Rebalancing Redox to Improve Biobutanol Production by Clostridium tyrobutyricum
Abstract
:1. Introduction
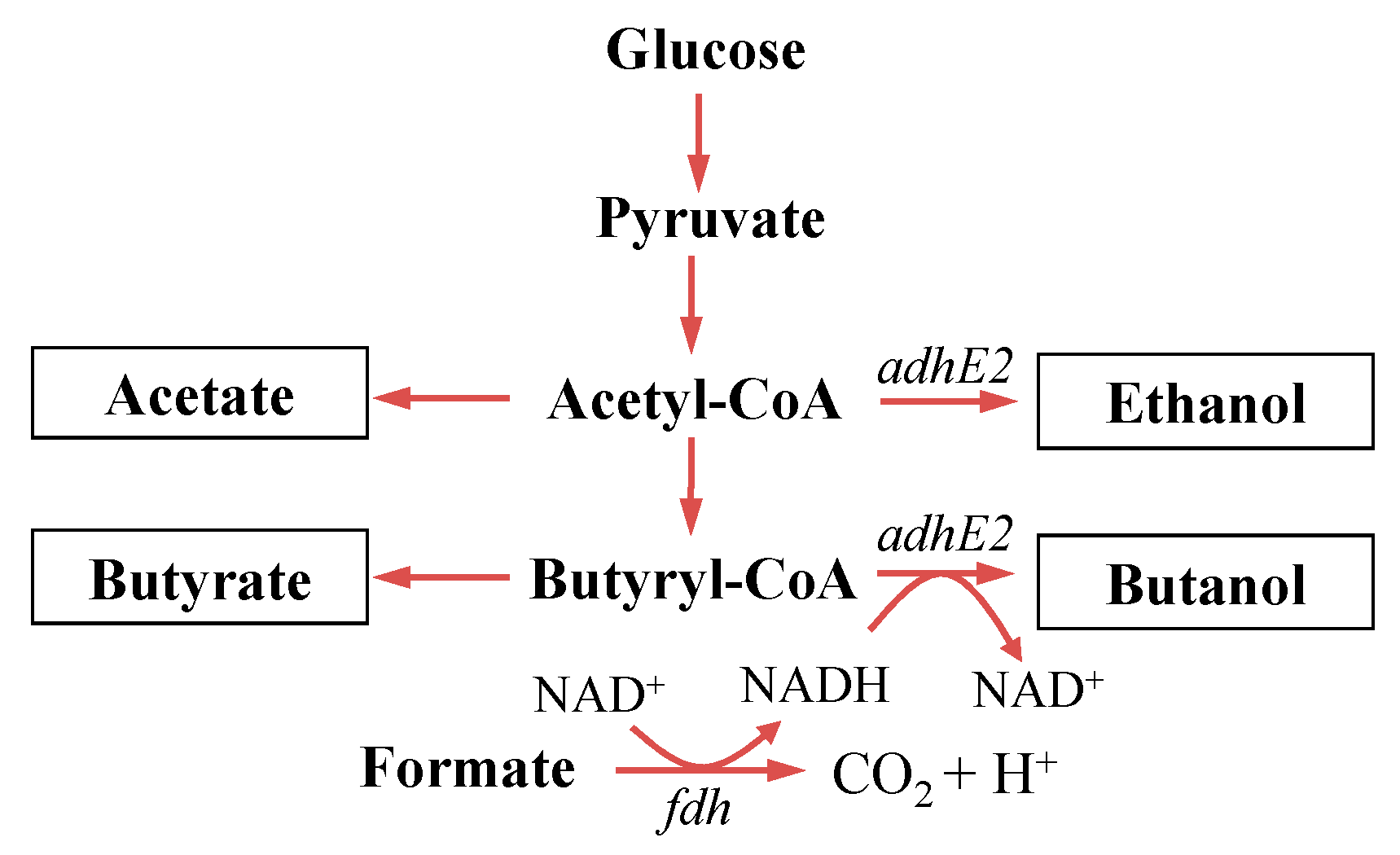
2. Materials and Methods
2.1. Strains and Media
| Plasmid/Strain | Relevant Characteristics | Reference/Source |
|---|---|---|
| Plasmids | ||
| pMTL-adhE2 | adhE2 overexpression with thl promoter | This study |
| pMTL-fdh-adhE2 | fdh and adhE2 overexpression with thl promoter | This study |
| Strains | ||
| C. tyrobutyricum | Clostridium, ATCC 25755, wild-type | ATCC |
| CTC-adhE2 | Clostridium with adhE2 overexpression, thiamphenicol resistant | [23] |
| CTC-fdh-adhE2 | Clostridium with fdh and adhE2 overexpression, thiamphenicol resistant | This study |
| E. coli CA434 | E. coli HB101 with plasmid R702, kanamycin resistant | [24] |
2.2. Plasmid Construction
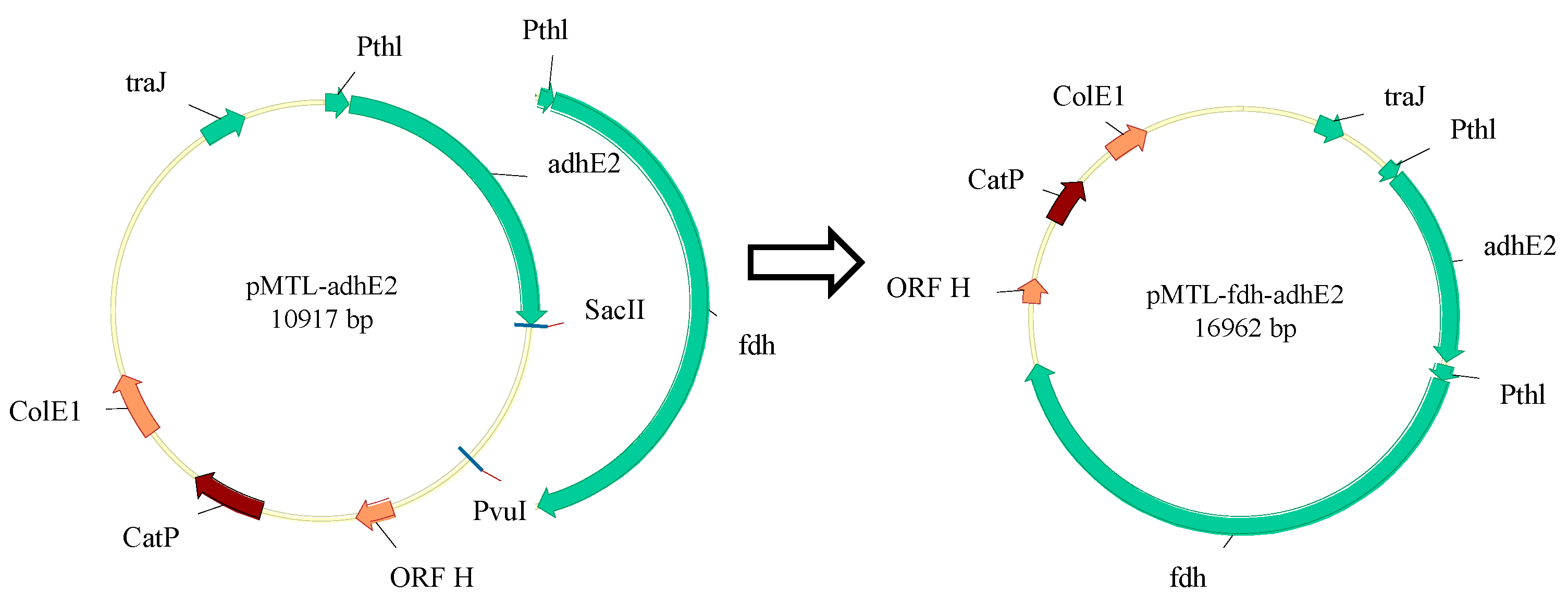
2.3. Mutant Construction
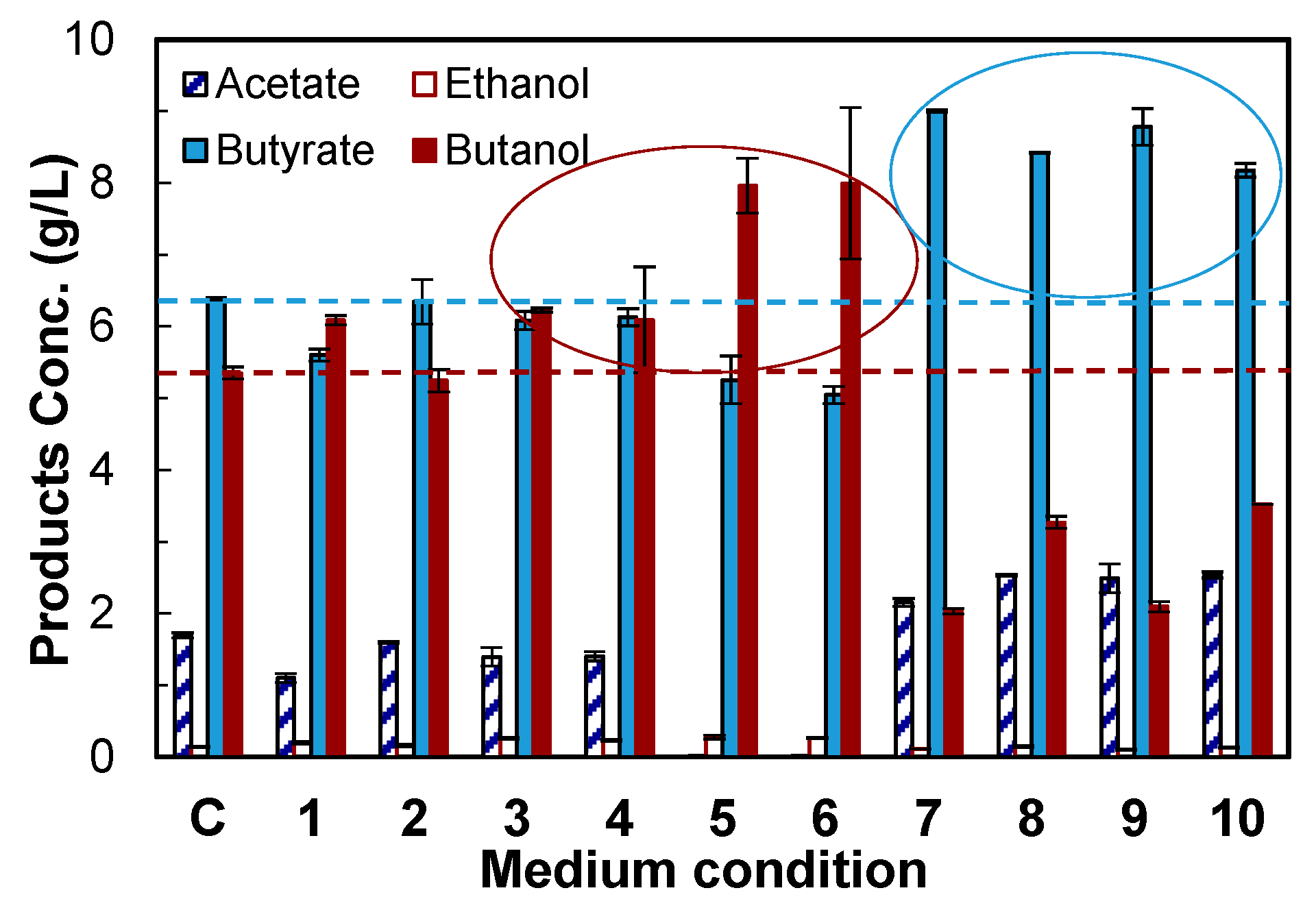
2.4. Medium Supplements Screening
| No. | Sodium Formate (1 g/L) | Vitamin B12 (0.001 g/L) | Methyl Viologen Hydrate (0.1 g/L) | Vanadate (1 g/L) | Acetamide (1 g/L) |
|---|---|---|---|---|---|
| C | − | − | − | − | − |
| 1 | + | − | − | − | − |
| 2 | − | + | − | − | − |
| 3 | − | − | + | − | − |
| 4 | + | + | − | − | − |
| 5 | + | − | + | − | − |
| 6 | + | + | + | − | − |
| 7 | + | − | − | + | − |
| 8 | + | − | − | − | + |
| 9 | + | + | − | + | − |
| 10 | + | + | − | − | + |
2.5. Butanol Fermentation
2.6. Activity Assay of Formate Dehydrogenase
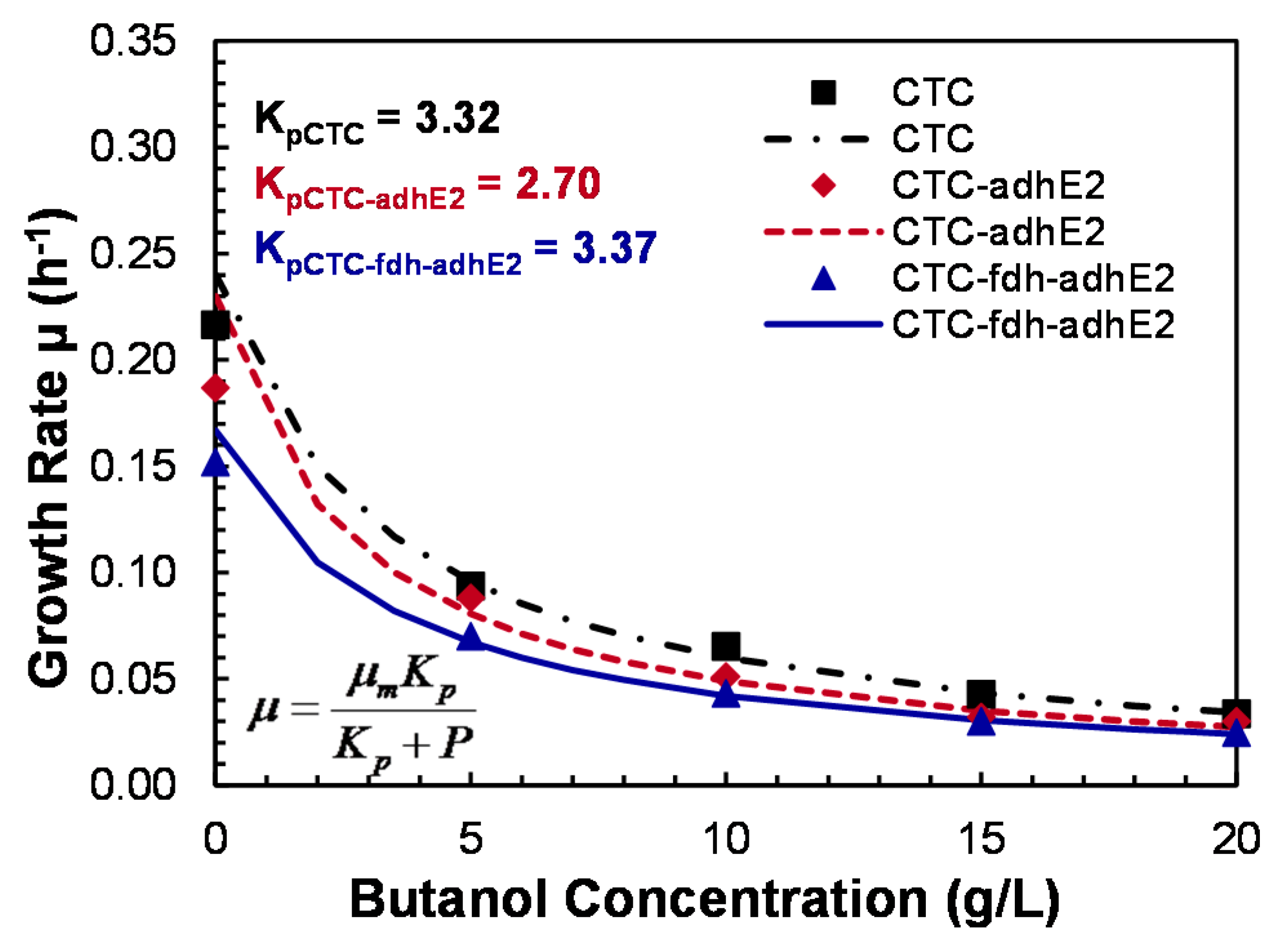
2.7. Butanol Tolerance
2.8. NADH Assay
3. Analytical Methods
4. Results
4.1. Construction and Evaluation of Redox-Engineered Mutant
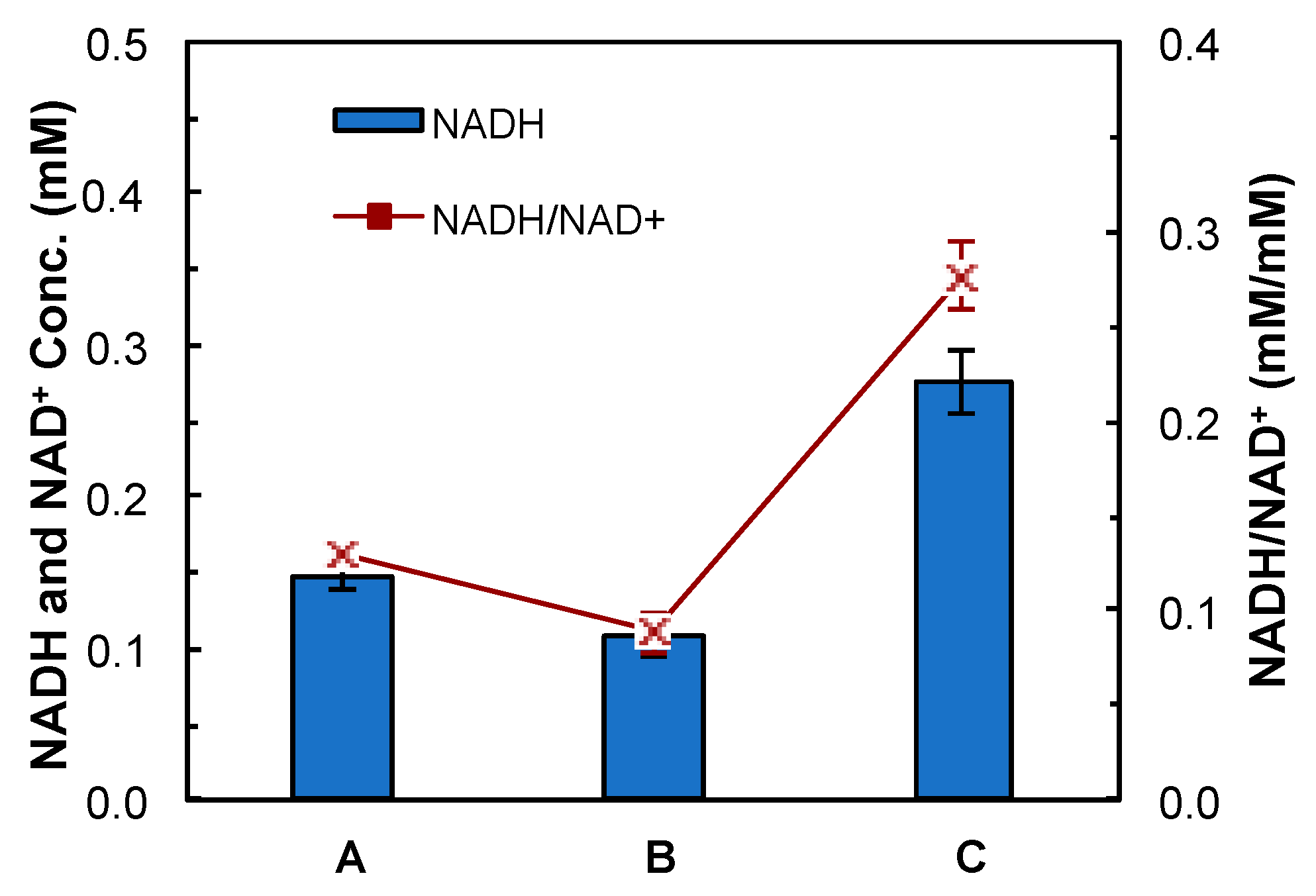
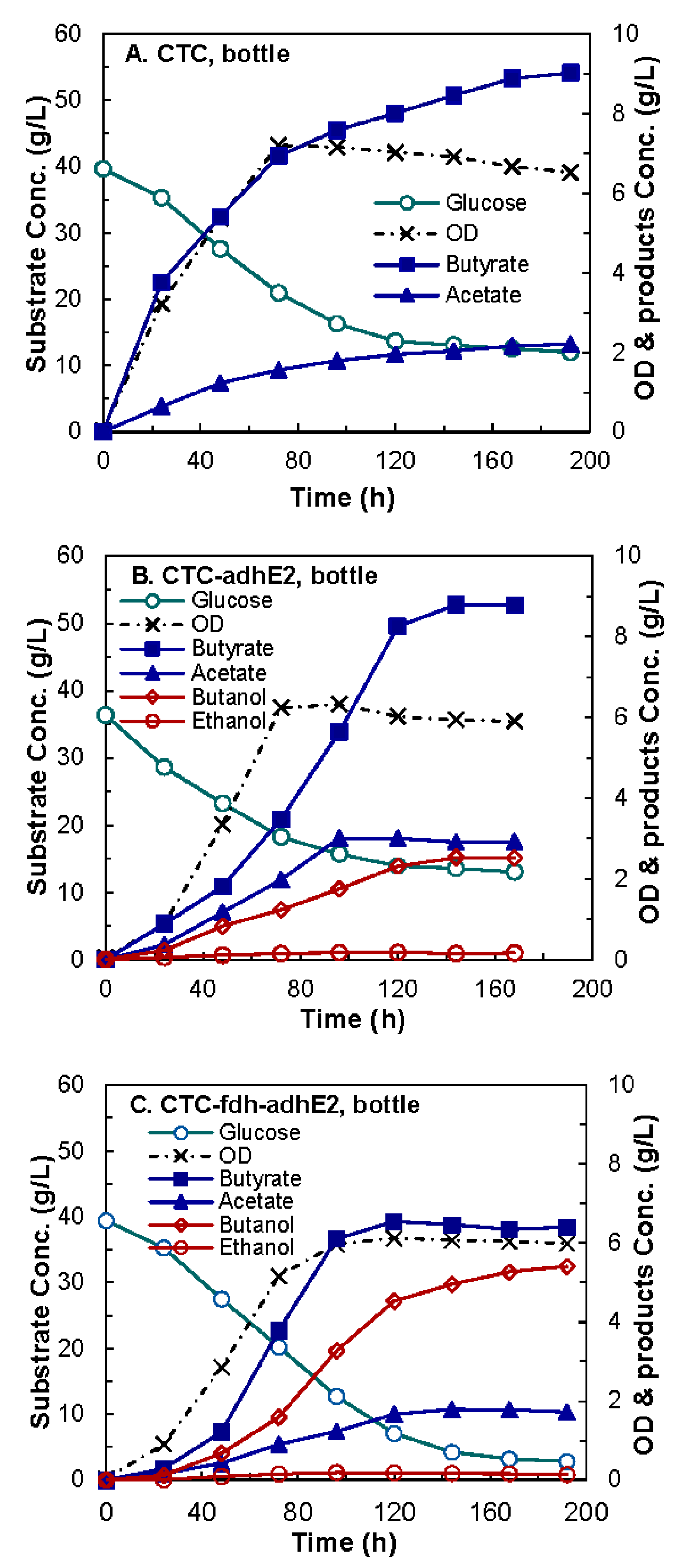
4.2. Evaluation of Medium Supplements
4.3. Butanol Fermentation
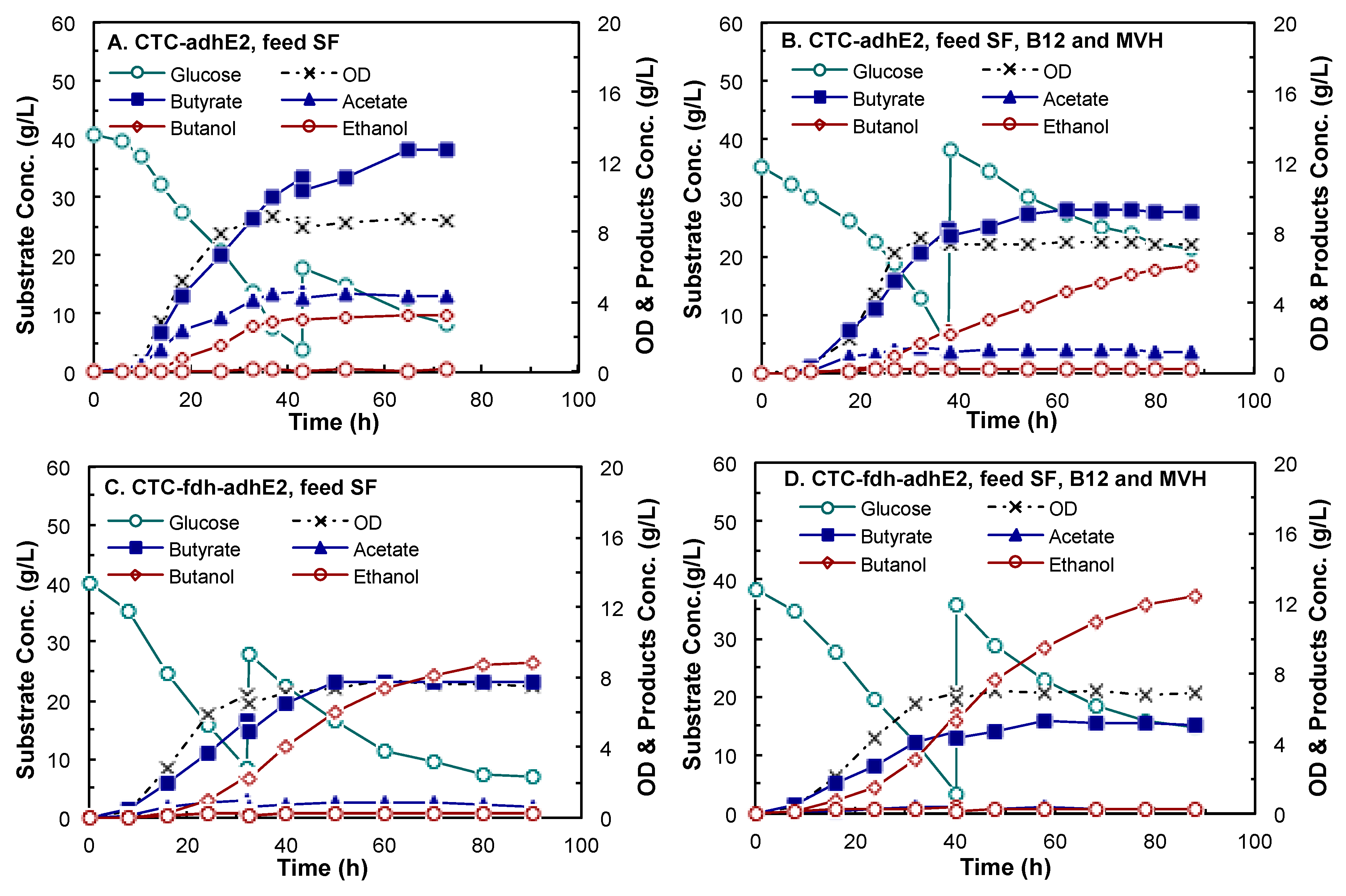
| Products | CTC-adhE2 (Control) | CTC-fdh-adhE2 | ||||
|---|---|---|---|---|---|---|
| SF | SF, B12 and MVH | SF | SF, B12 and MVH | |||
| Cell growth (h−1) | 0.18 ± 0.01 | 0.16 ± 0.0004 | 0.15 ± 0.001 | 0.15 ± 0.001 | ||
| Biomass yield (g/g) | 0.10 ± 0.01 | 0.09 ± 0.003 | 0.09 ± 0.001 | 0.08 ± 0.002 | ||
| Concentration (g/L) | Butanol | 3.18 ± 0.09 | 6.14 ± 0.05 | 8.83 ± 0.02 | 12.34 ± 0.02 | |
| Butyrate | 13.22 ± 0.73 | 9.32 ± 0.03 | 7.72 ± 0.05 | 5.05 ± 0.04 | ||
| Acetate | 4.15 ± 0.25 | 1.42 ± 0.07 | 0.69 ± 0.01 | 0.26 ± 0.03 | ||
| Ethanol | 0.11 ± 0.04 | 0.25 ± 0.02 | 0.24 ± 0.01 | 0.28 ± 0.07 | ||
| Yield (g/g-glucose) | Butanol | 0.05 ± 0.01 | 0.11 ± 0.003 | 0.16 ± 0.002 | 0.23 ± 0.002 | |
| Butyrate | 0.27 ± 0.02 | 0.20 ± 0.004 | 0.19 ± 0.01 | 0.10 ± 0.001 | ||
| Acetate | 0.06 ± 0.01 | 0.02 ± 0.002 | 0.02 ± 0.001 | 0.003 ± 0.0001 | ||
| Ethanol | 0.001 ± 0.0005 | 0.003 ± 0.0004 | 0.004 ± 0.0002 | 0.004 ± 0.0003 | ||
| Productivity (g/L/h) | Butanol | 0.12 ± 0.01 | 0.14 ± 0.01 | 0.17 ± 0.002 | 0.26 ± 0.01 | |
| Butyrate | 0.33 ± 0.01 | 0.26 ± 0.003 | 0.22 ± 0.004 | 0.15 ± 0.004 | ||
| Acetate | 0.14 ± 0.01 | 0.07 ± 0.004 | 0.05 ± 0.01 | 0.01 ± 0.002 | ||
| Ethanol | 0.006 ± 0.001 | 0.008 ± 0.0001 | 0.010 ± 0.0003 | 0.011 ± 0.0004 | ||
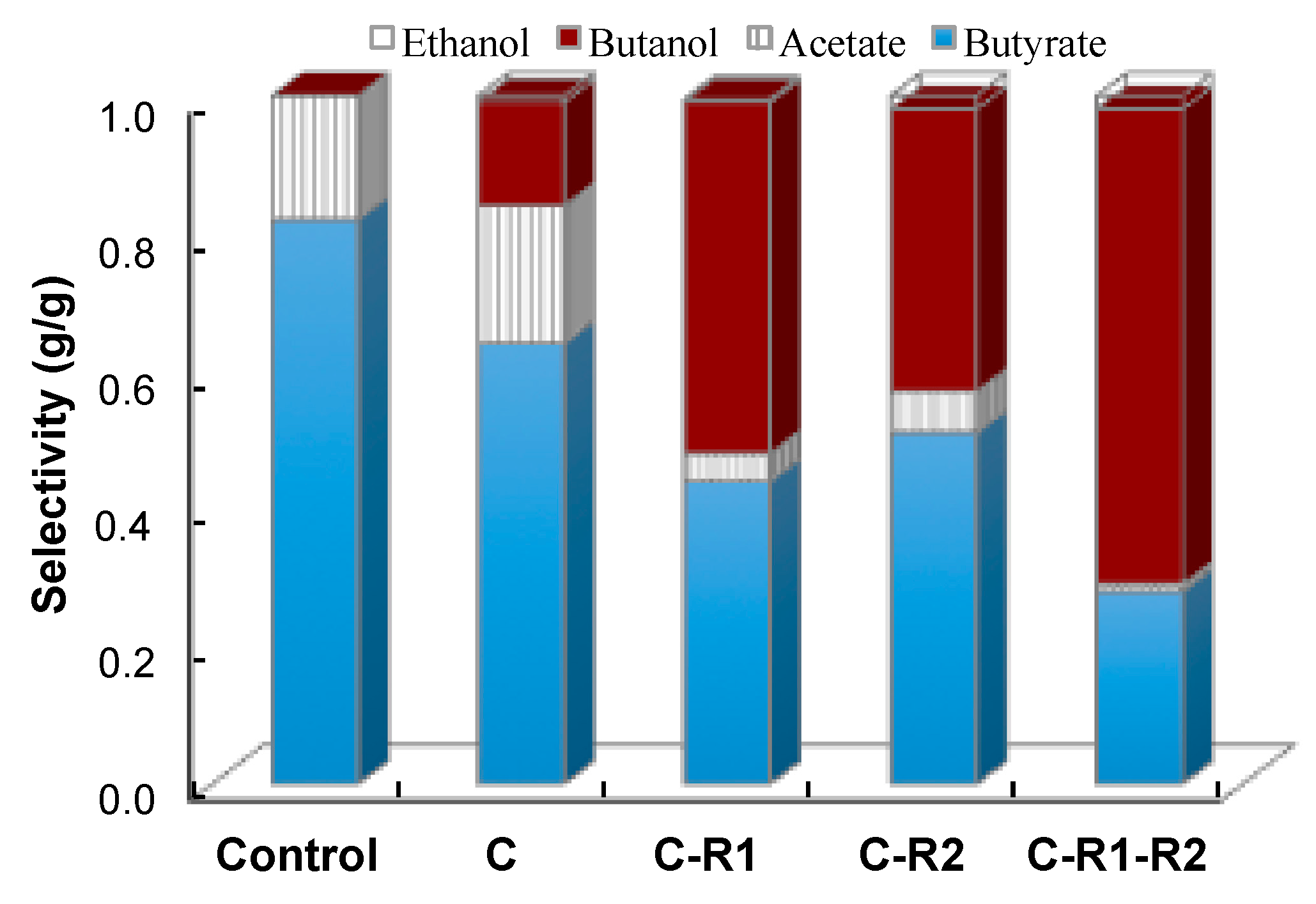
4.4. Butanol Tolerance
5. Discussion
5.1. Improvement of Butanol Production by Rebalancing Redox
5.2. Comparison with Previous Studies
| Strain | Characteristics | Mode | Carbon | Titer (g/L) | Yield (g/g) | Productivity (g/L/h) | Reference |
|---|---|---|---|---|---|---|---|
| CTC-adhE2 | +adhE2, pCB102 replicon | Bottle | Glucose | 2.5 | 0.15 * | 0.04 * | [16] |
| Bottle | Glucose | 2.6 | 0.14 | 0.03 | This study | ||
| CTpM2 | +adhE2, pBP1 replicon | Bottle | Glucose | 6.5 | 0.24 | 0.20 * | [16] |
| Bioreactor | Mannitol # | 20.0 # | 0.33 # | 0.32 # | [16] | ||
| CTC-fdh-adhE2 | +fdh+adhE2, pCB102 replicon | Bottle | Glucose | 6.9 | 0.21 | 0.20 | This study |
| Bioreactor | Glucose | 12.3 | 0.23 | 0.26 | This study |
6. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Zheng, Y.N.; Li, L.Z.; Xian, M.; Ma, Y.J.; Yang, J.M.; Xu, X.; He, D.Z. Problems with the microbial production of butanol. J. Ind. Microbiol. Biotechnol. 2009, 36, 1127–1138. [Google Scholar] [CrossRef] [PubMed]
- Milne, C.B.; Eddy, J.A.; Raju, R.; Ardekani, S.; Kim, P.J.; Senger, R.S.; Jin, Y.S.; Blaschek, H.P.; Price, N.D. Metabolic network reconstruction and genomescale model of butanol-producing strain Clostridium beijerinckii NCIMB 8052. BMC Syst. Biol. 2011, 5, 130–144. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.Y.; Park, J.H.; Jang, S.H.; Nielsen, L.K.; Kim, J.; Jung, K.S. Fermentative butanol production by Clostridia. Biotechnol. Bioeng. 2008, 101, 209–228. [Google Scholar] [CrossRef] [PubMed]
- Nicolaou, S.A.; Gaida, S.M.; Papoutsakis, E.T. A comparative view of metabolite and substrate stress and tolerance in microbial bioprocessing: From biofuels and chemicals, to biocatalysis and bioremediation. Metab. Eng. 2010, 12, 307–331. [Google Scholar] [CrossRef] [PubMed]
- Papoutsakis, E.T. Engineering solventogenic clostridia. Curr. Opin. Biotechnol. 2008, 19, 420–429. [Google Scholar] [CrossRef] [PubMed]
- Al-Hinai, M.A.; Fast, A.G.; Papoutsakis, E.T. Novel system for efficient isolation of Clostridium double-crossover allelic exchange mutants enabling markerless chromosomal gene deletions and DNA integration. Appl. Environ. Microbiol. 2012, 78, 8112–8121. [Google Scholar] [CrossRef] [PubMed]
- Atsumi, S.; Cann, A.F.; Connor, M.R.; Shen, C.R.; Smith, K.M.; Brynildsen, M.P.; Chou, K.J.; Hanai, T.; Liao, J.C. Metabolic engineering of Escherichia coli for 1-butanol production. Metab. Eng. 2008, 10, 305–311. [Google Scholar] [CrossRef] [PubMed]
- Berezina, O.V.; Zakharova, N.V.; Brandt, A.; Yarotsky, S.V.; Schwarz, W.H.; Zverlov, V.V. Reconstructing the clostridial n-butanol metabolic pathway in Lactobacillus brevis. Appl. Microbiol. Biotechnol. 2010, 87, 635–646. [Google Scholar] [CrossRef] [PubMed]
- Blombach, B.; Eikmanns, B.J. Current knowledge on isobutanol production with Escherichia coli, Bacillus subtilis and Corynebacterium glutamicum. Bioeng. Bugs. 2011, 2, 346–350. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, D.R.; Leonard, E.; Yoon, S.H.; Tseng, H.C.; Yuan, C.; Prather, K.L. Engineering alternative butanol production platforms in heterologous bacteria. Metab. Eng. 2009, 11, 262–273. [Google Scholar] [CrossRef] [PubMed]
- Shen, C.R.; Liao, J.C. Metabolic engineering of Escherichia coli for 1-butanol and 1-propanol production via the keto-acid pathways. Metab. Eng. 2008, 10, 312–320. [Google Scholar] [CrossRef] [PubMed]
- Steen, E.J.; Chan, R.; Prasad, N.; Myers, S.; Petzold, C.J.; Redding, A.; Ouellet, M.; Keasling, J.D. Metabolic engineering of Saccharomyces cerevisiae for the production of n-butanol. Microb. Cell. Fact. 2008, 7, 36. [Google Scholar] [CrossRef] [PubMed]
- Shen, C.R.; Lan, E.I.; Dekishima, Y.; Baez, A.; Cho, K.M.; Liao, J.C. Driving forces enable high-titer anaerobic 1-butanol synthesis in Escherichia coli. Appl. Environ. Microbiol. 2011, 77, 2905–2915. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Wang, J.; Liang, S.; Wang, X.; Cen, P.; Xu, Z. Production of butyric acid from glucose and xylose with immobilized cells of Clostridium tyrobutyricum in a fibrous-bed bioreactor. Appl. Biochem. Biotechnol. 2010, 160, 350–359. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Yang, S.T. Butyric acid and hydrogen production by Clostridium tyrobutyricum ATCC 25755 and mutants. Enzyme Microb. Technol. 2006, 38, 521–528. [Google Scholar] [CrossRef]
- Yu, M.; Du, Y.; Jiang, W.; Chang, W.L.; Yang, S.T.; Tang, I.C. Effects of different replicons in conjugative plasmids on transformation efficiency, plasmid stability, gene expression and n-butanol biosynthesis in Clostridium tyrobutyricum. Appl. Microbiol. Biotechnol. 2012, 93, 881–889. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.; Kojima, K.; Xu, N.; Mobley, J.; Zhou, L.; Yang, S.T.; Liu, X.M. Comparative proteomics analysis of high n-butanol producing metabolically engineered Clostridium tyrobutyricum. J. Biotechnol. 2015, 193, 108–119. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Jiang, W.; Yu, M.; Tang, I.C.; Yang, S.T. Metabolic process engineering of Clostridium tyrobutyricum Deltaack-adhE2 for enhanced n-butanol production from glucose: Effects of methyl viologen on NADH availability, flux distribution and fermentation kinetics. Biotechnol. Bioeng. 2015, 112, 705–715. [Google Scholar] [CrossRef] [PubMed]
- Ou, J.; Ma, C.; Xu, N.; Du, Y.; Liu, X.M. High Butanol Production by Regulating Carbon, Redox and Energy in Clostridia. Front. Chem. Sci. Eng. 2015. [Google Scholar] [CrossRef]
- Berrios-Rivera, S.J.; Bennett, G.N.; San, K.Y. Metabolic engineering of Escherichia coli: Increase of NADH availability by overexpressing an NAD(+)-dependent formate dehydrogenase. Metab. Eng. 2002, 4, 217–229. [Google Scholar] [CrossRef] [PubMed]
- Geertman, J.M.; van Dijken, J.P.; Pronk, J.T. Engineering NADH metabolism in Saccharomyces cerevisiae: formate as an electron donor for glycerol production by anaerobic, glucose-limited chemostat cultures. FEMS Yeast Res. 2006, 6, 1193–1203. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Hu, L.; Fan, L.; Tan, T. Enhanced 1-Butanol Production in Engineered Klebsiella pneumoniae by NADH Regeneration. Energy Fuels 2015, 29, 1823–1829. [Google Scholar] [CrossRef]
- Yu, M.; Zhang, Y.; Tang, I.C.; Yang, S.T. Metabolic engineering of Clostridium tyrobutyricum for n-butanol production. Metab. Eng. 2011, 13, 373–382. [Google Scholar] [CrossRef] [PubMed]
- Purdy, D.; O'Keeffe, T.A.; Elmore, M.; Herbert, M.; McLeod, A.; Bokori-Brown, M.; Ostrowski, A.; Minton, N.P. Conjugative transfer of clostridial shuttle vectors from Escherichia coli to Clostridium difficile through circumvention of the restriction barrier. Mol. Microbiol. 2002, 46, 439–452. [Google Scholar] [CrossRef] [PubMed]
- Dong, H.; Tao, W.; Zhang, Y.; Li, Y. Development of an anhydrotetracycline-inducible gene expression system for solvent-producing Clostridium acetobutylicum: A useful tool for strain engineering. Metab. Eng. 2012, 14, 59–67. [Google Scholar] [CrossRef] [PubMed]
- Heap, J.T.; Kuehne, S.A.; Ehsaan, M.; Cartman, S.T.; Cooksley, C.M.; Scott, J.C.; Minton, N.P. The ClosTron: Mutagenesis in Clostridium refined and streamlined. J. Microbiol. Methods. 2010, 80, 49–55. [Google Scholar] [CrossRef] [PubMed]
- Alissandratos, A.; Kim, H.K.; Matthews, H.; Hennessy, J.E.; Philbrook, A.; Easton, C.J. Clostridium carboxidivorans strain P7T recombinant formate dehydrogenase catalyzes reduction of CO(2) to formate. Appl. Environ. Microbiol. 2013, 79, 741–744. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, I.; Saiki, T.; Liu, S.M.; Ljungdahl, L.G. Purification and properties of NADP-dependent formate dehydrogenase from Clostridium thermoaceticum, a tungsten-selenium-iron protein. J. Biol. Chem. 1983, 258, 1826–1832. [Google Scholar] [PubMed]
- Heluane, H.; Evans, M.R.; Dagher, S.F.; Bruno-Barcena, J.M. Meta-analysis and functional validation of nutritional requirements of solventogenic Clostridia growing under butanol stress conditions and coutilization of D-glucose and D-xylose. Appl. Environ. Microbiol. 2011, 77, 4473–4485. [Google Scholar] [CrossRef] [PubMed]
- Gheshlaghi, R.; Scharer, J.M.; Moo-Young, M.; Chou, C.P. Metabolic pathways of clostridia for producing butanol. Biotechnol. Adv. 2009, 27, 764–781. [Google Scholar] [CrossRef] [PubMed]
- Amador-Noguez, D.; Brasg, I.A.; Feng, X.J.; Roquet, N.; Rabinowitz, J.D. Metabolome remodeling during the acidogenic-solventogenic transition in Clostridium acetobutylicum. Appl. Environ. Microbiol. 2011, 77, 7984–7997. [Google Scholar] [CrossRef] [PubMed]
- Nakayama, S.; Kosaka, T.; Hirakawa, H.; Matsuura, K.; Yoshino, S.; Furukawa, K. Metabolic engineering for solvent productivity by downregulation of the hydrogenase gene cluster hupCBA in Clostridium saccharoperbutylacetonicum strain N1–4. Appl. Microbiol. Biotechnol. 2008, 78, 483–493. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.; Ou, J.; McFann, S.; Miller, M.; Liu, X.M. High production of butyric acid by Clostridium tyrobutyricum mutant. Front. Chem. Sci. Eng. 2015. [Google Scholar] [CrossRef]
- Liao, C.; Seo, S.O.; Celik, V.; Liu, H.; Kong, W.; Wang, Y.; Blaschek., H.; Jin., Y.; Lu, T. Integrated, systems metabolic picture of acetone-butanol-ethanol fermentation by Clostridium acetobutylicum. Proc. Natl. Acad. Sci. USA 2015, 112, 8505–8510. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, C.; Ou, J.; Xu, N.; Fierst, J.L.; Yang, S.-T.; Liu, X. Rebalancing Redox to Improve Biobutanol Production by Clostridium tyrobutyricum. Bioengineering 2016, 3, 2. https://doi.org/10.3390/bioengineering3010002
Ma C, Ou J, Xu N, Fierst JL, Yang S-T, Liu X. Rebalancing Redox to Improve Biobutanol Production by Clostridium tyrobutyricum. Bioengineering. 2016; 3(1):2. https://doi.org/10.3390/bioengineering3010002
Chicago/Turabian StyleMa, Chao, Jianfa Ou, Ningning Xu, Janna L. Fierst, Shang-Tian Yang, and Xiaoguang Liu. 2016. "Rebalancing Redox to Improve Biobutanol Production by Clostridium tyrobutyricum" Bioengineering 3, no. 1: 2. https://doi.org/10.3390/bioengineering3010002
APA StyleMa, C., Ou, J., Xu, N., Fierst, J. L., Yang, S.-T., & Liu, X. (2016). Rebalancing Redox to Improve Biobutanol Production by Clostridium tyrobutyricum. Bioengineering, 3(1), 2. https://doi.org/10.3390/bioengineering3010002







