Modulation of Gut Microbiota and Antibiotic Resistance Genes by Heat-Killed Enterococcus faecalis EF-2001 in High-Fat Diet-Induced Obesity Mice: A Shotgun Metagenomics Study
Abstract
1. Introduction
2. Materials and Methods
2.1. Animal Experimental Design
2.2. Body Weight, Blood Chemistry, and Organ Weight Measurements
2.3. Serum Assay for Biochemical Parameters
2.4. Fecal DNA Extraction and Quality Control
2.5. Shotgun Metagenomics Sequencing
2.6. Gut Microbiota Analysis
2.7. Identification of ARGs and MGEs
2.8. Statistical Analysis
3. Results and Discussion
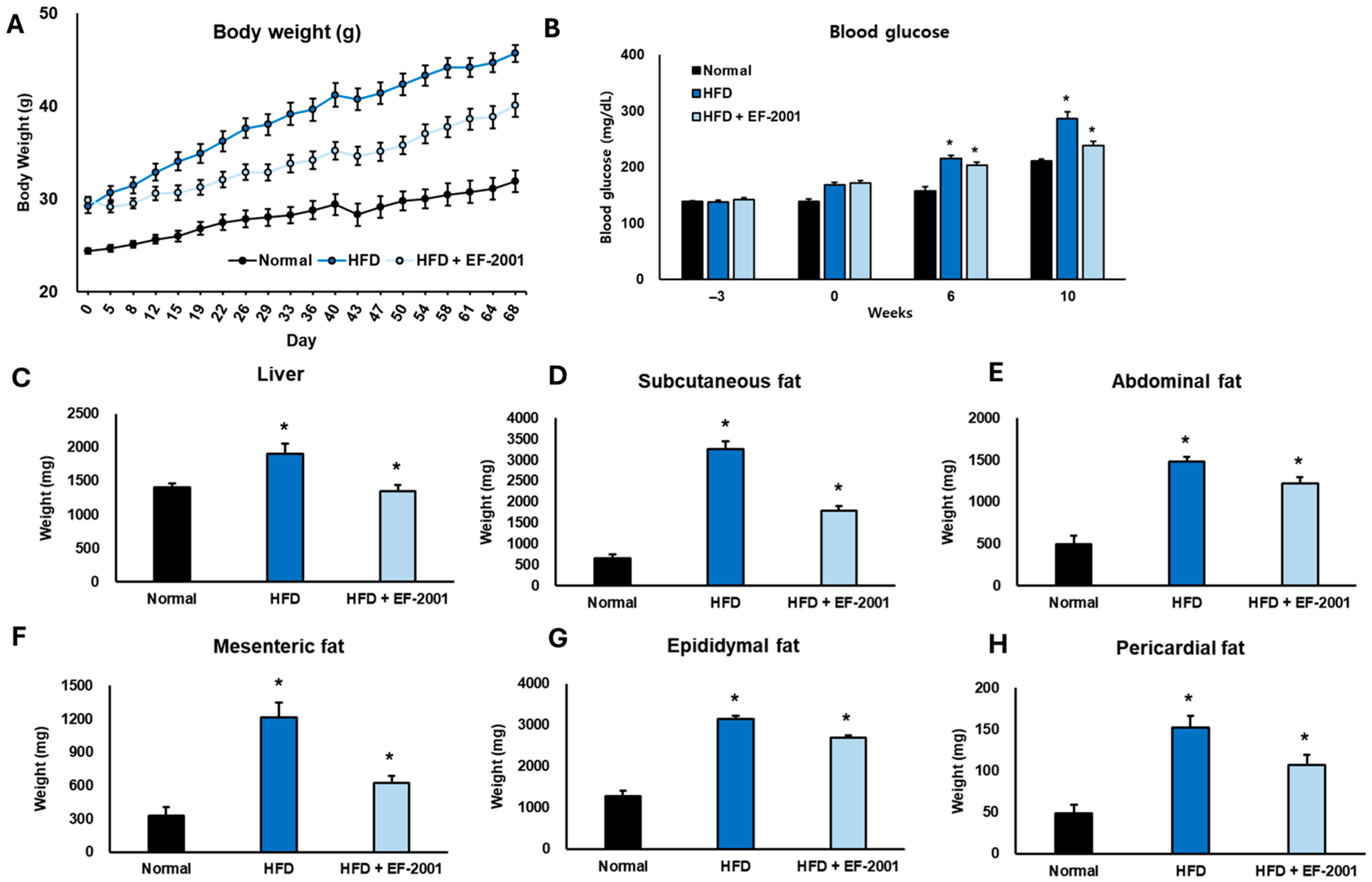
3.1. Effect of EF-2001 on Body Weight in HFD-Induced Obese Mice
3.2. Effect of EF-2001 on Blood Glucose in HFD-Induced Obese Mice
3.3. Effect of EF-2001 on Organ Weight in HFD-Induced Obese Mice
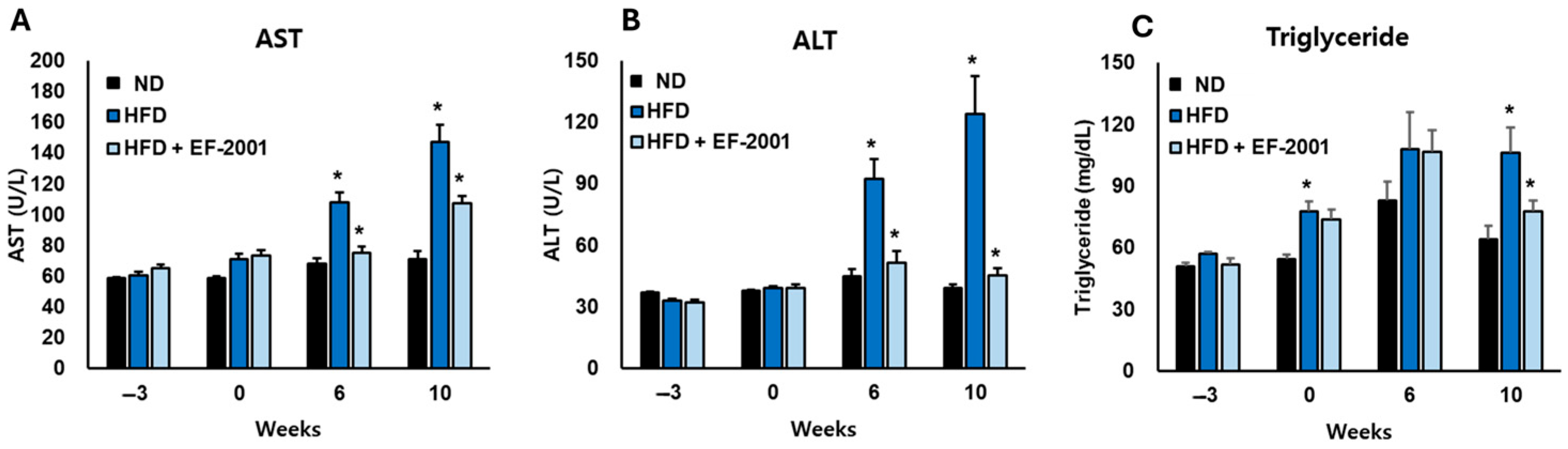
3.4. Effect of EF-2001 on Liver Function Markers in HFD-Induced Obese Mice
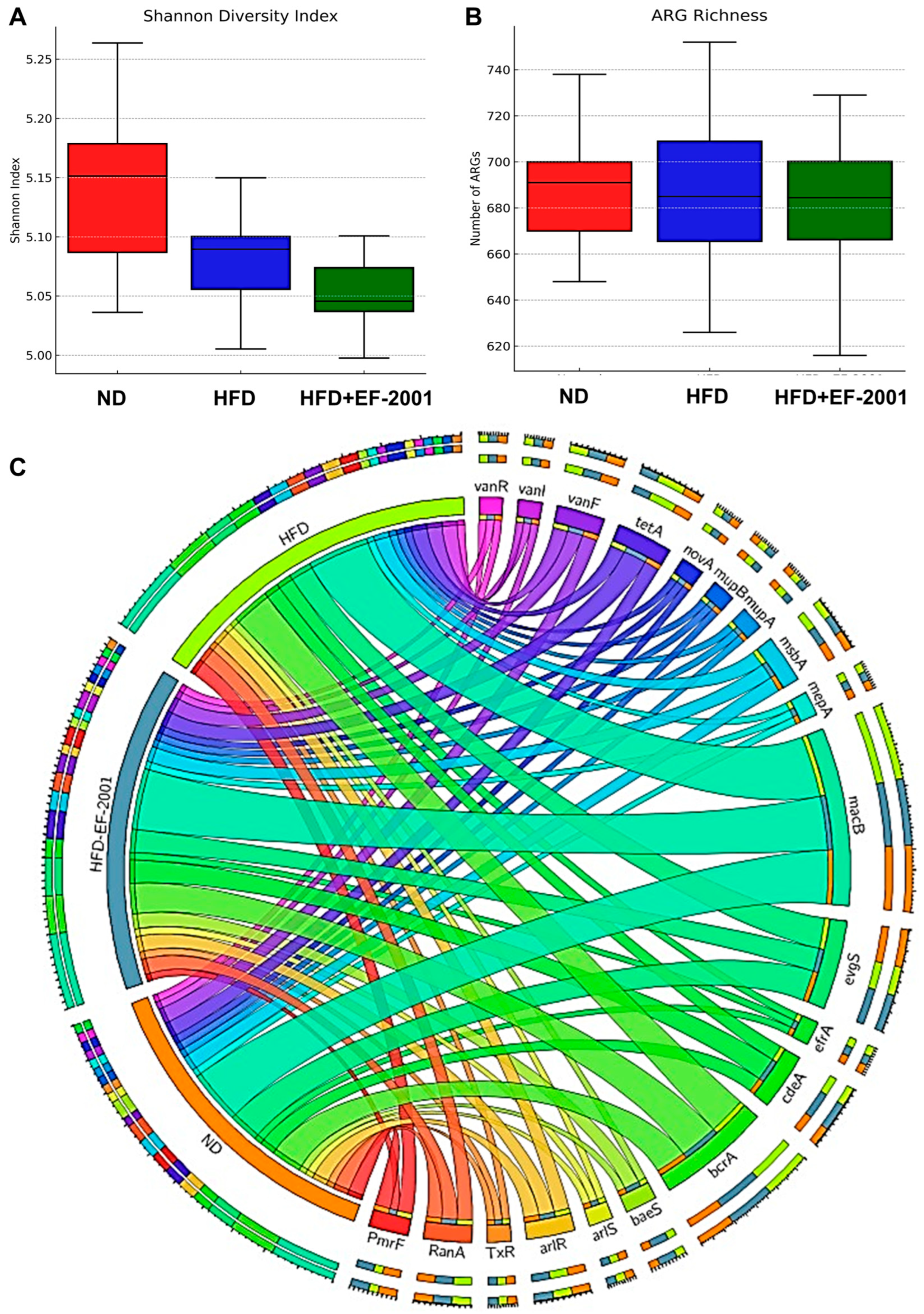
3.5. Metagenome Diversity Analysis
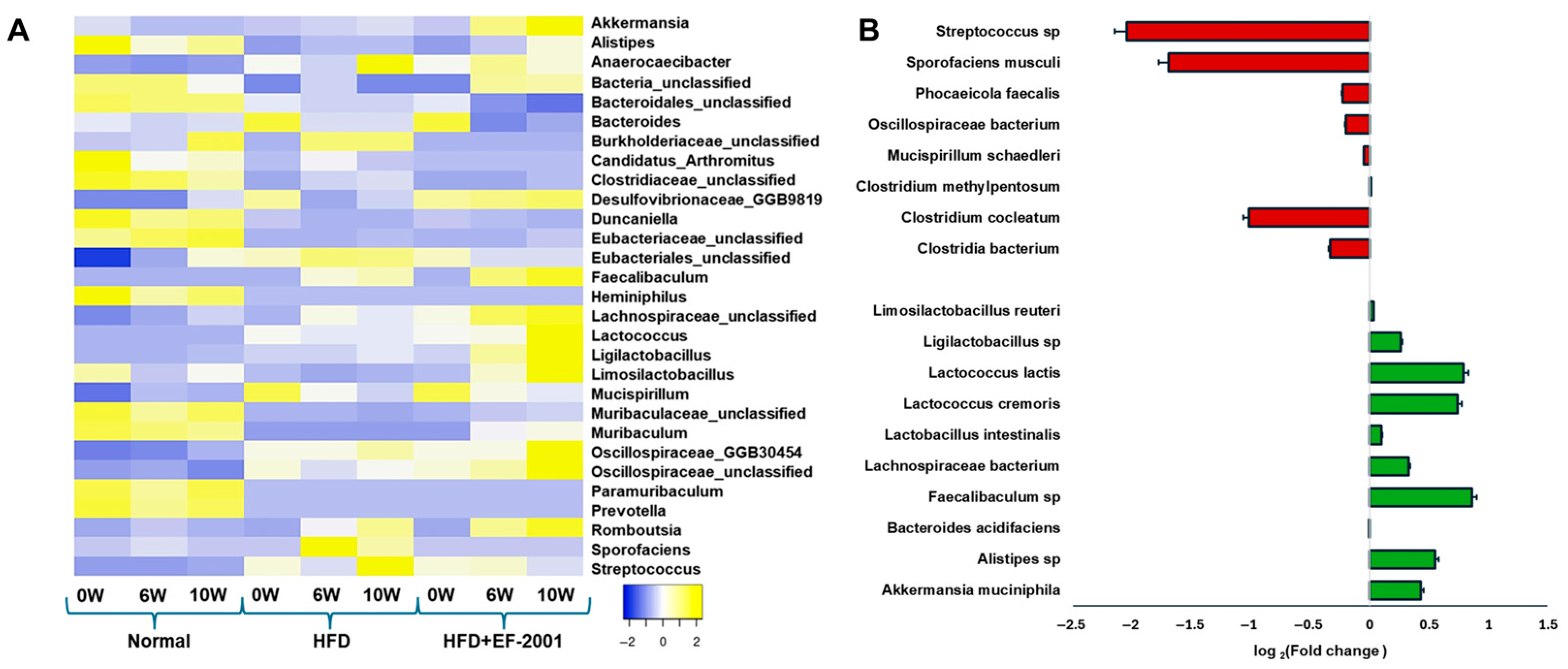
3.6. Gut Microbiota Variability
3.7. Gut Microbiota at the Phylum Level
3.8. Genus Level Analysis
3.9. Species-Level Analysis: HFD vs. HFD + EF-2001
3.10. ARG Abundance
4. Conclusions and Future Perspectives
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ryan, D.; Barquera, S.; Barata Cavalcanti, O.; Ralston, J. The Global Pandemic of Overweight and Obesity. In Handbook of Global Health; Kickbusch, I., Ganten, D., Moeti, M., Eds.; Springer International Publishing: Cham, Switzerland, 2021; pp. 739–773. [Google Scholar]
- Rosenbaum, M.; Knight, R.; Leibel, R.L. The gut microbiota in human energy homeostasis and obesity. Trends Endocrinol. Metab. 2015, 26, 493–501. [Google Scholar] [CrossRef] [PubMed]
- Enache, R.M.; Profir, M.; Roşu, O.A.; Creţoiu, S.M.; Gaspar, B.S. The Role of Gut Microbiota in the Onset and Progression of Obesity and Associated Comorbidities. Int. J. Mol. Sci. 2024, 25, 12321. [Google Scholar] [CrossRef]
- Sasidharan Pillai, S.; Gagnon, C.A.; Foster, C.; Ashraf, A.P. Exploring the Gut Microbiota: Key Insights Into Its Role in Obesity, Metabolic Syndrome, and Type 2 Diabetes. J. Clin. Endocrinol. Metab. 2024, 109, 2709–2719. [Google Scholar] [CrossRef]
- Stojanov, S.; Berlec, A.; Štrukelj, B. The influence of probiotics on the firmicutes/bacteroidetes ratio in the treatment of obesity and inflammatory bowel disease. Microorganisms 2020, 8, 1715. [Google Scholar] [CrossRef] [PubMed]
- Houtman, T.A.; Eckermann, H.A.; Smidt, H.; de Weerth, C. Gut microbiota and BMI throughout childhood: The role of firmicutes, bacteroidetes, and short-chain fatty acid producers. Sci. Rep. 2022, 12, 3140. [Google Scholar] [CrossRef]
- Kang, G.G.; Trevaskis, N.L.; Murphy, A.J.; Febbraio, M.A. Diet-induced gut dysbiosis and inflammation: Key drivers of obesity-driven NASH. iScience 2023, 26, 105905. [Google Scholar] [CrossRef]
- Song, R.; Hu, M.; Qin, X.; Qiu, L.; Wang, P.; Zhang, X.; Liu, R.; Wang, X. The roles of lipid metabolism in the pathogenesis of chronic diseases in the elderly. Nutrients 2023, 15, 3433. [Google Scholar] [CrossRef]
- Oudat, Q.; Okour, A. The Role of Probiotics in Modulating Gut Microbiota and Metabolic Health for Weight Management: A Mini Review. Acta Microbiol. Hell. 2025, 70, 5. [Google Scholar] [CrossRef]
- Song, X.; Liu, Y.; Zhang, X.; Weng, P.; Zhang, R.; Wu, Z. Role of intestinal probiotics in the modulation of lipid metabolism: Implications for therapeutic treatments. Food Sci. Hum. Wellness 2023, 12, 1439–1449. [Google Scholar] [CrossRef]
- Kocot, A.M.; Jarocka-Cyrta, E.; Drabińska, N. Overview of the importance of biotics in gut barrier integrity. Int. J. Mol. Sci. 2022, 23, 2896. [Google Scholar] [CrossRef]
- Song, E.-J.; Lee, E.-S.; Kim, Y.I.; Shin, D.-U.; Eom, J.-E.; Shin, H.S.; Lee, S.-Y.; Nam, Y.-D. Gut microbial change after administration of Lacticaseibacillus paracasei AO356 is associated with anti-obesity in a mouse model. Front. Endocrinol. 2023, 14, 1224636. [Google Scholar] [CrossRef] [PubMed]
- Guan, Y.; Zhu, R.; Zhao, W.; Wang, L.; You, L.; Zeng, Z.; Jiang, Q.; Zhu, Z.; Gou, J.; Zhang, Q.; et al. Effects of Lacticaseibacillus paracasei K56 on perceived stress among pregraduate students: A double-blind, randomized, placebo-controlled trial. Front. Nutr. 2025, 12, 1544713. [Google Scholar] [CrossRef]
- Cai, H.; Wang, Q.; Han, X.; Zhang, H.; Wang, N.; Huang, Y.; Yang, P.; Zhang, R.; Meng, K. In Vitro Evaluation of Probiotic Activities and Anti-Obesity Effects of Enterococcus faecalis EF-1 in Mice Fed a High-Fat Diet. Foods 2024, 13, 4095. [Google Scholar] [CrossRef]
- Sun, S.; Zhao, Y.; Pang, Z.; Wan, B.; Wang, J.; Wu, Z.; Wang, Q. Effects of Enterococcus faecalis Supplementation on Growth Performance, Hepatic Lipid Metabolism, and mRNA Expression of Lipid Metabolism Genes and Intestinal Flora in Geese. Animals 2025, 15, 268. [Google Scholar] [CrossRef] [PubMed]
- Liang, D.; Wu, F.; Zhou, D.; Tan, B.; Chen, T. Commercial probiotic products in public health: Current status and potential limitations. Crit. Rev. Food Sci. Nutr. 2024, 64, 6455–6476. [Google Scholar] [CrossRef] [PubMed]
- Saravanan, C.; Gopinath, N.K.; Ganesan, R.; Thirumurugan, D. Challenges and limitations in using bacterial metabolites as immunomodulators. Front. Cell. Infect. Microbiol. 2025, 15, 1535394. [Google Scholar] [CrossRef]
- Bourebaba, Y.; Marycz, K.; Mularczyk, M.; Bourebaba, L. Postbiotics as potential new therapeutic agents for metabolic disorders management. Biomed. Pharmacother. 2022, 153, 113138. [Google Scholar] [CrossRef]
- Mehta, J.P.; Ayakar, S.; Singhal, R.S. The potential of paraprobiotics and postbiotics to modulate the immune system: A Review. Microbiol. Res. 2023, 275, 127449. [Google Scholar] [CrossRef]
- Siciliano, R.A.; Reale, A.; Mazzeo, M.F.; Morandi, S.; Silvetti, T.; Brasca, M. Paraprobiotics: A new perspective for functional foods and nutraceuticals. Nutrients 2021, 13, 1225. [Google Scholar] [CrossRef]
- Lim, J.J.; Jung, A.H.; Joo Suh, H.; Choi, H.S.; Kim, H. Lactiplantibacillus plantarum K8-based paraprobiotics prevents obesity and obesity-induced inflammatory responses in high fat diet-fed mice. Food Res. Int. 2022, 155, 111066. [Google Scholar] [CrossRef]
- Park, M.; Joung, M.; Park, J.-H.; Ha, S.K.; Park, H.-Y. Role of postbiotics in diet-induced metabolic disorders. Nutrients 2022, 14, 3701. [Google Scholar] [CrossRef] [PubMed]
- Shin, Y.; Han, S.; Kwon, J.; Ju, S.; Choi, T.G.; Kang, I.; Kim, S.S. Roles of short-chain fatty acids in inflammatory bowel disease. Nutrients 2023, 15, 4466. [Google Scholar] [CrossRef]
- Shin, H.H.; Kim, J.-H.; Jung, Y.-J.; Kwak, M.-S.; Sung, M.-H.; Imm, J.-Y. Postbiotic potential of Bacillus velezensis KMU01 cell-free supernatant for the alleviation of obesity in mice. Heliyon 2024, 10, e25263. [Google Scholar] [CrossRef]
- İncili, G.K.; Akgöl, M.; Karatepe, P.; Tekin, A.; Kanmaz, H.; Kaya, B.; Hayaloğlu, A.A. Whole-cell postbiotics: An innovative approach for extending the shelf life and controlling major foodborne pathogens in chicken breast fillets. Food Bioprocess. Technol. 2023, 16, 1502–1524. [Google Scholar] [CrossRef]
- Park, S.-J.; Sharma, A.; Lee, H.-J. Postbiotics against obesity: Perception and overview based on pre-clinical and clinical studies. Int. J. Mol. Sci. 2023, 24, 6414. [Google Scholar] [CrossRef]
- Lee, J.-H.; Woo, K.-J.; Hong, J.; Han, K.-I.; Kim, H.S.; Kim, T.-J. Heat-killed enterococcus faecalis inhibit fl83b hepatic lipid accumulation and high fat diet-induced fatty liver damage in rats by activating lipolysis through the regulation the AMPK signaling pathway. Int. J. Mol. Sci. 2023, 24, 4486. [Google Scholar] [CrossRef]
- Fan, M.; Choi, Y.J.; Wedamulla, N.E.; Tang, Y.; Han, K.I.; Hwang, J.Y.; Kim, E.K. Heat-Killed Enterococcus faecalis EF-2001 Attenuate Lipid Accumulation in Diet-Induced Obese (DIO) Mice by Activating AMPK Signaling in Liver. Foods 2022, 11, 575. [Google Scholar] [CrossRef]
- Wu, Y.; Hu, A.; Shu, X.; Huang, W.; Zhang, R.; Xu, Y.; Yang, C. Lactobacillus plantarum postbiotics trigger AMPK-dependent autophagy to suppress Salmonella intracellular infection and NLRP3 inflammasome activation. J. Cell. Physiol. 2023, 238, 1336–1353. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Yu, C.; He, Y.; Zhu, S.; Wang, S.; Xu, Z.; You, S.; Jiao, Y.; Liu, S.-L.; Bao, H. Integrative metagenomic analysis reveals distinct gut microbial signatures related to obesity. BMC Microbiol. 2024, 24, 119. [Google Scholar] [CrossRef]
- Fan, X.; Lu, Y.; Zhao, Y.; Miao, H.; Qi, K.; Wang, R. An Insight into the Exploration of Antibiotic Resistance Genes in Calorie Restricted Diet Fed Mice. Nutrients 2023, 15, 3198. [Google Scholar] [CrossRef]
- Koorakula, R.; Schiavinato, M.; Ghanbari, M.; Wegl, G.; Grabner, N.; Koestelbauer, A.; Klose, V.; Dohm, J.C.; Domig, K.J. Metatranscriptomic Analysis of the Chicken Gut Resistome Response to In-Feed Antibiotics and Natural Feed Additives. Front. Microbiol. 2022, 13, 833790. [Google Scholar] [CrossRef] [PubMed]
- Pereira-Marques, J.; Ferreira, R.M.; Figueiredo, C. A metatranscriptomics strategy for efficient characterization of the microbiome in human tissues with low microbial biomass. Gut Microbes 2024, 16, 2323235. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Woo, K.J.; Kim, M.A.; Hong, J.; Kim, J.; Kim, S.H.; Han, K.I.; Iwasa, M.; Kim, T.J. Heat-Killed Enterococcus faecalis Prevents Adipogenesis and High Fat Diet-Induced Obesity by Inhibition of Lipid Accumulation through Inhibiting C/EBP-α and PPAR-γ in the Insulin Signaling Pathway. Nutrients 2022, 14, 1308. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, J.; Hashimoto, N.; Yin, T.; Sandagdorj, B.; Arakawa, C.; Inoue, T.; Suzuki, S. Heat-killed Lactobacillus brevis KB290 attenuates visceral fat accumulation induced by high-fat diet in mice. J. Appl. Microbiol. 2021, 131, 1998–2009. [Google Scholar] [CrossRef]
- Schellekens, H.; Torres-Fuentes, C.; van de Wouw, M.; Long-Smith, C.M.; Mitchell, A.; Strain, C.; Berding, K.; Bastiaanssen, T.F.S.; Rea, K.; Golubeva, A.V.; et al. Bifidobacterium longum counters the effects of obesity: Partial successful translation from rodent to human. EBioMedicine 2021, 63, 103176. [Google Scholar] [CrossRef]
- Kou, R.; Wang, J.; Li, A.; Wang, Y.; Zhang, B.; Liu, J.; Sun, Y.; Wang, S. Ameliorating Effects of Bifidobacterium longum subsp. infantis FB3-14 against High-Fat-Diet-Induced Obesity and Gut Microbiota Disorder. Nutrients 2023, 15, 4104. [Google Scholar] [CrossRef]
- Zhu, J.; Liu, X.; Liu, N.; Zhao, R.; Wang, S. Lactobacillus plantarum alleviates high-fat diet-induced obesity by altering the structure of mice intestinal microbial communities and serum metabolic profiles. Front. Microbiol. 2024, 15, 1425764. [Google Scholar] [CrossRef]
- Wang, J.; Tang, H.; Zhang, C.; Zhao, Y.; Derrien, M.; Rocher, E.; van-Hylckama Vlieg, J.E.T.; Strissel, K.; Zhao, L.; Obin, M.; et al. Modulation of gut microbiota during probiotic-mediated attenuation of metabolic syndrome in high fat diet-fed mice. ISME J. 2014, 9, 1–15. [Google Scholar] [CrossRef]
- Zhu, K.; Tan, F.; Mu, J.; Yi, R.; Zhou, X.; Zhao, X. Anti-Obesity Effects of Lactobacillus fermentum CQPC05 Isolated from Sichuan Pickle in High-Fat Diet-Induced Obese Mice through PPAR α Signaling Pathway. Microorganisms 2019, 7, 194. [Google Scholar] [CrossRef]
- Sun, S.; Zhang, Q.; Li, D.; Li, H.; Ma, H.; Wu, X.; Li, Y.; Wang, P.; Liu, R.; Feng, H.; et al. Heat-killed Bifidobacterium longum BBMN68 and inulin protect against high-fat diet-induced obesity by modulating gut microbiota. Front. Nutr. 2024, 11, 1406070. [Google Scholar] [CrossRef]
- Choroszy, M.; Litwinowicz, K.; Bednarz, R.; Roleder, T.; Lerman, A.; Toya, T.; Kamiński, K.; Sawicka-Śmiarowska, E.; Niemira, M.; Sobieszczańska, B. Human Gut Microbiota in Coronary Artery Disease: A Systematic Review and Meta-Analysis. Metabolites 2022, 12, 1165. [Google Scholar] [CrossRef]
- Yilmaz, Y. Postbiotics as Antiinflammatory and Immune-Modulating Bioactive Compounds in Metabolic Dysfunction-Associated Steatotic Liver Disease. Mol. Nutr. Food Res. 2024, 68, e2400754. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Zhang, S.; Tian, M.; Chen, J.; Chen, F.; Guan, W. Different Sources of High Fat Diet Induces Marked Changes in Gut Microbiota of Nursery Pigs. Front. Microbiol. 2020, 11, 859. [Google Scholar] [CrossRef]
- Kolodziejczyk, A.A.; Zheng, D.; Elinav, E. Diet–microbiota interactions and personalized nutrition. Nat. Rev. Microbiol. 2019, 17, 742–753. [Google Scholar] [CrossRef]
- Karwowska, Z.; Szczerbiak, P.; Kosciolek, T. Microbiome time series data reveal predictable patterns of change. Microbiol. Spectr. 2024, 12, e04109-23. [Google Scholar] [CrossRef]
- Hills, R.D., Jr.; Pontefract, B.A.; Mishcon, H.R.; Black, C.A.; Sutton, S.C.; Theberge, C.R. Gut Microbiome: Profound Implications for Diet and Disease. Nutrients 2019, 11, 1613. [Google Scholar] [CrossRef]
- Magne, F.; Gotteland, M.; Gauthier, L.; Zazueta, A.; Pesoa, S.; Navarrete, P.; Balamurugan, R. The Firmicutes/Bacteroidetes Ratio: A Relevant Marker of Gut Dysbiosis in Obese Patients? Nutrients 2020, 12, 1474. [Google Scholar] [CrossRef] [PubMed]
- Velazquez, K.T.; Enos, R.T.; Bader, J.E.; Sougiannis, A.T.; Carson, M.S.; Chatzistamou, I.; Carson, J.A.; Nagarkatti, P.S.; Nagarkatti, M.; Murphy, E.A. Prolonged high-fat-diet feeding promotes non-alcoholic fatty liver disease and alters gut microbiota in mice. World J. Hepatol. 2019, 11, 619–637. [Google Scholar] [CrossRef]
- Hamamah, S.; Amin, A.; Al-Kassir, A.L.; Chuang, J.; Covasa, M. Dietary Fat Modulation of Gut Microbiota and Impact on Regulatory Pathways Controlling Food Intake. Nutrients 2023, 15, 3365. [Google Scholar] [CrossRef] [PubMed]
- Hemarajata, P.; Versalovic, J. Effects of probiotics on gut microbiota: Mechanisms of intestinal immunomodulation and neuromodulation. Therap. Adv. Gastroenterol. 2013, 6, 39–51. [Google Scholar] [CrossRef]
- Ozma, M.A.; Abbasi, A.; Akrami, S.; Lahouty, M.; Shahbazi, N.; Ganbarov, K.; Pagliano, P.; Sabahi, S.; Kose, S.; Yousefi, M.; et al. Postbiotics as the key mediators of the gut microbiota-host interactions. Infez. Med. 2022, 30, 180–193. [Google Scholar] [CrossRef] [PubMed]
- Teame, T.; Wang, A.; Xie, M.; Zhang, Z.; Yang, Y.; Ding, Q.; Gao, C.; Olsen, R.E.; Ran, C.; Zhou, Z. Paraprobiotics and Postbiotics of Probiotic Lactobacilli, Their Positive Effects on the Host and Action Mechanisms: A Review. Front. Nutr. 2020, 7, 570344. [Google Scholar] [CrossRef] [PubMed]
- Jo, H.H.; Lee, M.Y.; Ha, S.E.; Yeom, D.H.; Kim, Y.S. Alteration in gut microbiota after colonoscopy: Proposed mechanisms and the role of probiotic interventions. Clin. Endosc. 2025, 58, 25–39. [Google Scholar] [CrossRef]
- Jalanka, J.; Salonen, A.; Salojarvi, J.; Ritari, J.; Immonen, O.; Marciani, L.; Gowland, P.; Hoad, C.; Garsed, K.; Lam, C.; et al. Effects of bowel cleansing on the intestinal microbiota. Gut 2015, 64, 1562–1568. [Google Scholar] [CrossRef] [PubMed]
- Song, W.; Sheng, Q.; Bai, Y.; Li, L.; Ning, X.; Liu, Y.; Song, C.; Wang, T.; Dong, X.; Luo, Y.; et al. Obesity, but not high-fat diet, is associated with bone loss that is reversed via CD4+CD25+Foxp3+ Tregs-mediated gut microbiome of non-obese mice. Npj Sci. Food 2023, 7, 14. [Google Scholar] [CrossRef]
- He, C.; Cheng, D.; Peng, C.; Li, Y.; Zhu, Y.; Lu, N. High-fat diet induces dysbiosis of gastric microbiota prior to gut microbiota in association with metabolic disorders in mice. Front. Microbiol. 2018, 9, 639. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, J.; Wu, C. Modulation of Gut Microbiota and Immune System by Probiotics, Pre-biotics, and Post-biotics. Front. Nutr. 2021, 8, 634897. [Google Scholar] [CrossRef]
- Parada Venegas, D.; De la Fuente, M.K.; Landskron, G.; Gonzalez, M.J.; Quera, R.; Dijkstra, G.; Harmsen, H.J.M.; Faber, K.N.; Hermoso, M.A. Short Chain Fatty Acids (SCFAs)-Mediated Gut Epithelial and Immune Regulation and Its Relevance for Inflammatory Bowel Diseases. Front. Immunol. 2019, 10, 277. [Google Scholar] [CrossRef]
- Niu, H.; Zhou, M.; Zogona, D.; Xing, Z.; Wu, T.; Chen, R.; Cui, D.; Liang, F.; Xu, X. Akkermansia muciniphila: A potential candidate for ameliorating metabolic diseases. Front. Immunol. 2024, 15, 1370658. [Google Scholar] [CrossRef]
- Park, B.H.; Kim, I.S.; Park, J.K.; Zhi, Z.; Lee, H.M.; Kwon, O.W.; Lee, B.C. Probiotic effect of Lactococcus lactis subsp. cremoris RPG-HL-0136 on intestinal mucosal immunity in mice. Appl. Biol. Chem. 2021, 64, 93. [Google Scholar] [CrossRef]
- Kaur, H.; Kaur, G.; Ali, S.A. Dairy-based probiotic-fermented functional foods: An update on their health-promoting properties. Fermentation 2022, 8, 425. [Google Scholar] [CrossRef]
- Darby, T.M.; Owens, J.A.; Saeedi, B.J.; Luo, L.; Matthews, J.D.; Robinson, B.S.; Naudin, C.R.; Jones, R.M. Lactococcus lactis subsp. cremoris is an efficacious beneficial bacterium that limits tissue injury in the intestine. Iscience 2019, 12, 356–367. [Google Scholar] [CrossRef]
- Zheng, Z.; Park, J.K.; Jiang, L.; Zhu, S.; Kwon, O.W.; Lee, B.C.; Lee, H.M.; Roh, Y.J.; Kang, J.H.; Park, B.H. Beneficial Effects of Fermentation of Red Chili Pepper Using Lactococcus lactis subs. Cremoris RPG-HL-0136 in High-Fat Diet-Induced Obese Mice. J. Med. Food 2023, 26, 81–92. [Google Scholar] [CrossRef] [PubMed]
- Raychaudhuri, S.; Shahinozzaman, M.; Fan, S.; Ogedengbe, O.; Subedi, U.; Obanda, D.N. Resistance to Diet Induced Visceral Fat Accumulation in C57BL/6NTac Mice Is Associated with an Enriched Lactococcus in the Gut Microbiota and the Phenotype of Immune B Cells in Intestine and Adipose Tissue. Microorganisms 2023, 11, 2153. [Google Scholar] [CrossRef] [PubMed]
- Naudin, C.R.; Maner-Smith, K.; Owens, J.A.; Wynn, G.M.; Robinson, B.S.; Matthews, J.D.; Reedy, A.R.; Luo, L.; Wolfarth, A.A.; Darby, T.M. Lactococcus lactis subspecies cremoris elicits protection against metabolic changes induced by a western-style diet. Gastroenterology 2020, 159, 639–651.e5. [Google Scholar] [CrossRef]
- Seo, K.-H.; Lee, H.G.; Eor, J.Y.; Jeon, H.J.; Yokoyama, W.; Kim, H. Effects of kefir lactic acid bacteria-derived postbiotic components on high fat diet-induced gut microbiota and obesity. Food Res. Int. 2022, 157, 111445. [Google Scholar] [CrossRef] [PubMed]
- Arellano-Garcia, L.I.; Portillo, M.P.; Martinez, J.A.; Courtois, A.; Milton-Laskibar, I. Postbiotics for the management of obesity, insulin resistance/type 2 diabetes and NAFLD. Beyond microbial viability. Crit. Rev. Food Sci. Nutr. 2024, 64, 1–24. [Google Scholar] [CrossRef]
- Kim, J.H.; Kwak, W.; Nam, Y.; Baek, J.; Lee, Y.; Yoon, S.; Kim, W. Effect of postbiotic Lactiplantibacillus plantarum LRCC5314 supplemented in powdered milk on type 2 diabetes in mice. J. Dairy Sci. 2024, 107, 5301–5315. [Google Scholar] [CrossRef]
- Dai, J.; Jiang, M.; Wang, X.; Lang, T.; Wan, L.; Wang, J. Human-derived bacterial strains mitigate colitis via modulating gut microbiota and repairing intestinal barrier function in mice. BMC Microbiol. 2024, 24, 96. [Google Scholar] [CrossRef]
- Lin, X.; Xu, M.; Lan, R.; Hu, D.; Zhang, S.; Zhang, S.; Lu, Y.; Sun, H.; Yang, J.; Liu, L.; et al. Gut commensal Alistipes shahii improves experimental colitis in mice with reduced intestinal epithelial damage and cytokine secretion. mSystems 2025, 10, e0160724. [Google Scholar] [CrossRef]
- Hakansson, A.; Molin, G. Gut microbiota and inflammation. Nutrients 2011, 3, 637–682. [Google Scholar] [CrossRef] [PubMed]
- Rasmussen, T.S.; Streidl, T.; Hitch, T.C.A.; Wortmann, E.; Deptula, P.; Kofoed, M.V.W.; Riedel, T.; Neumann-Schaal, M.; Hansen, M.; Nielsen, D.S.; et al. Sporofaciens musculi gen. nov., sp. nov., a novel bacterium isolated from the caecum of an obese mouse. Int. J. Syst. Evol. Microbiol. 2019, 71, 4673. [Google Scholar] [CrossRef]
- Zhu, H.; Hou, T. Modulatory Effects of Lactarius hatsudake on Obesity and Gut Microbiota in High-Fat Diet-Fed C57BL/6 Mice. Foods 2024, 13, 948. [Google Scholar] [CrossRef] [PubMed]
- Maya-Lucas, O.; Murugesan, S.; Nirmalkar, K.; Alcaraz, L.D.; Hoyo-Vadillo, C.; Pizano-Zárate, M.L.; García-Mena, J. The gut microbiome of Mexican children affected by obesity. Anaerobe 2019, 55, 11–23. [Google Scholar] [CrossRef]
- Xie, G.; Wang, X.; Liu, P.; Wei, R.; Chen, W.; Rajani, C.; Hernandez, B.Y.; Alegado, R.; Dong, B.; Li, D.; et al. Distinctly altered gut microbiota in the progression of liver disease. Oncotarget 2016, 7, 19355. [Google Scholar] [CrossRef]
- Hur, K.Y.; Lee, M.S. Gut Microbiota and Metabolic Disorders. Diabetes Metab. J. 2015, 39, 198–203. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Ko, G. Effect of metformin on metabolic improvement and gut microbiota. Appl. Environ. Microbiol. 2014, 80, 5935–5943. [Google Scholar] [CrossRef]
- Zhao, J.; Feng, T.; An, X.; Chen, X.; Han, N.; Wang, J.; Chang, G.; Hou, X. Livestock grazing is associated with the gut microbiota and antibiotic resistance genes in sympatric plateau pika (Ochotona curzoniae). Integr. Zool. 2024, 19, 646–661. [Google Scholar] [CrossRef]
- Zack, K.M.; Sorenson, T.; Joshi, S.G. Types and Mechanisms of Efflux Pump Systems and the Potential of Efflux Pump Inhibitors in the Restoration of Antimicrobial Susceptibility, with a Special Reference to Acinetobacter baumannii. Pathogens 2024, 13, 197. [Google Scholar] [CrossRef]
- Dridi, L.; Tankovic, J.; Petit, J.C. CdeA of Clostridium difficile, a new multidrug efflux transporter of the MATE family. Microb. Drug Resist. 2004, 10, 191–196. [Google Scholar] [CrossRef]
- Alcock, B.P.; Huynh, W.; Chalil, R.; Smith, K.W.; Raphenya, A.R.; Wlodarski, M.A.; Edalatmand, A.; Petkau, A.; Syed, S.A.; Tsang, K.K.; et al. CARD 2023: Expanded curation, support for machine learning, and resistome prediction at the Comprehensive Antibiotic Resistance Database. Nucleic Acids Res. 2023, 51, D690–D699. [Google Scholar] [CrossRef] [PubMed]
- Bai, J.; Zhu, X.; Zhao, K.; Yan, Y.; Xu, T.; Wang, J.; Zheng, J.; Huang, W.; Shi, L.; Shang, Y.; et al. The role of ArlRS in regulating oxacillin susceptibility in methicillin-resistant Staphylococcus aureus indicates it is a potential target for antimicrobial resistance breakers. Emerg. Microbes Infect. 2019, 8, 503–515. [Google Scholar] [CrossRef] [PubMed]
- Teo, J.W.; Tan, T.M.; Poh, C.L. Genetic determinants of tetracycline resistance in Vibrio harveyi. Antimicrob. Agents Chemother. 2002, 46, 1038–1045. [Google Scholar] [CrossRef] [PubMed]
- Seah, C.; Alexander, D.C.; Louie, L.; Simor, A.; Low, D.E.; Longtin, J.; Melano, R.G. MupB, a new high-level mupirocin resistance mechanism in Staphylococcus aureus. Antimicrob. Agents Chemother. 2012, 56, 1916–1920. [Google Scholar] [CrossRef]
- Mousa, W.K.; Mousa, S.; Ghemrawi, R.; Obaid, D.; Sarfraz, M.; Chehadeh, F.; Husband, S. Probiotics modulate host immune response and interact with the gut microbiota: Shaping their composition and mediating antibiotic resistance. Int. J. Mol. Sci. 2023, 24, 13783. [Google Scholar] [CrossRef]
- Choi, E.J.; Iwasa, M.; Han, K.I.; Kim, W.J.; Tang, Y.; Hwang, Y.J.; Chae, J.R.; Han, W.C.; Shin, Y.S.; Kim, E.K. Heat-Killed Enterococcus faecalis EF-2001 Ameliorates Atopic Dermatitis in a Murine Model. Nutrients 2016, 8, 146. [Google Scholar] [CrossRef]
- Han, K.I.; Shin, H.D.; Lee, Y.; Baek, S.; Moon, E.; Park, Y.B.; Cho, J.; Lee, J.H.; Kim, T.J.; Manoharan, R.K. Probiotic and Postbiotic Potentials of Enterococcus faecalis EF-2001: A Safety Assessment. Pharmaceuticals 2024, 17, 1383. [Google Scholar] [CrossRef]
- Choi, E.J.; Lee, H.J.; Kim, W.J.; Han, K.I.; Iwasa, M.; Kobayashi, K.; Debnath, T.; Tang, Y.; Kwak, Y.S.; Yoon, J.H.; et al. Enterococcus faecalis EF-2001 protects DNBS-induced inflammatory bowel disease in mice model. PLoS ONE 2019, 14, e0210854. [Google Scholar] [CrossRef]
- Panthee, S.; Paudel, A.; Hamamoto, H.; Ogasawara, A.A.; Iwasa, T.; Blom, J.; Sekimizu, K. Complete genome sequence and comparative genomic analysis of Enterococcus faecalis EF-2001, a probiotic bacterium. Genomics 2021, 113, 1534–1542. [Google Scholar] [CrossRef]
- Ma, L.; Tu, H.; Chen, T. Postbiotics in Human Health: A Narrative Review. Nutrients 2023, 15, 291. [Google Scholar] [CrossRef]
- Abd El-Ghany, W.A. Paraprobiotics and postbiotics: Contemporary and promising natural antibiotics alternatives and their applications in the poultry field. Open Vet. J. 2020, 10, 323–330. [Google Scholar] [CrossRef] [PubMed]
- Aggarwal, S.; Sabharwal, V.; Kaushik, P.; Joshi, A.; Aayushi, A.; Suri, M. Postbiotics: From emerging concept to application. Front. Sustain. Food Syst. 2022, 6, 887642. [Google Scholar] [CrossRef]
- Nezhadi, J.; Ahmadi, A. Assessing the efficacy of postbiotics derived from Lactobacillus plantarum on antibiotic resistance genes in nosocomial pathogens such as Enterococcus faecalis and Pseudomonas aeruginosa. Lett. Appl. Microbiol. 2024, 77, ovae127. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Manoharan, R.K.; Han, K.-I.; Shin, H.-D.; Lee, Y.; Baek, S.; Moon, E.; Park, Y.B.; Cho, J.; Srinivasan, S. Modulation of Gut Microbiota and Antibiotic Resistance Genes by Heat-Killed Enterococcus faecalis EF-2001 in High-Fat Diet-Induced Obesity Mice: A Shotgun Metagenomics Study. Bioengineering 2025, 12, 741. https://doi.org/10.3390/bioengineering12070741
Manoharan RK, Han K-I, Shin H-D, Lee Y, Baek S, Moon E, Park YB, Cho J, Srinivasan S. Modulation of Gut Microbiota and Antibiotic Resistance Genes by Heat-Killed Enterococcus faecalis EF-2001 in High-Fat Diet-Induced Obesity Mice: A Shotgun Metagenomics Study. Bioengineering. 2025; 12(7):741. https://doi.org/10.3390/bioengineering12070741
Chicago/Turabian StyleManoharan, Ranjith Kumar, Kwon-Il Han, Hyun-Dong Shin, Yura Lee, Sunhwa Baek, Eunjung Moon, Youn Bum Park, Junhui Cho, and Sathiyaraj Srinivasan. 2025. "Modulation of Gut Microbiota and Antibiotic Resistance Genes by Heat-Killed Enterococcus faecalis EF-2001 in High-Fat Diet-Induced Obesity Mice: A Shotgun Metagenomics Study" Bioengineering 12, no. 7: 741. https://doi.org/10.3390/bioengineering12070741
APA StyleManoharan, R. K., Han, K.-I., Shin, H.-D., Lee, Y., Baek, S., Moon, E., Park, Y. B., Cho, J., & Srinivasan, S. (2025). Modulation of Gut Microbiota and Antibiotic Resistance Genes by Heat-Killed Enterococcus faecalis EF-2001 in High-Fat Diet-Induced Obesity Mice: A Shotgun Metagenomics Study. Bioengineering, 12(7), 741. https://doi.org/10.3390/bioengineering12070741







