Defining a Simplified Process in Yeast for Production of Enveloped VLP Dengue Vaccine
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Line Construction and Research Cell Bank Generation
2.2. Fermentation Process
Batch and Fed-Batch Cultivation
2.3. Purification of Virus-like Particles (VLPs)
2.3.1. Cell Lysis
2.3.2. Sucrose Density Gradient (SDG)
2.3.3. Chromatographic Platform
Primary Chromatography
Polishing Chromatography
2.4. Analytical Characterization
2.4.1. Biochemical Assays
2.4.2. Biophysical Assays
2.4.3. Purity Assays
PicoGreen dsDNA
Host Cell Protein (HCP)
Total Protein
2.5. Statistical Analysis
3. Results and Discussion
- (i)
- Demonstration of the versatility of using the Komagataella phaffii platform in production of nanoparticles (dengue viral-like particles (VLPs)).
- (ii)
- Culture optimization using DoE approaches in small-scale bioreactors.
- (iii)
- Development of a novel VLP purification platform.
- (iv)
- Creation of an alternative biophysical characterization of the dengue vaccine prototype.
3.1. Cultivation Process Improvement to Produce the Dengue VLP Vaccine Prototype (GS115/pAO815_Den1Den2)
3.1.1. Host and Strain Selection
3.1.2. Optimization of the Fermentation Process at Small Scale, Using a Design of Experiments (DoE) Approach
3.2. VLP Purification Process
3.3. VLP Characterization
3.3.1. Biochemical Characterization
3.3.2. Biophysical Characterization
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- WHO. Media Centre Dengue—Global Situation. Available online: https://www.who.int/emergencies/disease-outbreak-news/item/2024-DON518 (accessed on 11 October 2024).
- Bayani, F.; Hashkavaei, N.S.; Arjmand, S.; Rezaei, S.; Uskoković, V.; Alijanianzadeh, M.; Uversky, V.N.; Ranaei Siadat, S.O.; Mozaffari-Jovin, S.; Sefidbakht, Y. An Overview of the Vaccine Platforms to Combat COVID-19 with a Focus on the Subunit Vaccines. Prog. Biophys. Mol. Biol. 2023, 178, 32–49. [Google Scholar] [CrossRef]
- Friedler, A. Sociocultural, Behavioural and Political Factors Shaping the COVID-19 Pandemic: The Need for a Biocultural Approach to Understanding Pandemics and (Re)Emerging Pathogens. Glob. Public Health 2021, 16, 17–35. [Google Scholar] [CrossRef]
- Gully, P.R. Pandemics, Regional Outbreaks, and Sudden-Onset Disasters. Healthc. Manag. Forum 2020, 33, 164–169. [Google Scholar] [CrossRef] [PubMed]
- Pollard, A.J.; Bijker, E.M. A Guide to Vaccinology: From Basic Principles to New Developments. Nat. Rev. Immunol. 2021, 21, 83–100. [Google Scholar] [CrossRef]
- Kozak, M.; Hu, J. The Integrated Consideration of Vaccine Platforms, Adjuvants, and Delivery Routes for Successful Vaccine Development. Vaccines 2023, 11, 695. [Google Scholar] [CrossRef]
- Henríquez, R.; Muñoz-Barroso, I. Viral Vector- and Virus-like Particle-Based Vaccines against Infectious Diseases: A Minireview. Heliyon 2024, 10, e34927. [Google Scholar] [CrossRef]
- Mohsen, M.O.; Bachmann, M.F. Virus-like Particle Vaccinology, from Bench to Bedside. Cell. Mol. Immunol. 2022, 19, 993–1011. [Google Scholar] [CrossRef] [PubMed]
- Schiller, J.T.; Lowy, D.R. Understanding and Learning from the Success of Prophylactic Human Papillomavirus Vaccines. Nat. Rev. Microbiol. 2012, 10, 681–692. [Google Scholar] [CrossRef] [PubMed]
- Narayan, R.; Tripathi, S. Intrinsic ADE: The Dark Side of Antibody Dependent Enhancement During Dengue Infection. Front. Cell. Infect. Microbiol. 2020, 10, 580096. [Google Scholar] [CrossRef]
- Torres-Flores, J.M.; Reyes-Sandoval, A.; Salazar, M.I. Dengue Vaccines: An Update. BioDrugs 2022, 36, 325–336. [Google Scholar] [CrossRef]
- Wilder-Smith, A.; Hombach, J.; Ferguson, N.; Selgelid, M.; O’Brien, K.; Vannice, K.; Barrett, A.; Ferdinand, E.; Flasche, S.; Guzman, M.; et al. Deliberations of the Strategic Advisory Group of Experts on Immunization on the Use of CYD-TDV Dengue Vaccine. Lancet Infect. Dis. 2019, 19, e31–e38. [Google Scholar] [CrossRef]
- Silva, J.P.; Fernandez-Sesma, A. Challenges on the Development of a Dengue Vaccine: A Comprehensive Review of the State of the Art. J. Gen. Virol. 2023, 104, 001831. [Google Scholar] [CrossRef]
- WHO. Vaccines and Immunization: Dengue. Available online: https://www.who.int/news-room/questions-and-answers/item/dengue-vaccines (accessed on 10 June 2024).
- Balingit, J.C.; Abe, M.; Suzuki, R.; Xayavong, D.; Ngwe Tun, M.M.; Takamatsu, Y.; Sonoda, K.; Morita, K. Cross-Genotype Immunogenicity and Antibody-Dependent Enhancement of KD-382 Dengue Vaccine in Flavivirus-Naïve Adults. npj Vaccines 2025, 10, 148. [Google Scholar] [CrossRef]
- Srivastava, V.; Nand, K.N.; Ahmad, A.; Kumar, R. Yeast-Based Virus-like Particles as an Emerging Platform for Vaccine Development and Delivery. Vaccines 2023, 11, 479. [Google Scholar] [CrossRef] [PubMed]
- Patra, P.; Das, M.; Kundu, P.; Ghosh, A. Recent Advances in Systems and Synthetic Biology Approaches for Developing Novel Cell-Factories in Non-Conventional Yeasts. Biotechnol. Adv. 2021, 47, 107695. [Google Scholar] [CrossRef] [PubMed]
- Fassola, L.A.; Rupil, L.L.; Martínez, F.; Albrieu-Llinás, G.; Serradell, M.d.C. Induction of Systemic and Mucosal Immune Response against Zika Virus by Vaccination with Non-Infectious Chimeric VLPs. Sci. Rep. 2025, 15, 24834. [Google Scholar] [CrossRef]
- Nooraei, S.; Bahrulolum, H.; Hoseini, Z.S.; Katalani, C.; Hajizade, A.; Easton, A.J.; Ahmadian, G. Virus-like Particles: Preparation, Immunogenicity and Their Roles as Nanovaccines and Drug Nanocarriers. J. Nanobiotechnol. 2021, 19, 59. [Google Scholar] [CrossRef] [PubMed]
- Chavda, V.P.; Ghali, E.N.H.K.; Balar, P.C.; Chauhan, S.C.; Tiwari, N.; Shukla, S.; Athalye, M.; Patravale, V.; Apostolopoulos, V.; Yallapu, M.M. Protein Subunit Vaccines: Promising Frontiers against COVID-19. J. Control. Release 2024, 366, 761–782. [Google Scholar] [CrossRef]
- de Sá Magalhães, S.; Keshavarz-Moore, E.P. Pastoris (Komagataella phaffii) as a Cost-effective Tool for Vaccine Production for Low- and Middle-income Countries (Lmics). Bioengineering 2021, 8, 119. [Google Scholar] [CrossRef]
- Karbalaei, M.; Rezaee, S.A.; Farsiani, H. Pichia pastoris: A Highly Successful Expression System for Optimal Synthesis of Heterologous Proteins. J. Cell. Physiol. 2020, 235, 5867–5881. [Google Scholar] [CrossRef]
- Tir, N.; Heistinger, L.; Grünwald-Gruber, C.; Jakob, L.A.; Dickgiesser, S.; Rasche, N.; Mattanovich, D. From Strain Engineering to Process Development: Monoclonal Antibody Production with an Unnatural Amino Acid in Pichia pastoris. Microb. Cell Fact. 2022, 21, 157. [Google Scholar] [CrossRef]
- De Sa Magalhaes, S.; Morris, S.; Kusumawardani, S.; Wijayadikusumah Riza, A.; Nurainy, N.; Keshavarz-Moore, E. Technical Transfer of a Rapid Microbial Platform for Vaccine Production. Vaccine Insights 2023, 02, 133–146. [Google Scholar] [CrossRef]
- Invitrogen Life Technologies. Invitrogen Corporation Pichia Fermentation Process Guidelines Overview Overview, Continued. Prog. Bot. 2002, 67, 1–11. [Google Scholar]
- De Sá Magalhães, S.; De Santis, E.; Hussein-Gore, S.; Colomb-Delsuc, M.; Keshavarz-Moore, E. Quality Assessment of Virus-like Particle: A New Transmission Electron Microscopy Approach. Front. Mol. Biosci. 2022, 9, 975054. [Google Scholar] [CrossRef]
- Capone, S.; Horvat, J.; Herwig, C.; Spadiut, O. Development of a Mixed Feed Strategy for a Recombinant Pichia pastoris Strain Producing with a De-Repression Promoter. Microb. Cell Fact. 2015, 14, 101. [Google Scholar] [CrossRef] [PubMed]
- Prielhofer, R.; Maurer, M.; Klein, J.; Wenger, J.; Kiziak, C.; Gasser, B.; Mattanovich, D. Induction without Methanol: Novel Regulated Promoters Enable High-Level Expression in Pichia pastoris. Microb. Cell Fact. 2013, 12, 5. [Google Scholar] [CrossRef] [PubMed]
- Niu, H.; Jost, L.; Pirlot, N.; Sassi, H.; Daukandt, M.; Rodriguez, C.; Fickers, P. A Quantitative Study of Methanol/Sorbitol Co-Feeding Process of a Pichia pastoris Mut+/PAOX1-LacZ Strain. Microb. Cell Fact. 2013, 12, 33. [Google Scholar] [CrossRef]
- Jungo, C.; Marison, I.; von Stockar, U. Mixed Feeds of Glycerol and Methanol Can Improve the Performance of Pichia pastoris Cultures: A Quantitative Study Based on Concentration Gradients in Transient Continuous Cultures. J. Biotechnol. 2007, 128, 824–837. [Google Scholar] [CrossRef]
- Bill, R.M. (Ed.) Recombinant Protein Production in Yeast: Methods and Protocols; Humana Press: Totowa, NJ, USA, 2012. [Google Scholar]
- Fang, Z.; Lyu, J.; Li, J.; Li, C.; Zhang, Y.; Guo, Y.; Wang, Y.; Zhang, Y.; Chen, K. Application of Bioreactor Technology for Cell Culture-Based Viral Vaccine Production: Present Status and Future Prospects. Front. Bioeng. Biotechnol. 2022, 10, 921755. [Google Scholar] [CrossRef] [PubMed]
- Bowden, G.D.; Pichler, B.J.; Maurer, A. A Design of Experiments (DoE) Approach Accelerates the Optimization of Copper-Mediated 18F-Fluorination Reactions of Arylstannanes. Sci. Rep. 2019, 9, 11370. [Google Scholar] [CrossRef]
- Peng, X.; Yang, G.; Shi, Y.; Zhou, Y.; Zhang, M.; Li, S. Box–Behnken Design Based Statistical Modeling for the Extraction and Physicochemical Properties of Pectin from Sunflower Heads and the Comparison with Commercial Low-Methoxyl Pectin. Sci. Rep. 2020, 10, 3595. [Google Scholar] [CrossRef] [PubMed]
- Velez-Suberbie, M.L.; Morris, S.A.; Kaur, K.; Hickey, J.M.; Joshi, S.B.; Volkin, D.B.; Bracewell, D.G.; Mukhopadhyay, T.K. Holistic Process Development to Mitigate Proteolysis of a Subunit Rotavirus Vaccine Candidate Produced in Pichia pastoris by Means of an Acid PH Pulse during Fed-Batch Fermentation. Biotechnol. Prog. 2020, 36, e2966. [Google Scholar] [CrossRef]
- Samandoulgou, I.; Hammami, R.; Rayas, R.M.; Fliss, I.; Jean, J. Stability of Secondary and Tertiary Structures of Virus-like Particles Representing Noroviruses: Effects of PH, Ionic Strength, and Temperature and Implications for Adhesion to Surfaces. Appl. Environ. Microbiol. 2015, 81, 7680–7686. [Google Scholar] [CrossRef]
- De Brabander, P.; Uitterhaegen, E.; Delmulle, T.; De Winter, K.; Soetaert, W. Challenges and Progress towards Industrial Recombinant Protein Production in Yeasts: A Review. Biotechnol. Adv. 2023, 64, 108121. [Google Scholar] [CrossRef]
- Negrete, A.; Pai, A.; Shiloach, J. Use of Hollow Fiber Tangential Flow Filtration for the Recovery and Concentration of HIV Virus-like Particles Produced in Insect Cells. J. Virol. Methods 2014, 195, 240–246. [Google Scholar] [CrossRef]
- Pedro, L.; Soares, S.S.; Ferreira, G.N.M. Purification of Bionanoparticles. Chem. Eng. Technol. 2008, 31, 815–825. [Google Scholar] [CrossRef]
- Morenweiser, R. Downstream Processing of Viral Vectors and Vaccines. Gene Ther. 2005, 12, S103–S110. [Google Scholar] [CrossRef]
- Rajpoot, R.K.; Shukla, R.; Arora, U.; Swaminathan, S.; Khanna, N. Dengue Envelope-Based “four-in-One” Virus-like Particles Produced Using Pichia pastoris Induce Enhancement-Lacking, Domain III-Directed Tetravalent Neutralising Antibodies in Mice. Sci. Rep. 2018, 8, 8643. [Google Scholar] [CrossRef] [PubMed]
- Mukhopadhyay, E.; Brod, F.; Angell-Manning, P.; Green, N.; Tarrant, R.D.; Detmers, F.J.; Bolam, E.J.; Baleanu, I.N.; Hobson, M.; Whale, G.; et al. Production of a High Purity, C-Tagged Hepatitis B Surface Antigen Fusion Protein VLP Vaccine for Malaria Expressed in Pichia pastoris under CGMP Conditions. Biotechnol. Bioeng. 2022, 119, 2784–2793. [Google Scholar] [CrossRef]
- Frank Detmers, F.; Hermans, P.; Clasen, R.; de Rooij, J. C-Tag Affinity Tag, from Routine Protein Purification to Use in a CGMP Production Process. Thermosci. Poster 2018, 47, 435–446. [Google Scholar]
- ThermoFisher Affinity Tags Head Toward the Vaccination Clinic. Available online: https://go.technologynetworks.com/affinity-tags-head-toward-the-vaccination-clinic (accessed on 17 June 2025).
- Bornemann, S.; Herzog, M.; Roling, L.; Paulisch, T.O.; Brandis, D.; Kriegler, S.; Galla, H.J.; Glorius, F.; Winter, R. Interaction of Imidazolium-Based Lipids with Phospholipid Bilayer Membranes of Different Complexity. Phys. Chem. Chem. Phys. 2020, 22, 9775–9788. [Google Scholar] [CrossRef]
- Aaron, M.; Andy, P.; Kelley, K.; Tom, S.; Kett, W. Development of an Affinity Chromatography Process for Lentiviral Vectors Lentiviral Vector Affinity Purification Western Blot. 2023. Available online: https://www.repligen.com/Products/Resins/avipure/SP_AVIpure_lenti_affinity_resins_31JAN2024.pdf (accessed on 17 June 2025).
- Azadi, S.; Sadjady, S.; Mortazavi, S.; Naghdi, N.; Mahboubi, A.; Solaimanian, R. Bioprocess and Downstream Optimization of Recombinant Human Growth Hormone in Pichia pastoris. Res. Pharm. Sci. 2018, 13, 222–238. [Google Scholar] [CrossRef]
- González-Domínguez, I.; Puente-Massaguer, E.; Cervera, L.; Gòdia, F. Quality Assessment of Virus-like Particles at Single Particle Level: A Comparative Study. Viruses 2020, 12, 223. [Google Scholar] [CrossRef] [PubMed]
- Rolland, D.; Gauthier, M.; Dugua, J.M.; Fournier, C.; Delpech, L.; Watelet, B.; Letourneur, O.; Arnaud, M.; Jolivet, M. Purification of Recombinant HBc Antigen Expressed in Escherichia coli and Pichia pastoris: Comparison of Size-Exclusion Chromatography and Ultracentrifugation. J. Chromatogr. B Biomed. Sci. Appl. 2001, 753, 51–65. [Google Scholar] [CrossRef] [PubMed]
- Heider, S.; Metzner, C. Quantitative Real-Time Single Particle Analysis of Virions. Virology 2014, 462–463, 199–206. [Google Scholar] [CrossRef] [PubMed]
- Lua, L.H.L.; Connors, N.K.; Sainsbury, F.; Chuan, Y.P.; Wibowo, N.; Middelberg, A.P.J. Bioengineering Virus-like Particles as Vaccines. Biotechnol. Bioeng. 2014, 111, 425–440. [Google Scholar] [CrossRef] [PubMed]
- Chulanetra, M.; Punnakitikashem, P.; Mahasongkram, K.; Chaicumpa, W.; Glab-ampai, K. Immunogenicity of Intraperitoneal and Intranasal Liposome Adjuvanted VLP Vaccines against SARS-CoV-2 Infection. Sci. Rep. 2024, 14, 27311. [Google Scholar] [CrossRef]
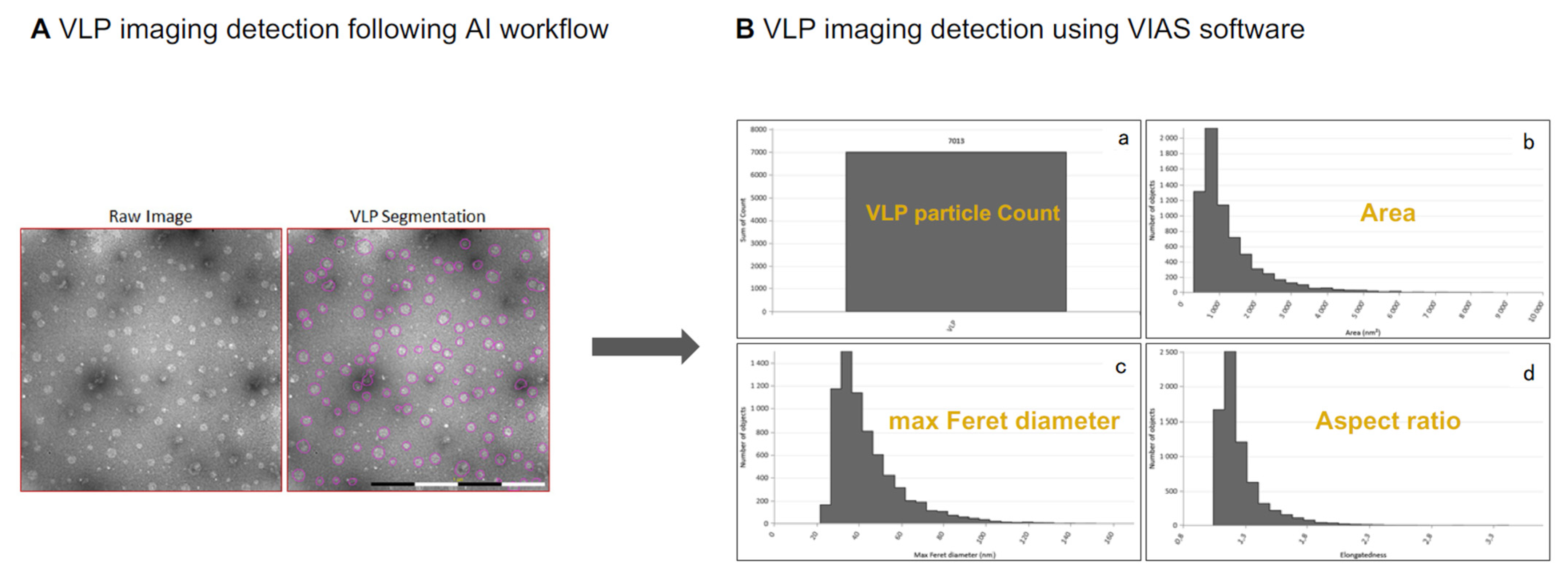
- Ryner, M.; Forsberg, N.; Carvalho, V.; Peixoto, C.; Carvalho, S.B. Using TEM-Based Image Analysis to Validate the Presence of HA Spikes on Influenza VLPs. BioPharm Int. 2019, 32, 35–41. [Google Scholar]
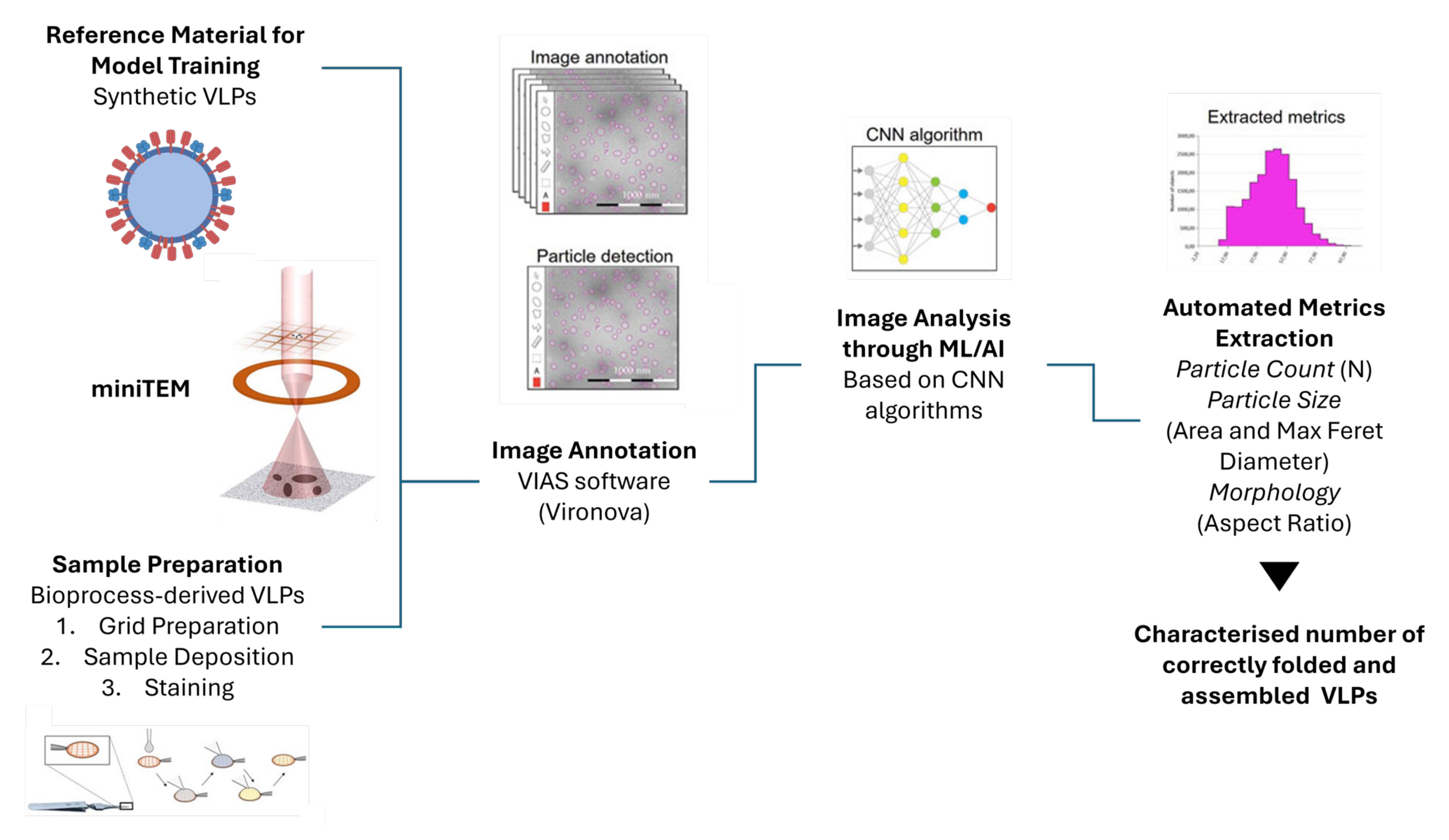
- Colomb-Delsuc, M.; Härmark, J.; Eriksson, B.; Nordström, R.; Carvalho, V.; Kylberg, G.; Sintorn, I.-M. Advancing TEM Based Biomedical Nanoparticle Characterization: GMP Compliant TEM Workflow in a BSL2 Environment and Automation Using MiniTEM. Microsc. Microanal. 2019, 25, 1066–1067. [Google Scholar] [CrossRef]
- Laxmi, B.; Devi, P.U.M.; Thanjavur, N.; Buddolla, V. The Applications of Artificial Intelligence (AI)-Driven Tools in Virus-Like Particles (VLPs) Research. Curr. Microbiol. 2024, 81, 234. [Google Scholar] [CrossRef]
- Västberg, A.; Bolinsson, H.; Leeman, M.; Nilsson, L.; Nylander, T.; Sejwal, K.; Sintorn, I.M.; Lidayová, K.; Sjögren, H.; Wahlgren, M.; et al. Investigating Thermally Induced Aggregation of Somatropin- New Insights Using Orthogonal Techniques. Int. J. Pharm. 2023, 637, 122829. [Google Scholar] [CrossRef] [PubMed]
- Fu, X.; Chen, W.C.; Argento, C.; Clarner, P.; Bhatt, V.; Dickerson, R.; Bou-Assaf, G.; Bakhshayeshi, M.; Lu, X.; Bergelson, S.; et al. Analytical Strategies for Quantification of Adeno-Associated Virus Empty Capsids to Support Process Development. Hum. Gene Ther. Methods 2019, 30, 144–152. [Google Scholar] [CrossRef]
- Aynekulu Mersha, D.G.; van der Sterren, I.; van Leeuwen, L.P.M.; Langerak, T.; Hakim, M.S.; Martina, B.; van Lelyveld, S.F.L.; van Gorp, E.C.M. The Role of Antibody-Dependent Enhancement in Dengue Vaccination. Trop. Dis. Travel Med. Vaccines 2024, 10, 22. [Google Scholar] [CrossRef]
- Bachmann, M.F.; van Damme, P.; Lienert, F.; Schwarz, T.F. Virus-like Particles: A Versatile and Effective Vaccine Platform. Expert Rev. Vaccines 2025, 24, 444–456. [Google Scholar] [CrossRef]
- Parra-González, M.; Nájera-Maldonado, L.; Peralta-Cuevas, E.; Gutierrez-Onofre, A.J.; Garcia-Atutxa, I.; Villanueva-Flores, F. Overcoming Dengue Vaccine Challenges through Next-Generation Virus-like Particle Immunization Strategies. Front. Cell. Infect. Microbiol. 2025, 15, 1614805. [Google Scholar] [CrossRef] [PubMed]
- Thoresen, D.; Matsuda, K.; Urakami, A.; Ngwe Tun, M.M.; Nomura, T.; Moi, M.L.; Watanabe, Y.; Ishikawa, M.; Hau, T.T.T.; Yamamoto, H.; et al. A Tetravalent Dengue Virus-like Particle Vaccine Induces High Levels of Neutralizing Antibodies and Reduces Dengue Replication in Non-Human Primates. J. Virol. 2024, 98, e00239-24. [Google Scholar] [CrossRef] [PubMed]
- Nagasaki University; VLP Therapeutics. Phase I Clinical Trial of Novel Dengue Virus-like Particle (VLP) Vaccines. Available online: https://www.ghitfund.org/investment/portfoliodetail/detail/237/en (accessed on 30 July 2025).
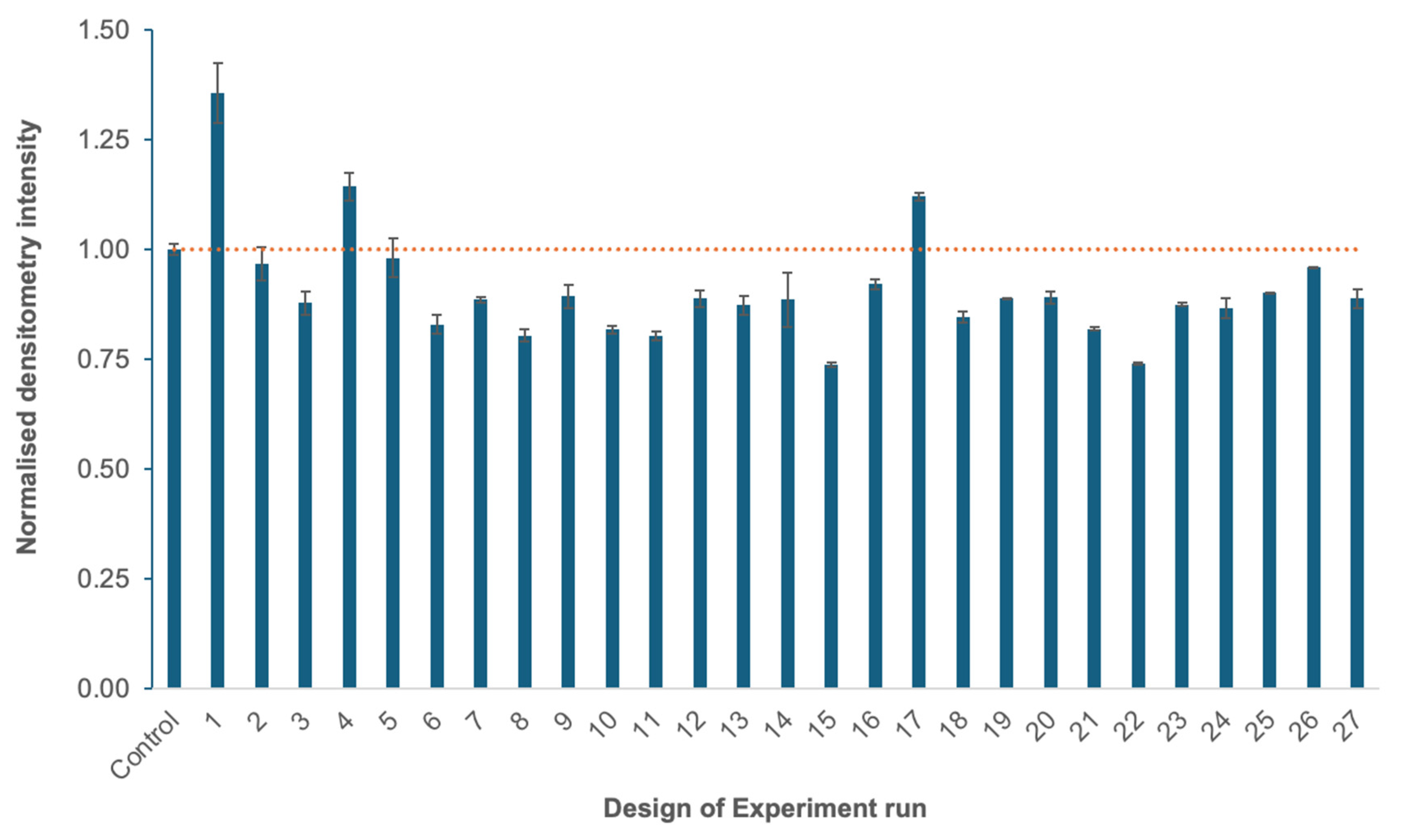
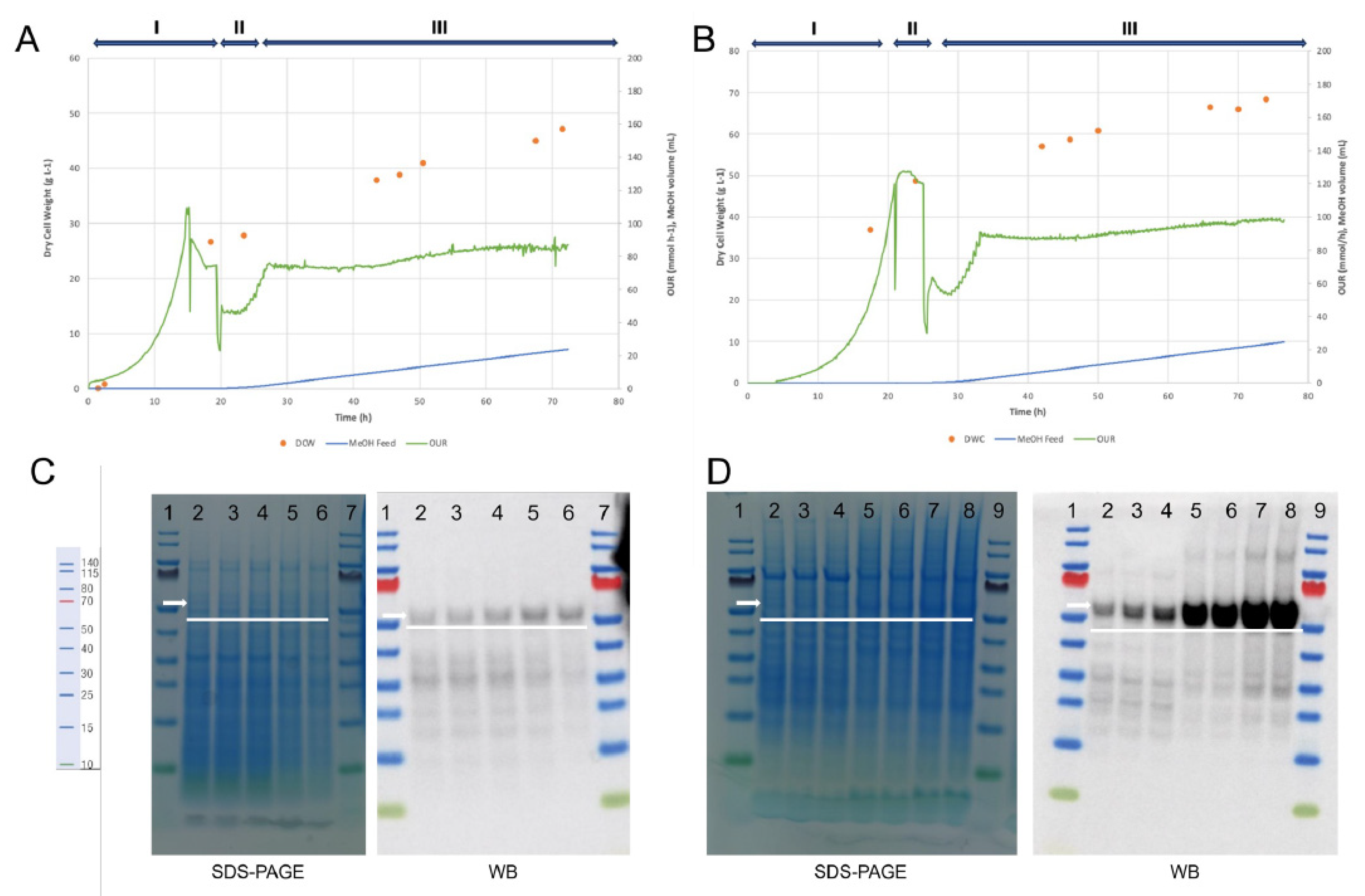
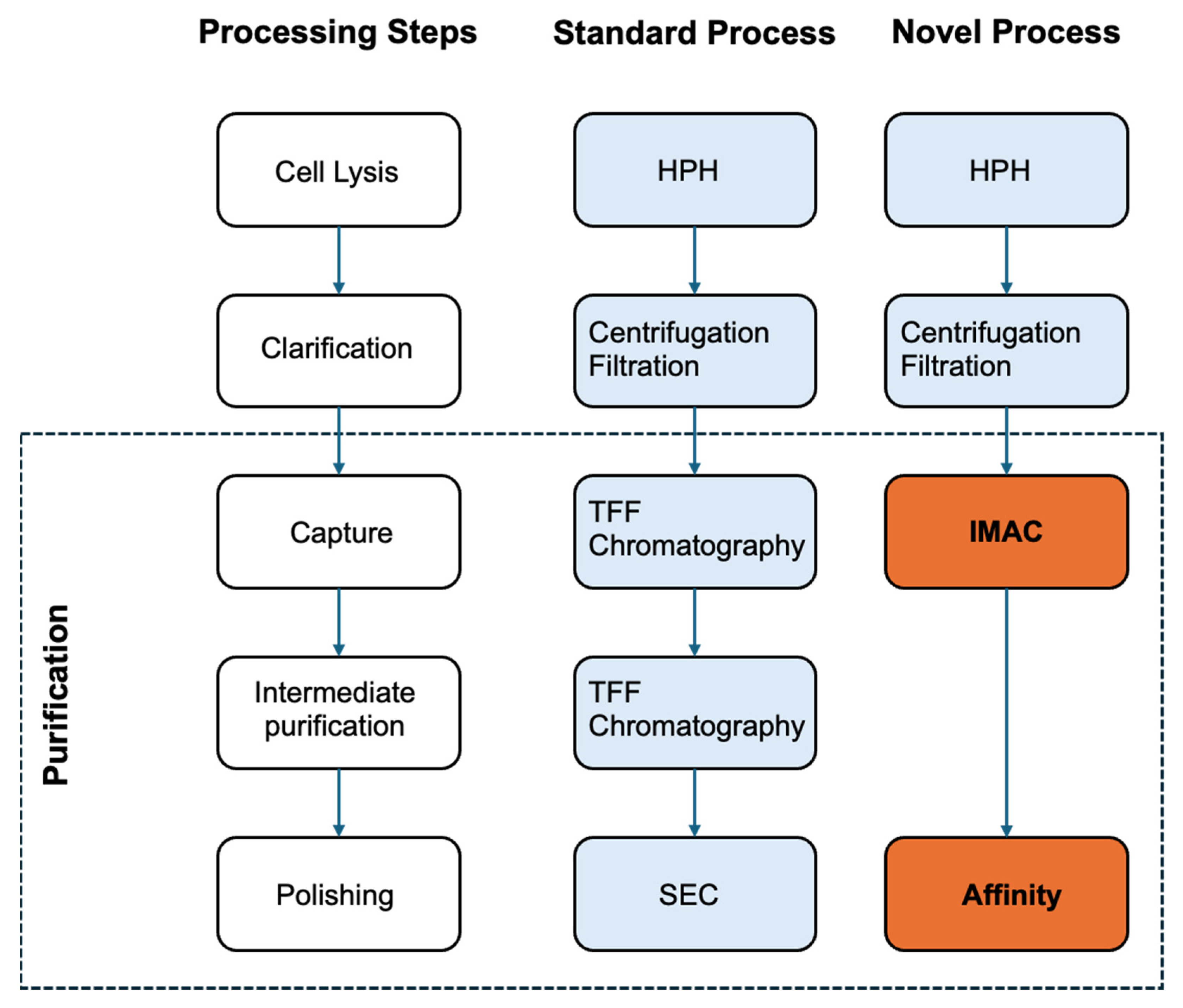

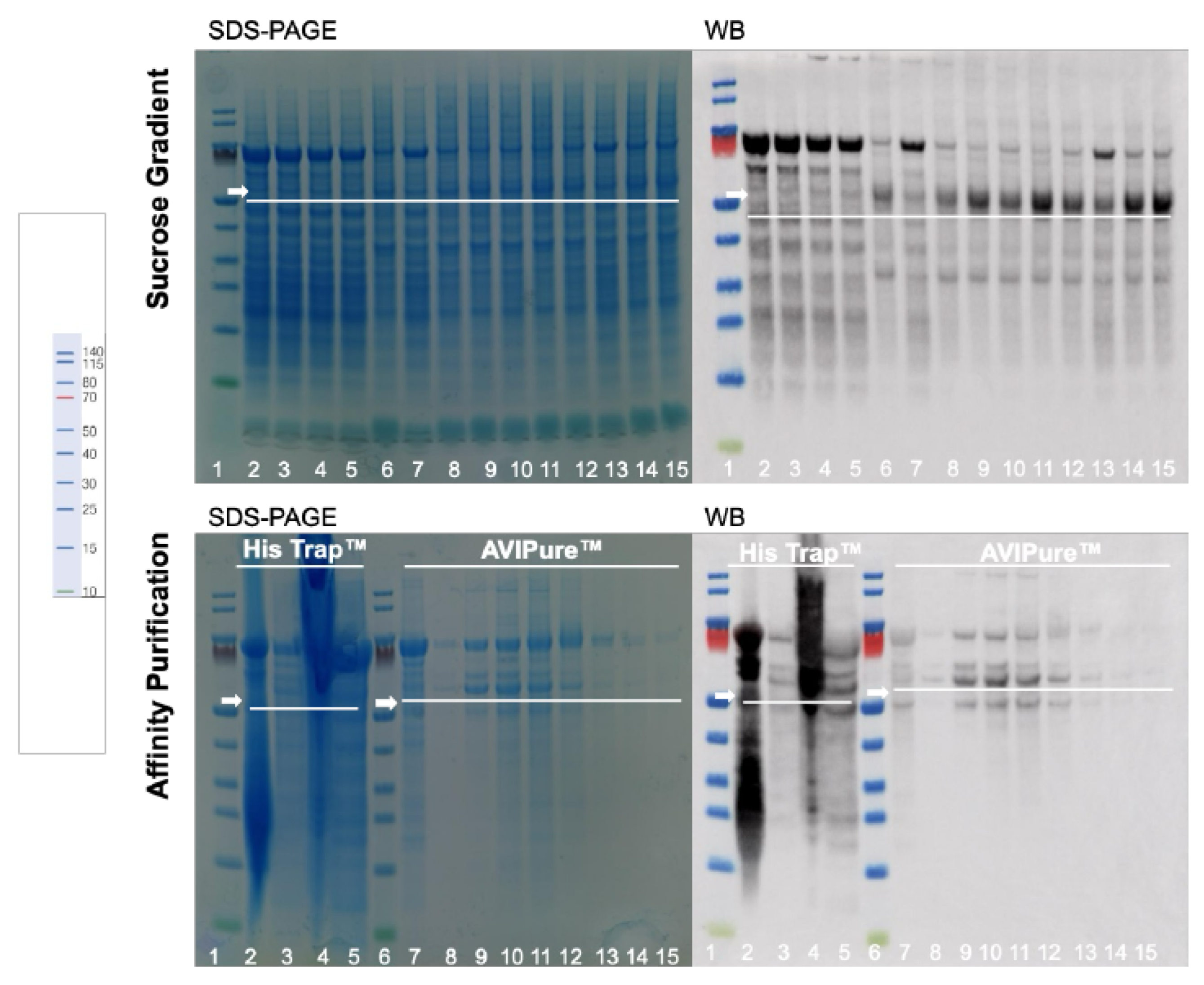
| Feature | Yeast (Komagataella phaffii) | Insect Cells (e.g., Sf9, High Five) |
|---|---|---|
| Growth rate | Fast | Moderate |
| Cost of cultivation | Low | Higher |
| Scalability | High | Moderate |
| PTM fidelity | Limited (e.g., hypermannosylation) | Better than yeast, but not fully human-like |
| Glycosylation | High mannose-type, lacks sialylation | Complex-type, lacks terminal sialic acids |
| Protein folding | May be suboptimal for complex proteins | Better suited for complex protein folding |
| Immunogenicity of VLPs | Often strong, but PTMs may affect epitopes | Strong, with a more native-like antigen structure |
| Regulatory acceptance | Established (e.g., HBV, HPV vaccines) | Increasing, used in licensed vaccines (e.g., FluBlok) |
| Production yield | High | Moderate |
| Use in licensed vaccines | Yes (e.g., HBV, HPV) | Yes (e.g., FluBlok for influenza) |
| Variables | Levels | ||
|---|---|---|---|
| −1 (min.) | 0 | 1 (max.) | |
| Temperature (°C) | 26 | 28 | 30 |
| pH (-) | 3.5 | 5.0 | 6.5 |
| Dissolved oxygen (%) | 20 | 30 | 40 |
| Methanol feed (mL/h/L) | 2.5 | 3 | 3.5 |
| Chromatography Method | Removal Yield (%) | |
|---|---|---|
| Total Protein | Total DNA | |
| Histrap™ | 96 ± 0.1 | 97 ± 3.0 |
| Avipure™ | 94 ± 0.03 | 93 ± 1.0 |
| Chromatography Method | Proportion % | |
|---|---|---|
| Ratio DENV1 | Ratio DENV2 | |
| Histrap™ | 57 ± 0.02 | 43 ± 0.02 |
| Avipure™ | 62 ± 0.04 | 38 ± 0.04 |
| DENV1 (ng/mL) | DENV2 (ng/mL) | HCP (μg/mL) | Total DNA (ng/mL) |
|---|---|---|---|
| 118 ± 3.0 | 70 ± 2.0 | 2.7 ± 0.4 | 81 ± 3.0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
de Sá Magalhães, S.; Morris, S.A.; Kusumawardani, S.; Wijayadikusumah, A.R.; Nurainy, N.; Keshavarz-Moore, E. Defining a Simplified Process in Yeast for Production of Enveloped VLP Dengue Vaccine. Bioengineering 2025, 12, 956. https://doi.org/10.3390/bioengineering12090956
de Sá Magalhães S, Morris SA, Kusumawardani S, Wijayadikusumah AR, Nurainy N, Keshavarz-Moore E. Defining a Simplified Process in Yeast for Production of Enveloped VLP Dengue Vaccine. Bioengineering. 2025; 12(9):956. https://doi.org/10.3390/bioengineering12090956
Chicago/Turabian Stylede Sá Magalhães, Salomé, Stephen A. Morris, Shinta Kusumawardani, Acep Riza Wijayadikusumah, Neni Nurainy, and Eli Keshavarz-Moore. 2025. "Defining a Simplified Process in Yeast for Production of Enveloped VLP Dengue Vaccine" Bioengineering 12, no. 9: 956. https://doi.org/10.3390/bioengineering12090956
APA Stylede Sá Magalhães, S., Morris, S. A., Kusumawardani, S., Wijayadikusumah, A. R., Nurainy, N., & Keshavarz-Moore, E. (2025). Defining a Simplified Process in Yeast for Production of Enveloped VLP Dengue Vaccine. Bioengineering, 12(9), 956. https://doi.org/10.3390/bioengineering12090956







