–87. [Google Scholar]
- Prabhu, A.A.; Mandal, B.; Dasu, V.V. Medium optimization for high yield production of extracellular human interferon-γ from Pichia pastoris: A statistical optimization and neural network-based approach. Korean J. Chem. Eng. 2017, 34, 1109–1121. [Google Scholar] [CrossRef]
- Aghaeepoor, M.; Kobarfard, F.; Akbari Eidgahi, M.R.; Shabani, A.; Dehnavi, E.; Ahmadi, M. Optimization of culture media for extracellular expression of streptokinase in Escherichia coli using response surface methodology in combination with plackett-burman design. Trop. J. Pharm. Res. 2017, 16, 2567–2576. [Google Scholar] [CrossRef]
- Puri, M.; Kaur, A.; Singh, R.S.; Singh, A. Response surface optimization of medium components for naringinase production from staphylococcus xylosus MAK2. Appl. Biochem. Biotechnol. 2010, 162, 181–191. [Google Scholar] [CrossRef] [PubMed]
- Pandey, R.K.; Senthil, P.; Boriwal, L.; Malviya, A. Experimental investigation on influence of ECM process parameters on responses using full factorial design. Mater. Today Proc. 2017, 4, 3666–3671. [Google Scholar] [CrossRef]
- Nam, S.H.; Park, J.; Jun, W.; Kim, D.; Ko, J.A.; Abd El-Aty, A.M. Transglycosylation of gallic acid by using Leuconostoc glucansucrase and its characterization as a functional cosmetic agent. AMB Express 2017, 7, 224. [Google Scholar] [CrossRef] [PubMed]
- Wahid, Z.; Nadir, N. Improvement of one factor at a time through design of experiments. World Appl. Sci. J. 2013, 21, 56–61. [Google Scholar]
- Anbu, P.; Annadurai, G.; Hur, B. Production of alkaline protease from a newly isolated Exiguobacterium profundum BK-P23 evaluated using the response surface methodology. Biologia 2013, 68, 186–193. [Google Scholar] [CrossRef]
- Roohi, M.K. Statistical optimization of cold-active chitinase production by mutagenized cells of multi-enzyme producing Bacillus cereus GA6. Rend Lincei. 2015, 26, 271–280. [Google Scholar] [CrossRef]
- Mahajan, P.M.; Gokhale, S.V.; Lele, S.S. Production of Nattokinase Using Bacillus natto NRRL 3666: Media optimization, scale Up, and kinetic modeling. Food Sci. Biotechnol. 2010, 9, 1593–1603. [Google Scholar] [CrossRef]
- Purama, R.K.; Goyal, A. Application of response surface methodology for maximizing dextransucrase production from leuconostoc mesenteroides NRRL B-640 in a bioreactor. Appl. Biochem. Biotechnol. 2008, 151, 182–192. [Google Scholar] [CrossRef] [PubMed]
- Haider, M.A.; Pakshirajan, K. Screening and optimization of media constituents for enhancing lipolytic activity by a soil microorganism using statistically designed experiments. Appl. Biochem. Biotechnol. 2007, 141, 377–390. [Google Scholar] [CrossRef] [PubMed]
- Samarin, Z.E.; Abolghasemi, S.; Dehnavi, E.; Akbarzadeh, A.; Hadian, A.; Khodabandeh, M. Response Surface Optimization of the Expression Conditions for Synthetic Human Interferon α -2b Gene in Escherichia coli. Indian J. Pharm. Sci. 2018, 80, 470–479. [Google Scholar] [CrossRef]
- Saravanan, P.; Muthuvelayudham, R.; Rajesh Kannan, R.; Viruthagiri, T. Optimization of cellulase production using Trichoderma reesei by RSM and comparison with genetic algorithm. Front. Chem. Sci. Eng. 2012, 6, 443–452. [Google Scholar] [CrossRef]
- Graslund, S.; Nordlund, P.; Weigelt, J.; Hallberg, B.M.; Bray, J.; Gileadi, O.; Knapp, S. Protein production and purification. Nat. Methods 2008, 5, 135–146. [Google Scholar] [CrossRef] [PubMed]
- Naili, B.; Sahnoun, M.; Bejar, S.; Kammoun, R. Optimization of submerged Aspergillus oryzae S2 α-amylase production. Food Sci. Biotechnol. 2016, 25, 185–192. [Google Scholar] [CrossRef] [PubMed]
- Goswami, R.; Mukherjee, S.; Chakraborty, A.K.; Balachandran, S.; Sinha Babu, S.P.; Chaudhury, S. Optimization of growth determinants of a potent cellulolytic bacterium isolated from lignocellulosic biomass for enhancing biogas production. Clean Technol. Environ. Policy 2016, 18, 1565–1583. [Google Scholar] [CrossRef]
- Zhao, Y.; Liang, Y.; Liu, L.; Cheng, J.; Yuan, Y. Medium Optimization for Antifungal Active Substance Production from Streptomyces Lydicus Using Response Surface Methodology. Trans. Tianjin Univ. 2017, 23, 78–86. [Google Scholar] [CrossRef]
- Manivasagan, P.; Sivasankar, P.; Venkatesan, J.; Sivakumar, K.; Kim, S.K. Optimization, production and characterization of glycolipid biosurfactant from the marine actinobacterium, Streptomyces sp. MAB36. Bioprocess Biosyst. Eng. 2014, 37, 783–797. [Google Scholar] [CrossRef] [PubMed]
- Ramadas, N.V.; Soccol, C.R.; Pandey, A. A statistical approach for optimization of polyhydroxybutyrate production by Bacillus sphaericus NCIM 5149 under submerged fermentation using central composite design. Appl. Biochem. Biotechnol. 2010, 162, 996–1007. [Google Scholar] [CrossRef] [PubMed]
- Kandasamy, S.; Muthusamy, G.; Balakrishnan, S.; Duraisamy, S.; Thangasamy, S.; Seralathan, K.K. Optimization of protease production from surface-modified coffee pulp waste and corncobs using Bacillus sp. by SSF. 3 Biotech 2016, 6, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Jones, B.; Nachtsheim, C.J. A Class of Three-Level Designs for Definitive Screening in the Presence of Second-Order Effects. J. Qual. Technol. 2011, 43, 1–15. [Google Scholar] [CrossRef]
- Lee, C.W.; Wang, H.J.; Hwang, J.K.; Tseng, C.P. Protein thermal stability enhancement by designing salt bridges: A combined computational and experimental study. PLoS ONE 2014, 9, e112751. [Google Scholar] [CrossRef] [PubMed]
- Mittal, D.; Srivatsava, A.; Govindasamy, S.; Chandrasekaran, M. Optimization of Critical Medium Components for Protein Production by Nostoc ellipsosporum Using Response Surface Methodology. Arab. J. Sci. Eng. 2015, 40, 875–880. [Google Scholar] [CrossRef]
- Adinarayana, K.; Ellaiah, P. Response surface optimization of the critical medium components for the production of alkaline protease by a newly isolated Bacillus Sp. J. Pharm. Pharm. Sci. 2002, 5, 272–278. [Google Scholar] [PubMed]
- Bhagwat, P.K.; Jhample, S.B.; Dandge, P.B. Statistical medium optimization for the production of collagenolytic protease by Pseudomonas sp. SUK using response surface methodology. Microbiology 2015, 84, 520–530. [Google Scholar] [CrossRef]
- Chandel, M.; Azmi, W. Bioresource Technology Optimization of process parameters for the production of tyrosine phenol lyase by Citrobacter freundii MTCC 2424. Bioresour. Technol. 2009, 100, 1840–1846. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Liu, M.; Liu, J.; Dai, H.; Zhou, X.; Liu, X. Medium optimization for the production of avermectin B1a by Streptomyces avermitilis 14-12A using response surface methodology. Bioresour. Technol. 2009, 100, 4012–4016. [Google Scholar] [CrossRef] [PubMed]
- El-Ahmady El-Naggar, N.; Moawad, H.; Abdelwahed, N.A.M. Optimization of fermentation conditions for enhancing extracellular production of L-asparaginase, an anti-leukemic agent, by newly isolated Streptomyces brollosae NEAE-115 using solid state fermentation. Ann. Microbiol. 2017, 67, 1–15. [Google Scholar] [CrossRef]
- Wass, J. First Steps in Experimental Design–The Screening Experiment. J. Valid. Technol. 2010, 49–57. [Google Scholar]
- Pham, P.; Taillandier, P.; Delmas, M.; Strehaiano, P. Optimization of a culture medium for xylanase production by Bacillus sp. using statistical experimental designs. World J. Microbiol. Biotechnol. 1998, 14, 185–190. [Google Scholar] [CrossRef]
- Vijayaraghavan, P.; Vincent, S.G.P. Statistical optimization of fibrinolytic enzyme production by Pseudoalteromonas sp. IND11 using cow dung substrate by response surface methodology. Springerplus 2014, 3, 60. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.L.; Lin, T.H.; Juang, R.S. Optimization of recombinant hexaoligochitin-producing chitinase production with response surface methodology. Int. J. Biol. Macromol. 2013, 62, 518–522. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.H.; Lu, F.P.; He, Y.N.; Zhang, J.K.; Du, L.X. Statistical optimization of medium components for avilamycin production by Streptomyces viridochromogenes Tü57-1 using response surface methodology. J. Ind. Microbiol. Biotechnol. 2007, 34, 271–278. [Google Scholar] [CrossRef] [PubMed]
- Jia, B.; Jin, Z.H.; Mei, L.H. Medium optimization based on statistical methodologies for pristinamycins production by Streptomyces Pristinaespiralis. Appl. Biochem. Biotechnol. 2008, 144, 133–143. [Google Scholar] [CrossRef] [PubMed]
- Kammoun, R.; Naili, B.; Bejar, S. Application of a statistical design to the optimization of parameters and culture medium for α-amylase production by Aspergillus oryzae CBS 819.72 grown on gruel (wheat grinding by-product). Bioresour. Technol. 2008, 99, 5602–5609. [Google Scholar] [CrossRef] [PubMed]
- Niladevi, K.N.; Sukumaran, R.K.; Jacob, N.; Anisha, G.S.; Prema, P. Optimization of laccase production from a novel strain-Streptomyces psammoticus using response surface methodology. Microbiol. Res. 2009, 164, 105–113. [Google Scholar] [CrossRef] [PubMed]
- Sai-Ut, S.; Benjakul, S.; Sumpavapol, P.; Kishimura, H. Optimization of gelatinolytic enzyme production by B. amyloliquefaciens sp. H11 through Plackett-Burman design and response surface methodology. Int. Aquat. Res. 2014, 6, 59. [Google Scholar] [CrossRef]
- Thenmozhi, C.; Sankar, R.; Karuppiah, V.; Sampathkumar, P. L-asparaginase production by mangrove derived Bacillus cereus MAB5: Optimization by response surface methodology. Asian Pac. J. Trop. Med. 2011, 4, 486–491. [Google Scholar] [CrossRef]
- Jones, B.; Nachtsheim, C.J. Definitive screening designs with added two-level categorical factors. J. Qual. Technol. 2013, 45, 121–129. [Google Scholar] [CrossRef]
- Swalley, S.E.; Fulghum, J.R.; Chambers, S.P. Screening factors effecting a response in soluble protein expression: Formalized approach using design of experiments. Anal. Biochem. 2006, 351, 122–127. [Google Scholar] [CrossRef] [PubMed]
- Cao, W.; Li, H.; Zhang, J.; Li, D.; Acheampong, D.O.; Chen, Z. Periplasmic expression optimization of VEGFR2 D3 adopting response surface methodology: Antiangiogenic activity study. Protein Expr. Purif. 2013, 90, 55–66. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Fattah, Y.R.; Olama, Z.A. L-asparaginase production by Pseudomonas aeruginosa in solid-state culture: Evaluation and optimization of culture conditions using factorial designs. Process Biochem. 2002, 38, 115–122. [Google Scholar] [CrossRef]
- Meriem, G.; Mahmoud, K. Optimization of chitinase production by a new Streptomyces griseorubens C9 isolate using response surface methodology. Ann. Microbiol. 2017, 67, 175–183. [Google Scholar] [CrossRef]
- Ghosh, S.; Murthy, S.; Govindasamy, S.; Chandrasekaran, M. Optimization of L-asparaginase production by Serratia marcescens (NCIM 2919) under solid state fermentation using coconut oil cake. Sustain. Chem. Process 2013, 1, 9. [Google Scholar] [CrossRef]
- Woods, D.C.; Lewis, S.M. Design of Experiments for Screening. In Handbook of Uncertainty Quantification; Springer: Cham, Switzerland, 2015; pp. 1–43. [Google Scholar]
- Dey, S.S.; Dora, K.C. Optimization of the production of shrimp waste protein hydrolysate using microbial proteases adopting response surface methodology. J. Food Sci. Technol. 2014, 51, 16–24. [Google Scholar] [CrossRef] [PubMed]
- Myers, R.; Montgomery, D.; Vining, G.G.; Borror, C.; Kowalski, S. Response Surface Methodology: A Retrospective and Literature Survey. J. Qual. Technol. 2004, 36, 53–77. [Google Scholar] [CrossRef]
- Dadzie, R.G.; Ma, H.; Abano, E.E.; Qu, W.; Mao, S. Optimization of process conditions for production of angiotensin I-converting enzyme (ACE) inhibitory peptides from vital wheat gluten using response surface methodology. Food Sci. Biotechnol. 2013, 22, 1531–1537. [Google Scholar] [CrossRef]
- Thakkar, A.; Saraf, M. Application of Statistically Based Experimental Designs to Optimize Cellulase Production and Identification of Gene. Nat. Prod. Bioprospect. 2014, 4, 341–351. [Google Scholar] [CrossRef] [PubMed]
- Garai, D.; Kumar, V. Response surface optimization for xylanase with high volumetric productivity by indigenous alkali tolerant Aspergillus candidus under submerged cultivation. 3 Biotech 2012, 3, 127–136. [Google Scholar] [CrossRef] [PubMed]
- Khurana, S.; Kapoor, M.; Gupta, S.; Kuhad, R.C. Statistical optimization of alkaline xylanase production from Streptomyces violaceoruber under submerged fermentation using response surface methodology. Indian J. Microbiol. 2007, 47, 144–152. [Google Scholar] [CrossRef] [PubMed]
- Morowvat, M.H.; Memari, H.R. Optimization of Fermentation Conditions for Recombinant Human Interferon Beta Production by Escherichia coli Using the Response Surface Methodology. Jundishapur J. Microbiol. 2015, 8, 1–8. [Google Scholar] [CrossRef]
- Tabandeh, F.; Khodabandeh, M.; Yakhchali, B.; Habib-Ghomi, H.; Shariati, P. Response surface methodology for optimizing the induction conditions of recombinant interferon beta during high cell density culture. Chem. Eng. Sci. 2008, 63, 2477–2483. [Google Scholar] [CrossRef]
- Anderson-Cook, C.M.; Borror, C.M.; Montgomery, D.C. Response surface design evaluation and comparison. J. Stat. Plan. Inference 2009, 139, 629–641. [Google Scholar] [CrossRef]
- Liu, C.; Sun, Z.T.; Du, J.H.; Wang, J. Response surface optimization of fermentation conditions for producing xylanase by Aspergillus niger SL-05. J. Ind. Microbiol. Biotechnol. 2008, 35, 703–711. [Google Scholar] [CrossRef] [PubMed]
- Behera, S.K.; Meena, H.; Chakraborty, S.; Meikap, B.C. Application of response surface methodology (RSM) for optimization of leaching parameters for ash reduction from low-grade coal. Int. J. Min. Sci. Technol. 2018, 28, 621–629. [Google Scholar] [CrossRef]
- Rani, V.; Dash, S.; Nain, L.; Arora, A. Expression of novel glucose tolerant β-glucosidase on cell surface by Rhodotorula glutinis isolate. Biocatal. Agric. Biotechnol. 2015, 4, 380–387. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, Q.; Wang, Y.; Han, H.; Hou, Y.; Shi, Y. Statistical optimization for the production of recombinant cold-adapted superoxide dismutase in E. coli using response surface methodology. Bioengineered 2017, 5979, 1–7. [Google Scholar]
- Maldonado, L.P.; Hernández, V.E.B.; Rivero, E.M.; Barba de la Rosa, A.P.; Flores, J.L.F.; Acevedo, L.G.O. Optimization of culture conditions for a synthetic gene expression in Escherichia coli using response surface methodology: The case of human interferon beta. Biomol. Eng. 2007, 24, 217–222. [Google Scholar] [CrossRef] [PubMed]
- Balderas, H.V.E.; Paz Maldonado, L.M.T.; Rivero, E.M.; Barba de la Rosa, A.P.; Ordoñez Acevedo, L.G.; Rodríguez, A.D.L. Optimization of human interferon gamma production in Escherichia coli by response surface methodology. Biotechnol. Bioprocess Eng. 2008, 13, 7–13. [Google Scholar] [CrossRef]
- Batra, J.; Beri, D.; Mishra, S. Response surface methodology based optimization of β-glucosidase production from Pichia Pastor. Appl. Biochem. Biotechnol. 2014, 172, 380–393. [Google Scholar] [CrossRef] [PubMed]
- Dey, G.; Mitra, A.; Banerjee, R.; Maiti, B.R. Enhanced production of amylase by optimization of nutritional constituents using response surface methodology. Biochem. Eng. J. 2001, 7, 227–231. [Google Scholar] [CrossRef]
- Tanyildizi, M.S.; Özer, D.; Elibol, M. Optimization of α-amylase production by Bacillus sp. using response surface methodology. Process Biochem. 2005, 40, 2291–2296. [Google Scholar] [CrossRef]
- Low, K.O.; Muhammad Mahadi, N.; Abdul Rahim, R.; Rabu, A.; Abu Bakar, F.D.; Murad, A.M.A. An effective extracellular protein secretion by an ABC transporter system in Escherichia coli: Statistical modeling and optimization of cyclodextrin glucanotransferase secretory production. J. Ind. Microbiol. Biotechnol. 2011, 38, 1587–1597. [Google Scholar] [CrossRef] [PubMed]
- Hao, D.C.; Zhu, P.H.; Yang, S.L.; Yang, L. Optimization of recombinant cytochrome P450 2C9 protein production in Escherichia coli DH5α by statistically-based experimental design. World J. Microbiol. Biotechnol. 2006, 22, 1169–1176. [Google Scholar] [CrossRef]
- Ghoshoon, M.B.; Berenjian, A.; Hemmati, S.; Dabbagh, F.; Karimi, Z.; Negahdaripour, M. Extracellular Production of Recombinant l-Asparaginase II in Escherichia coli: Medium Optimization Using Response Surface Methodology. Int. J. Pept. Res. Ther. 2015, 21, 487–495. [Google Scholar] [CrossRef]
- Nikerel, I.E.; Toksoy, E.; Kirdar, B.; Yildirim, R. Optimizing medium composition for TaqI endonuclease production by recombinant Escherichia coli cells using response surface methodology. Process Biochem. 2005, 40, 1633–1639. [Google Scholar] [CrossRef]
- Farliahati, M.R.; Ramanan, R.N.; Mohamad, R.; Puspaningsih, N.N.T.; Ariff, A.B. Enhanced production of xylanase by recombinant Escherichia coli DH5α through optimization of medium composition using response surface methodology. Ann. Microbiol. 2010, 60, 279–285. [Google Scholar] [CrossRef]
- Muntari, B.; Amid, A.; Mel, M.; Jami, M.S.; Salleh, H.M. Recombinant bromelain production in Escherichia coli: Process optimization in shake flask culture by response surface methodology. AMB Express 2012, 2. [Google Scholar] [CrossRef] [PubMed]
- Sunitha, K.; Lee, J.K.; Oh, T.K. Optimization of medium components for phytase production by E. coli using response surface methodology. Bioprocess Eng. 1999, 21, 477–481. [Google Scholar] [CrossRef]
- Lee, Y.J.; Kim, H.J.; Gao, W.; Chung, C.H.; Lee, J.W. Statistical optimization for production of carboxymethylcellulase of Bacillus amyloliquefaciens DL-3 by a recombinant Escherichia coli JM109/DL-3 from rice bran using response surface method. Biotechnol. Bioprocess Eng. 2012, 17, 227–235. [Google Scholar] [CrossRef]
- Joshi, S.; Satyanarayana, T. Optimization of heterologous expression of the phytase (PPHY) of Pichia anomala in P. pastoris and its applicability in fractionating allergenic glycinin from soy protein. J. Ind. Microbiol. Biotechnol. 2014, 41, 977–987. [Google Scholar] [PubMed]
- Chen, Y.; Xing, X.H.; Ye, F.; Kuang, Y.; Luo, M. Production of MBP–HepA fusion protein in recombinant Escherichia coli by optimization of culture medium. Biochem. Eng. J. 2007, 34, 114–121. [Google Scholar] [CrossRef]
- Pan, H.; Xie, Z.; Bao, W.; Zhang, J. Optimization of culture conditions to enhance cis-epoxysuccinate hydrolase production in Escherichia coli by response surface methodology. Biochem. Eng. J. 2008, 42, 133–138. [Google Scholar] [CrossRef]
- Karimineghlani, P.; Neghlani, P.K.; Azadmehr, A. Optimization of lead ions adsorption on hydrolyzed polyacrylonitrile fibers using central composite design. Desalination Water Treat. 2017, 83, 133–143. [Google Scholar] [CrossRef]
- Almeida, J.M.; Lima, V.A.; Giloni-Lima, P.C.; Knob, A. Passion fruit peel as novel substrate for enhanced beta-glucosidases production by Penicillium verruculosum: Potential of the crude extract for biomass hydrolysis. Biomass Bioenergy 2015, 72, 216–226. [Google Scholar] [CrossRef]
- Singh, A.K.; Chhatpar, H.S. Optimization of protease production by Streptomyces sp. A6 using statistical approach for reclamation of shellfish waste. World J. Microbiol. Biotechnol. 2010, 26, 1631–1639. [Google Scholar] [CrossRef]
- Khataee, A.R. Application of central composite design for the optimization of photo-destruction of a textile dye using UV/S2O82-process. Pol. J. Chem. Technol. 2009, 11, 38–45. [Google Scholar] [CrossRef]
- Nelofer, R.; Ramanan, R.N.; Rahman, R.A.; Basri, M.; Ariff, A.B. Comparison of the estimation capabilities of response surface methodology and artificial neural network for the optimization of recombinant lipase production by E. coli BL21. J. Ind. Microbiol. Biotechnol. 2012, 39, 243–254. [Google Scholar] [CrossRef] [PubMed]
- Vellanki, R.N.; Potumarthi, R.; Mangamoori, L.N. Constitutive expression and optimization of nutrients for streptokinase production by pichia pastoris using statistical methods. Appl. Biochem. Biotechnol. 2009, 158, 25–40. [Google Scholar] [CrossRef] [PubMed]
- Sharma, L.; Kumar, S.A.; Panda, B.; Mallick, N. Process optimization for poly-β-hydroxybutyrate production in a nitrogen fixing cyanobacterium, Nostoc muscorum using response surface methodology. Bioresour. Technol. 2007, 98, 987–993. [Google Scholar] [CrossRef] [PubMed]
- Tatineni, R.; Doddapaneni, K.K.; Potumarthi, R.C.; Mangamoori, L.N. Optimization of keratinase production and enzyme activity using response surface methodology with streptomyces sp7. Appl. Biochem. Biotechnol. 2007, 141, 187–201. [Google Scholar] [CrossRef] [PubMed]
- Pareek, N.; Ghosh, S.; Singh, R.P.; Vivekanand, V. Mustard oil cake as an inexpensive support for production of chitin deacetylase by Penicillium oxalicum SAEM-51 under solid-state fermentation. Biocatal. Agric. Biotechnol. 2014, 3, 212–217. [Google Scholar] [CrossRef]
- Tavakkoli, M.; Hamidi-Esfahani, Z.; Azizi, M.H. Optimization of Corynebacterium glutamicum Glutamic Acid Production by Response Surface Methodology. Food Bioprocess Technol. 2012, 5, 92–99. [Google Scholar] [CrossRef]
- Almeida, M.; Erthal, R.; Padua, E.; Silveira, L.; Am, L. Response surface methodology (RSM) as a tool for optimization in analytical chemistry. Talanta 2008, 76, 965–977. [Google Scholar]
- Nagar, S.; Gupta, V.K.; Kumar, D.; Kumar, L.; Kuhad, R.C. Production and optimization of cellulase-free, alkali-stable xylanase by Bacillus pumilus SV-85S in submerged fermentation. J. Ind. Microbiol. Biotechnol. 2010, 37, 71–83. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Xu, T.; Ma, X.; Guo, K.; Kai, L.; Zhao, Y. Optimization of culture conditions for production of cis-epoxysuccinic acid hydrolase using response surface methodology. Bioresour. Technol. 2008, 99, 5391–5396. [Google Scholar] [CrossRef] [PubMed]
- Ray, R.C.; Kar, S. Statistical optimization of [alpha]-amylase production by Bacillus brevis MTCC 7521 in solid-state fermentation using cassava bagasse. Biologia 2009, 64, 864–870. [Google Scholar] [CrossRef]
- Rekha, V.P.B.; Ghosh, M.; Adapa, V.; Oh, S.J.; Pulicherla, K.K.; Sambasiva Rao, K.R.S. Optimization of polygalacturonase production from a newly isolated Thalassospira frigidphilosprofundus to use in pectin hydrolysis: Statistical approach. Biomed Res. Int. 2013. [Google Scholar] [CrossRef] [PubMed]
- Deswal, A.; Deora, N.S.; Mishra, H.N. Optimization of Enzymatic Production Process of Oat Milk Using Response Surface Methodology. Food Bioprocess Technol. 2014, 7, 610–618. [Google Scholar] [CrossRef]
- Halmi, M.I.E.; Abdullah, S.R.S.; Wasoh, H.; Johari, W.L.W.; Ali, M.S.; Shaharuddin, N.A. Optimization and maximization of hexavalent molybdenum reduction to Mo-blue by Serratia sp. strain MIE2 using response surface methodology. Rend. Lincei 2016, 27, 697–709. [Google Scholar] [CrossRef]
- Khoshayand, F.; Goodarzi, S.; Shahverdi, A.R.; Khoshayand, M.R. Optimization of Culture Conditions for Fermentation of Soymilk Using Lactobacillus casei by Response Surface Methodology. Probiotics Antimicrob. Proteins 2011, 3, 159–167. [Google Scholar] [CrossRef] [PubMed]
- Cui, F.; Liu, Z.; Li, Y.; Ping, L.; Ping, L.; Zhang, Z. Production of mycelial biomass and exo-polymer by Hericium erinaceus CZ-2: Optimization of nutrients levels using response surface methodology. Biotechnol. Bioprocess Eng. 2010, 15, 299–2307. [Google Scholar] [CrossRef]
- Partida-Sedas, G.; Montes-García, N.; Carvajal-Zarrabal, O.; López-Zamora, L.; Gómez-Rodríguez, J.; Aguilar-Uscanga, M.G. Optimization of Hydrolysis Process to Obtain Fermentable Sugars from Sweet Sorghum Bagasse Using a Box–Behnken Design. Sugar Tech 2017, 19, 317–325. [Google Scholar] [CrossRef]
- Rajendran, A.; Thangavelu, V. Optimization and Modeling of Process Parameters for Lipase Production by Bacillus Brevis. Food Bioprocess Technol. 2012, 5, 310–322. [Google Scholar] [CrossRef]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).