Medical LLMs: Fine-Tuning vs. Retrieval-Augmented Generation
Abstract
1. Introduction
1.1. Objective
1.2. Contributions
- It measures the impact on several models of three adaptation strategies: fine-tuning (FT), retrieval-augmented generation (RAG), and a hybrid FT+RAG approach.
- It addresses four central research questions: (1) Which model demonstrates the best overall performance? (2) Which adaptation method—fine-tuning, RAG, or FT+RAG—is the most effective overall? (3) Which model performs optimally with each specific adaptation method? (4) Which adaptation method yields the best results for each model?
- It applies a comprehensive evaluation framework employing both lexical and semantic metrics. The metrics used include BLEU, GLEU, ROUGE, METEOR, Precision, Recall, F1, BERTScore, SBERT cosine similarity, and Negation-Aware Semantic Similarity.
- Finally, it offers an overall perspective of model performance across many adaptation approaches.
1.3. Organizations
2. Related Work
3. Proposed Framework
4. System Design Architecture
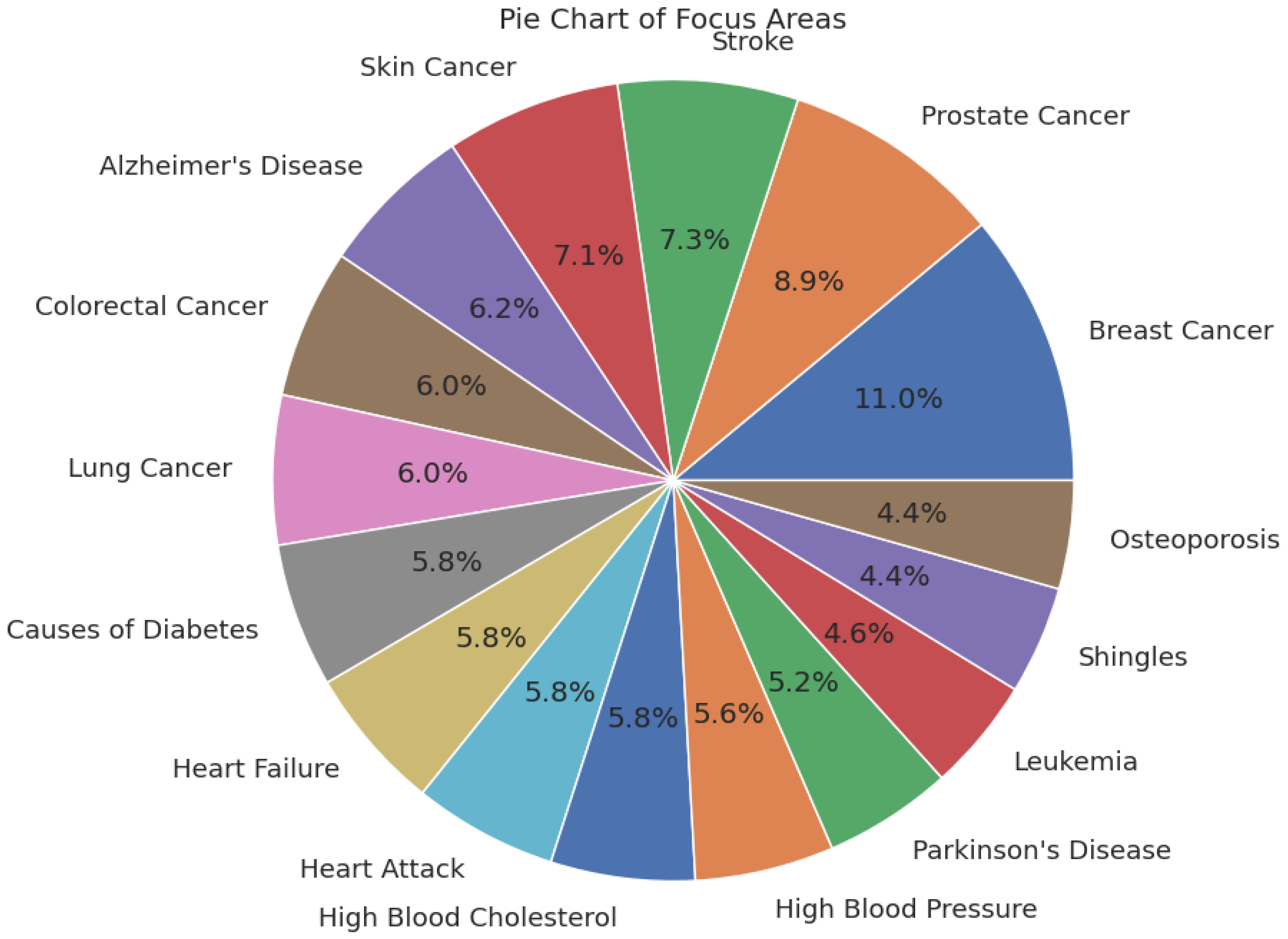
4.1. Dataset
4.2. Large Language Models
4.3. ChromaDB
4.4. System Configuration
5. Methodology
5.1. Preprocessing of Dataset
5.2. Fine-Tuning and RAG Implementation
5.2.1. Fine-Tuning
- Rank (r): 16, which controlled the number of parameters in the low-rank matrices.
- Target Modules: Used on query, key, value, and output projection layers.
- Gradient Checkpointing: Used to limit the use of memory when training.
- Batch size: 2 (per device).
- Gradient accumulation steps: 4.
- Learning rate: 2 × .
- Optimizer: AdamW with 8-bit precision.
- Mixed precision training: FP16.
5.2.2. Retrieval-Augmented Generation (RAG)
5.2.3. Retrieval-Augmented Generation on Fine-Tuned Models (FT+RAG)
5.3. Evaluation Using a Query Set
Evaluation Metrics
6. Results and Discussion
6.1. Model Training Outcomes
6.1.1. Training Duration
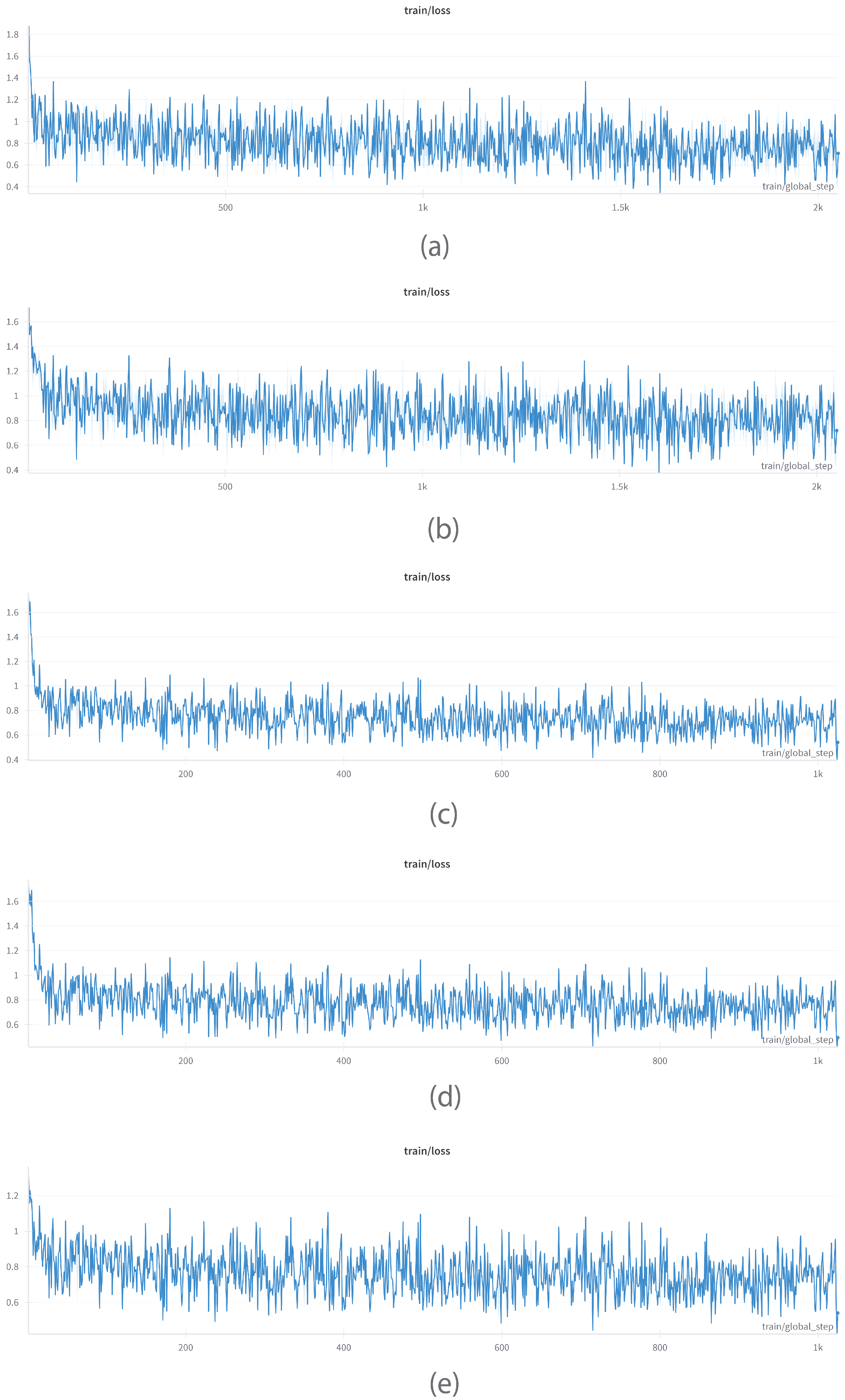
6.1.2. Train/Loss
6.1.3. GPU Power Consumption
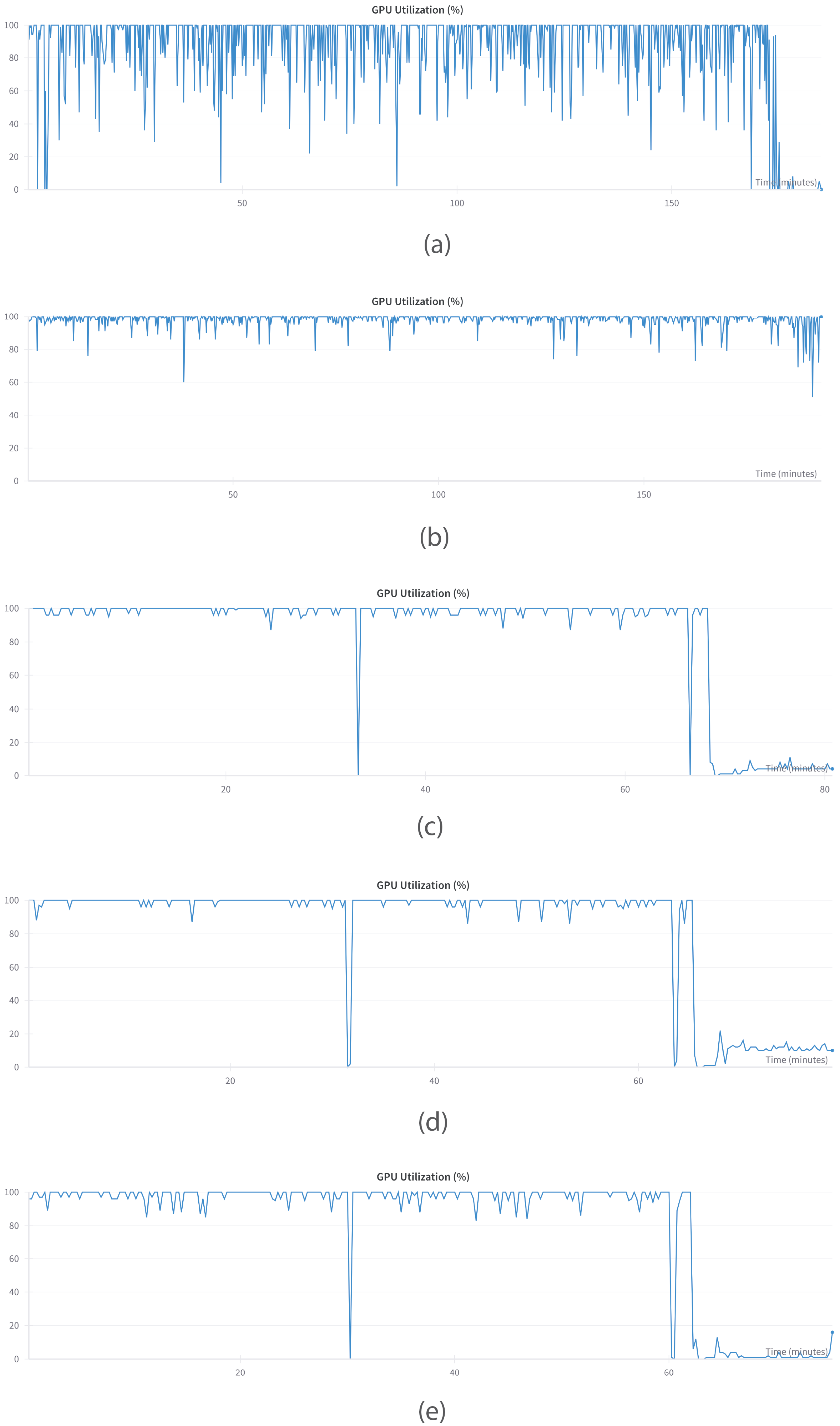
6.1.4. GPU Performance
6.2. Perplexity as a Benchmark for LLMs
6.3. Lexical and Semantic Evaluation (Quantitative Results)
6.4. Statistical Analysis
6.5. Key Findings
6.6. Performance Comparison with Other Models
6.7. Comparative Evaluation with Published Baselines
6.8. Carbon Emission
7. Limitations and Future Work
7.1. Limitations
7.2. Future Work
8. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| LLM | Large Language Model |
| GEMMA | Gemma-2-9B |
| LLAMA | Llama-3.1-8B |
| QWEN | Qwen2.5-7B |
| MISTRAL | Mistral-7B-Instruct |
| PHI | Phi-3.5-mini-instruct |
| FT | Fine-Tuning |
| RAG | Retrieval-Augmented Generation |
| FT+RAG | Fine-Tuning & Retrieval-Augmented Generation |
| LoRA | Low-Rank Adaptation |
| QLoRA | Quantized Low-Rank Adaptation |
| PPL | Perplexity |
| ROUGE | Recall-Oriented Understudy for Gisting Evaluation |
| BLEU | Bilingual Evaluation Understudy |
| GLEU | Google-Bilingual Evaluation Understudy |
| R-1 | ROUGE-1 |
| R-2 | ROUGE-2 |
| R-L | ROUGE-L |
| BERT | Bidirectional Encoder Representations from Transformers |
| SBERT | Sentence-BERT |
| BS_P | BERTScore Precision |
| BS_R | BERTScore Recall |
| BS_F1 | BERTScore F1 |
| SB_CS | SBERT Cosine Similarity |
| NASS | Negation Aware Semantic Similarity |
References
- Anisuzzaman, D.; Malins, J.G.; Friedman, P.A.; Attia, Z.I. Fine-Tuning Large Language Models for Specialized Use Cases. Mayo Clin. Proc. Digit. Health 2025, 3, 100184. [Google Scholar] [CrossRef] [PubMed]
- Grattafiori, A.; Dubey, A.; Jauhri, A.; Pandey, A.; Kadian, A.; Al-Dahle, A.; Letman, A.; Mathur, A.; Schelten, A.; Vaughan, A.; et al. The llama 3 herd of models. arXiv 2024, arXiv:2407.21783. [Google Scholar]
- Team, G.; Riviere, M.; Pathak, S.; Sessa, P.G.; Hardin, C.; Bhupatiraju, S.; Hussenot, L.; Mesnard, T.; Shahriari, B.; Ramé, A.; et al. Gemma 2: Improving open language models at a practical size. arXiv 2024, arXiv:2408.00118. [Google Scholar]
- Jiang, A.Q.; Sablayrolles, A.; Mensch, A.; Bamford, C.; Chaplot, D.S.; de las Casas, D.; Bressand, F.; Lengyel, G.; Lample, G.; Saulnier, L.; et al. Mistral 7B. arXiv 2023, arXiv:2310.06825. [Google Scholar]
- Yang, A.; Yang, B.; Zhang, B.; Hui, B.; Zheng, B.; Yu, B.; Li, C.; Liu, D.; Huang, F.; Wei, H.; et al. Qwen2. 5 technical report. arXiv 2024, arXiv:2412.15115. [Google Scholar]
- Abdin, M.; Aneja, J.; Awadalla, H.; Awadallah, A.; Awan, A.A.; Bach, N.; Bahree, A.; Bakhtiari, A.; Bao, J.; Behl, H.; et al. Phi-3 technical report: A highly capable language model locally on your phone. arXiv 2024, arXiv:2404.14219. [Google Scholar]
- Hu, E.J.; Shen, Y.; Wallis, P.; Allen-Zhu, Z.; Li, Y.; Wang, S.; Wang, L.; Chen, W. Lora: Low-rank adaptation of large language models. ICLR 2022, 1, 3. [Google Scholar]
- Dettmers, T.; Pagnoni, A.; Holtzman, A.; Zettlemoyer, L. Qlora: Efficient finetuning of quantized llms. Adv. Neural Inf. Process. Syst. 2023, 36, 10088–10115. [Google Scholar]
- Lewis, P.; Perez, E.; Piktus, A.; Petroni, F.; Karpukhin, V.; Goyal, N.; Küttler, H.; Lewis, M.; Yih, W.t.; Rocktäschel, T.; et al. Retrieval-augmented generation for knowledge-intensive nlp tasks. Adv. Neural Inf. Process. Syst. 2020, 33, 9459–9474. [Google Scholar]
- Balaguer, A.; Benara, V.; Cunha, R.L.d.F.; Hendry, T.; Holstein, D.; Marsman, J.; Mecklenburg, N.; Malvar, S.; Nunes, L.O.; Padilha, R.; et al. RAG vs. fine-tuning: Pipelines, tradeoffs, and a case study on agriculture. arXiv 2024, arXiv:2401.08406. [Google Scholar]
- Ovadia, O.; Brief, M.; Mishaeli, M.; Elisha, O. Fine-tuning or retrieval? comparing knowledge injection in llms. arXiv 2023, arXiv:2312.05934. [Google Scholar]
- Soudani, H.; Kanoulas, E.; Hasibi, F. Fine tuning vs. retrieval augmented generation for less popular knowledge. In Proceedings of the 2024 Annual International ACM SIGIR Conference on Research and Development in Information Retrieval in the Asia Pacific Region, Tokyo, Japan, 9–12 December 2024; pp. 12–22. [Google Scholar]
- Bassamzadeh, N.; Methani, C. A Comparative Study of DSL Code Generation: Fine-Tuning vs. Optimized Retrieval Augmentation. arXiv 2024, arXiv:2407.02742. [Google Scholar]
- Bora, A.; Cuayáhuitl, H. Systematic Analysis of Retrieval-Augmented Generation-Based LLMs for Medical Chatbot Applications. Mach. Learn. Knowl. Extr. 2024, 6, 2355–2374. [Google Scholar] [CrossRef]
- da Costa, L.Y.; de Oliveira, J.B. Adapting LLMs to New Domains: A Comparative Study of Fine-Tuning and RAG strategies for Portuguese QA Tasks. In Proceedings of the Simpósio Brasileiro de Tecnologia da Informação e da Linguagem Humana (STIL), SBC, Pará, Brazil, 20–23 May 2024; pp. 267–277. [Google Scholar]
- Lakatos, R.; Pollner, P.; Hajdu, A.; Joo, T. Investigating the performance of Retrieval-Augmented Generation and fine-tuning for the development of AI-driven knowledge-based systems. arXiv 2024, arXiv:2403.09727. [Google Scholar]
- Shen, M.; Umar, M.; Maeng, K.; Suh, G.E.; Gupta, U. Towards Understanding Systems Trade-offs in Retrieval-Augmented Generation Model Inference. arXiv 2024, arXiv:2412.11854. [Google Scholar]
- Dodgson, J.; Nanzheng, L.; Peh, J.; Pattirane, A.R.J.; Alhajir, A.D.; Dinarto, E.R.; Lim, J.; Ahmad, S.D. Establishing performance baselines in fine-tuning, retrieval-augmented generation and soft-prompting for non-specialist llm users. arXiv 2023, arXiv:2311.05903. [Google Scholar]
- Ke, Y.; Jin, L.; Elangovan, K.; Abdullah, H.R.; Liu, N.; Sia, A.T.H.; Soh, C.R.; Tung, J.Y.M.; Ong, J.C.L.; Ting, D.S.W. Development and Testing of Retrieval Augmented Generation in Large Language Models–A Case Study Report. arXiv 2024, arXiv:2402.01733. [Google Scholar]
- Zhao, S.; Yang, Y.; Wang, Z.; He, Z.; Qiu, L.K.; Qiu, L. Retrieval augmented generation (rag) and beyond: A comprehensive survey on how to make your llms use external data more wisely. arXiv 2024, arXiv:2409.14924. [Google Scholar]
- Ben Abacha, A.; Demner-Fushman, D. A question-entailment approach to question answering. BMC Bioinform. 2019, 20, 511. [Google Scholar] [CrossRef]
- Han, D.; Han, M.; Unsloth Team. Unsloth 2023. Available online: http://github.com/unslothai/unsloth (accessed on 14 April 2025).
- Pingua, B.; Murmu, D.; Kandpal, M.; Rautaray, J.; Mishra, P.; Barik, R.K.; Saikia, M.J. Mitigating adversarial manipulation in LLMs: A prompt-based approach to counter Jailbreak attacks (Prompt-G). PeerJ Comput. Sci. 2024, 10, e2374. [Google Scholar] [CrossRef]
- Liu, H.; Tam, D.; Muqeeth, M.; Mohta, J.; Huang, T.; Bansal, M.; Raffel, C.A. Few-shot parameter-efficient fine-tuning is better and cheaper than in-context learning. Adv. Neural Inf. Process. Syst. 2022, 35, 1950–1965. [Google Scholar]
- Ilapaka, A.; Ghosh, R. A Comprehensive RAG-Based LLM for AI-Driven Mental Health Chatbot. In Proceedings of the 2025 7th International Congress on Human-Computer Interaction, Optimization and Robotic Applications (ICHORA), Ankara, Turkiye, 23–24 May 2025; pp. 1–5. [Google Scholar]
- Keerthichandra, M.; Vihidun, T.; Lakshan, S.; Perera, I. Large Language Model-Based Student Intent Classification for Intelligent Tutoring Systems. In Proceedings of the 2024 9th International Conference on Information Technology Research (ICITR), Colombo, Sri Lanka, 5–6 December 2024; pp. 1–6. [Google Scholar]
- Kumar, S. Overriding Safety protections of Open-source Models. arXiv 2024, arXiv:2409.19476. [Google Scholar]
- Xiong, G.; Jin, Q.; Lu, Z.; Zhang, A. Benchmarking retrieval-augmented generation for medicine. In Proceedings of the Findings of the Association for Computational Linguistics ACL 2024, Bangkok, Thailand, 11–16 August 2024; pp. 6233–6251. [Google Scholar]
- Ahmed, B.S.; Baader, L.O.; Bayram, F.; Jagstedt, S.; Magnusson, P. Quality Assurance for LLM-RAG Systems: Empirical Insights from Tourism Application Testing. arXiv 2025, arXiv:2502.05782. [Google Scholar]
- Nguyen, P.V.; Tran, M.N.; Nguyen, L.; Dinh, D. Advancing Vietnamese Information Retrieval with Learning Objective and Benchmark. arXiv 2025, arXiv:2503.07470. [Google Scholar]
- Anschütz, M.; Lozano, D.M.; Groh, G. This is not correct! negation-aware evaluation of language generation systems. arXiv 2023, arXiv:2307.13989. [Google Scholar]
- Nazi, Z.A.; Peng, W. Large language models in healthcare and medical domain: A review. Informatics 2024, 11, 57. [Google Scholar] [CrossRef]
- Chaubey, H.K.; Tripathi, G.; Ranjan, R. Comparative analysis of RAG, fine-tuning, and prompt engineering in chatbot development. In Proceedings of the 2024 International Conference on Future Technologies for Smart Society (ICFTSS), Kuala Lumpur, Malaysia, 7–8 August 2024; pp. 169–172. [Google Scholar]
- Barnett, S.; Brannelly, Z.; Kurniawan, S.; Wong, S. Fine-tuning or fine-failing? debunking performance myths in large language models. arXiv 2024, arXiv:2406.11201. [Google Scholar]
- Lyu, Y.; Yan, L.; Wang, S.; Shi, H.; Yin, D.; Ren, P.; Chen, Z.; de Rijke, M.; Ren, Z. Knowtuning: Knowledge-aware fine-tuning for large language models. arXiv 2024, arXiv:2402.11176. [Google Scholar]
- Budakoglu, G.; Emekci, H. Unveiling the Power of Large Language Models: A Comparative Study of Retrieval-Augmented Generation, Fine-Tuning and Their Synergistic Fusion for Enhanced Performance. IEEE Access 2025, 13, 30936–90951. [Google Scholar] [CrossRef]
- Vrdoljak, J.; Boban, Z.; Males, I.; Skrabic, R.; Kumric, M.; Ottosen, A.; Clemencau, A.; Bozic, J.; Völker, S. Evaluating large language and large reasoning models as decision support tools in emergency internal medicine. Comput. Biol. Med. 2025, 192, 110351. [Google Scholar] [CrossRef]






| Author(s) | Title | Method(s) | Key Takeaways |
|---|---|---|---|
| [11] | Fine-Tuning or Retrieval? Comparing Knowledge Injection in LLMs | FT, RAG | RAG was more effective at injecting both novel and known knowledge. |
| [12] | Fine Tuning vs. Retrieval Augmented Generation for Less Popular Knowledge | FT, RAG | RAG outperformed FT for rare knowledge; FT improved with data augmentation. |
| [13] | A Comparative Study of DSL Code Generation: Fine-Tuning vs. Optimized Retrieval Augmentation | FT, RAG | RAG performed better for unseen APIs; FT had higher code similarity. |
| [14] | Systematic Analysis of Retrieval-Augmented Generation-Based Medical Chatbots | FT, RAG, FT+RAG | FT+RAG achieved best results for medical QA under resource constraints. |
| [15] | Adapting LLMs to New Domains: A Comparative Study of Fine-Tuning and RAG Strategies for Portuguese QA Tasks | FT, RAG, FT+RAG | Hybrid FT+RAG approach provided best domain adaptation for Portuguese QA. |
| [16] | Investigating the Performance of Retrieval- Augmented Generation and Fine-Tuning for the Development of AI-Driven Knowledge-Based Systems | FT, RAG | RAG showed higher BLEU and ROUGE; FT had slightly better creativity. |
| [17] | Towards Understanding Systems Trade-offs in Retrieval-Augmented Generation Model Inference | RAG | RAG offers no retraining needs but incurs latency and memory costs. |
| [18] | Establishing Performance Baselines in Fine-Tuning, Retrieval-Augmented Generation and Soft-Prompting for Non-Specialist LLM Users | FT, RAG, Soft Prompting | RAG had best accuracy for post-2021 queries; soft prompting improved outcomes. |
| [19] | Development and Testing of Retrieval Augmented Generation in Large Language Models | RAG, FT | GPT-4 with RAG achieved 91.4% accuracy, surpassing human experts. |
| [20] | Retrieval Augmented Generation (RAG) and Beyond: A Comprehensive Survey on How to Make your LLMs use External Data More Wisely | FT, RAG, Hybrid | Survey emphasizes hybrid strategies to meet complex real-world LLM tasks. |
| Model | Original (Unsloth) PPL (↓) | Fine Tuned (MedQuAD) PPL (↓) | ΔPPL |
|---|---|---|---|
| PHI | 34.79 ± 0.8 | 33.64 ± 0.7 | −1.15 |
| QWEN | 38.15 ± 1.2 | 41.15 ± 1.5 | +3.00 |
| LLAMA | 51.52 ± 2.1 | 48.48 ± 1.9 | −3.04 |
| MISTRAL | 39.73 ± 1.4 | 47.84 ± 2.0 | +8.11 |
| GEMMA | 186.43 ± 5.7 | 86.79 ± 3.2 | −99.64 |
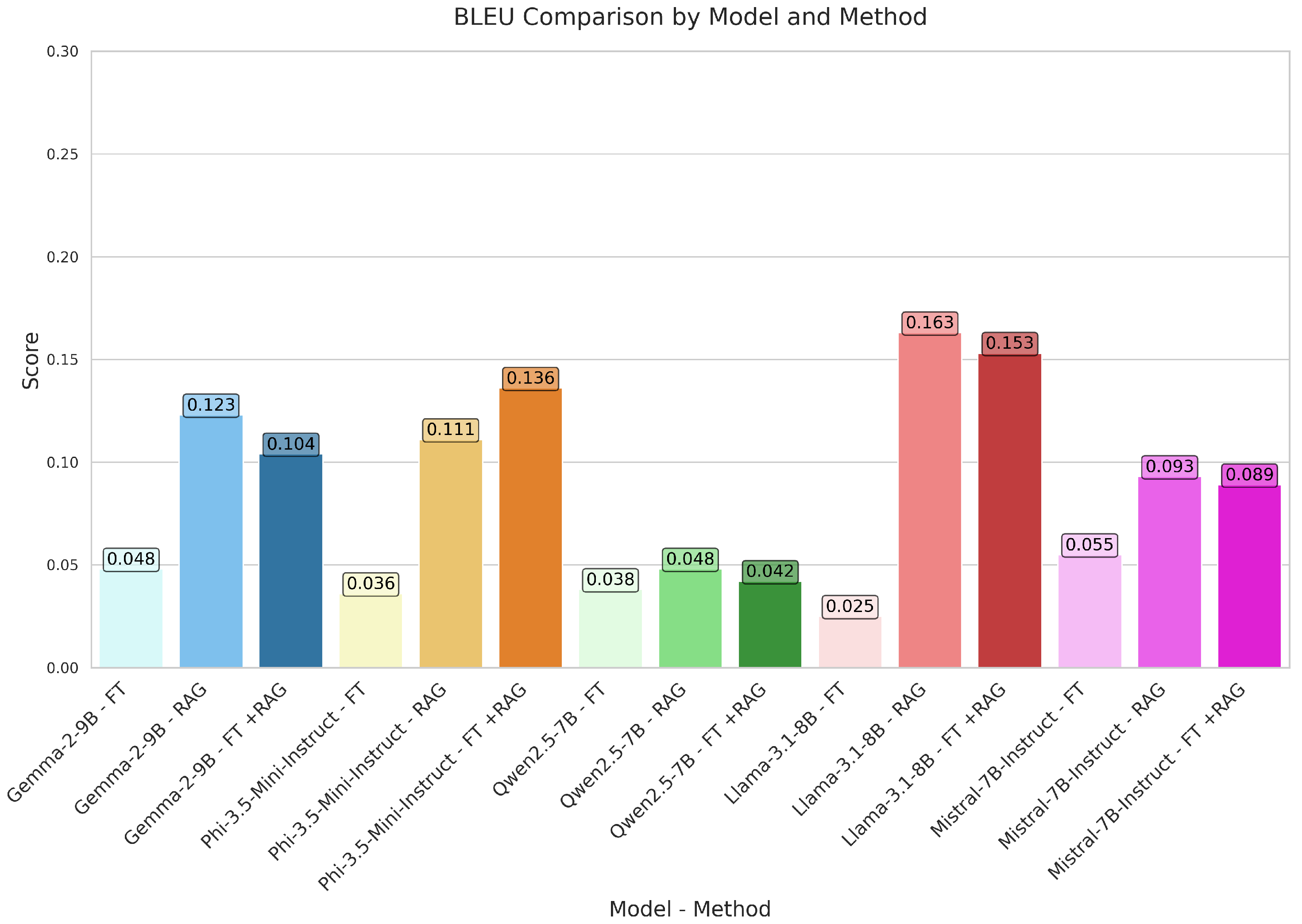
| Model | Method | BLEU | GLEU | R-1 | R-2 | R-L | METEOR | Precision | Recall | F1 | BS_P | BS_R | BS_F1 | SB_CS | NASS |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
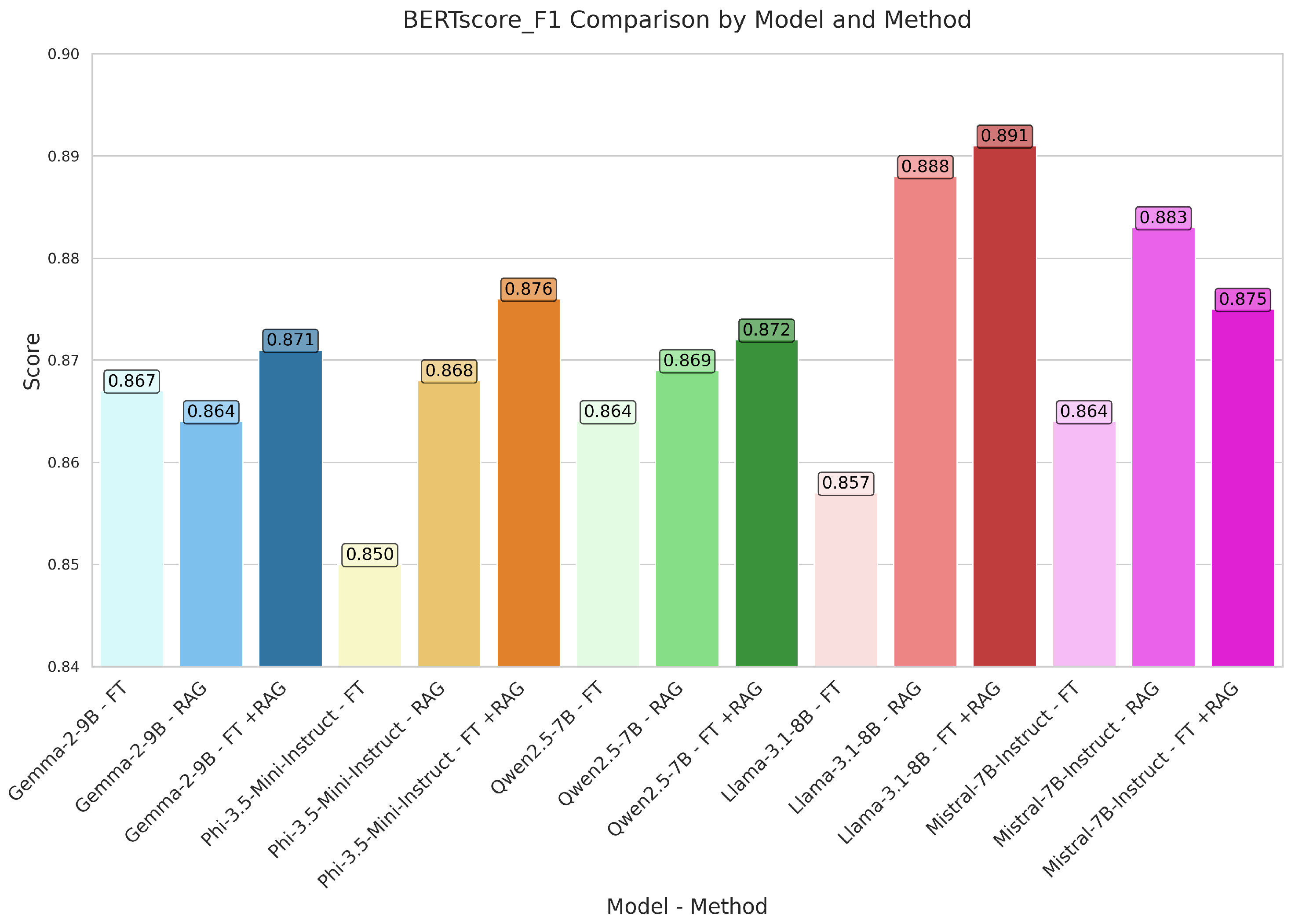
| GEMMA | FT | 0.048 | 0.129 | 0.313 | 0.118 | 0.296 | 0.196 | 0.569 | 0.336 | 0.392 | 0.888 | 0.848 | 0.867 | 0.802 | 0.857 |
| RAG | 0.123 | 0.190 | 0.354 | 0.212 | 0.342 | 0.235 | 0.585 | 0.357 | 0.391 | 0.881 | 0.849 | 0.864 | 0.760 | 0.819 | |
| FT+RAG | 0.104 | 0.181 | 0.351 | 0.193 | 0.338 | 0.240 | 0.586 | 0.367 | 0.405 | 0.890 | 0.854 | 0.871 | 0.791 | 0.841 | |
| PHI | FT | 0.036 | 0.122 | 0.267 | 0.076 | 0.247 | 0.200 | 0.433 | 0.387 | 0.390 | 0.855 | 0.846 | 0.850 | 0.781 | 0.824 |
| RAG | 0.111 | 0.190 | 0.366 | 0.212 | 0.354 | 0.243 | 0.602 | 0.367 | 0.417 | 0.891 | 0.846 | 0.868 | 0.810 | 0.875 | |
| FT+RAG | 0.136 | 0.217 | 0.391 | 0.250 | 0.380 | 0.258 | 0.651 | 0.366 | 0.435 | 0.903 | 0.852 | 0.876 | 0.815 | 0.881 | |
| QWEN | FT | 0.038 | 0.109 | 0.276 | 0.106 | 0.263 | 0.162 | 0.610 | 0.265 | 0.334 | 0.892 | 0.838 | 0.864 | 0.788 | 0.856 |
| RAG | 0.048 | 0.128 | 0.306 | 0.168 | 0.294 | 0.161 | 0.720 | 0.215 | 0.304 | 0.908 | 0.835 | 0.869 | 0.773 | 0.837 | |
| FT+RAG | 0.042 | 0.130 | 0.302 | 0.166 | 0.290 | 0.160 | 0.716 | 0.232 | 0.316 | 0.911 | 0.838 | 0.872 | 0.787 | 0.859 | |
| LLAMA | FT | 0.025 | 0.089 | 0.262 | 0.080 | 0.245 | 0.146 | 0.571 | 0.248 | 0.315 | 0.881 | 0.834 | 0.857 | 0.766 | 0.816 |
| RAG | 0.163 | 0.237 | 0.418 | 0.280 | 0.407 | 0.279 | 0.711 | 0.369 | 0.436 | 0.916 | 0.862 | 0.888 | 0.822 | 0.898 | |
| FT+RAG | 0.153 | 0.236 | 0.423 | 0.299 | 0.413 | 0.268 | 0.756 | 0.342 | 0.419 | 0.925 | 0.860 | 0.891 | 0.819 | 0.908 | |
| MISTRAL | FT | 0.055 | 0.136 | 0.315 | 0.116 | 0.297 | 0.206 | 0.527 | 0.355 | 0.401 | 0.879 | 0.851 | 0.864 | 0.803 | 0.845 |
| RAG | 0.093 | 0.186 | 0.386 | 0.235 | 0.368 | 0.220 | 0.762 | 0.290 | 0.389 | 0.918 | 0.850 | 0.883 | 0.821 | 0.878 | |
| FT+RAG | 0.089 | 0.172 | 0.346 | 0.205 | 0.331 | 0.207 | 0.694 | 0.291 | 0.370 | 0.907 | 0.846 | 0.875 | 0.796 | 0.857 |
| Model | Method | BLEU | GLEU | R-1 | R-2 | R-L | METEOR | Precision | Recall | F1 | BS_P | BS_R | BS_F1 | SB_CS | NASS |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GEMMA | FT | (0.039, 0.058) | (0.118, 0.140) | — | (0.107, 0.130) | — | — | — | — | — | — | — | — | (0.784, 0.820) | — |
| RAG | (0.088, 0.165) | (0.155, 0.226) | — | (0.166, 0.258) | — | — | — | — | — | — | — | — | (0.730, 0.789) | — | |
| FT+RAG | — | (0.151, 0.214) | — | — | — | — | — | — | — | — | — | — | (0.766, 0.816) | — | |
| PHI | FT | (0.028, 0.045) | (0.113, 0.131) | (0.253, 0.280) | (0.067, 0.085) | (0.234, 0.262) | — | (0.408, 0.458) | — | (0.374, 0.405) | (0.851, 0.859) | — | (0.845, 0.855) | (0.760, 0.801) | (0.803, 0.842) |
| RAG | (0.084, 0.142) | (0.164, 0.219) | (0.332, 0.399) | (0.176, 0.253) | (0.320, 0.389) | — | (0.560, 0.644) | — | — | (0.884, 0.899) | (0.839, 0.853) | (0.862, 0.874) | — | (0.844, 0.900) | |
| FT+RAG | (0.103, 0.172) | (0.186, 0.252) | (0.350, 0.433) | (0.201, 0.295) | (0.338, 0.422) | — | (0.606, 0.696) | — | (0.402, 0.469) | (0.893, 0.911) | (0.844, 0.860) | (0.869, 0.885) | (0.794, 0.835) | (0.854, 0.911) | |
| QWEN | FT | — | — | — | (0.093, 0.117) | — | — | (0.580, 0.641) | (0.234, 0.296) | — | (0.887, 0.897) | — | (0.859, 0.869) | — | — |
| RAG | — | — | — | (0.129, 0.204) | — | — | (0.676, 0.767) | (0.186, 0.248) | — | (0.900, 0.917) | — | — | — | — | |
| FT+RAG | — | — | — | (0.135, 0.199) | — | — | (0.669, 0.762) | — | — | (0.902, 0.921) | — | (0.867, 0.880) | — | — | |
| LLAMA | FT | (0.019, 0.033) | (0.080, 0.100) | (0.246, 0.277) | (0.071, 0.091) | (0.231, 0.260) | (0.129, 0.163) | (0.542, 0.600) | (0.220, 0.274) | (0.293, 0.336) | (0.875, 0.886) | (0.828, 0.841) | (0.852, 0.862) | (0.747, 0.787) | (0.794, 0.835) |
| RAG | (0.120, 0.204) | (0.197, 0.279) | (0.372, 0.463) | (0.233, 0.329) | (0.363, 0.448) | (0.236, 0.323) | (0.669, 0.751) | (0.319, 0.418) | (0.392, 0.479) | (0.907, 0.926) | (0.852, 0.873) | (0.878, 0.896) | (0.797, 0.843) | (0.873, 0.922) | |
| FT+RAG | (0.113, 0.194) | (0.196, 0.278) | (0.381, 0.465) | (0.249, 0.347) | (0.370, 0.457) | (0.226, 0.309) | (0.708, 0.803) | (0.297, 0.390) | (0.379, 0.463) | (0.915, 0.936) | (0.850, 0.870) | (0.882, 0.899) | (0.795, 0.840) | (0.884, 0.932) | |
| MISTRAL | FT | — | — | (0.298, 0.332) | (0.105, 0.128) | (0.282, 0.313) | — | (0.496, 0.557) | (0.329, 0.381) | — | (0.873, 0.885) | — | (0.859, 0.869) | (0.787, 0.818) | (0.829, 0.861) |
| RAG | — | — | (0.350, 0.424) | (0.194, 0.271) | (0.332, 0.408) | — | (0.720, 0.802) | (0.254, 0.329) | — | (0.910, 0.928) | — | (0.875, 0.891) | (0.805, 0.839) | (0.852, 0.901) | |
| FT+RAG | — | — | — | (0.169, 0.245) | — | — | (0.649, 0.739) | (0.253, 0.328) | — | (0.898, 0.917) | — | (0.868, 0.883) | — | — |
| Model | Comparison | BLEU | GLEU | R-1 | R-2 | R-L | METEOR | Precision | Recall | F1 | BS_P | BS_R | BS_F1 | SB_CS | NASS |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GEMMA | FT vs. RAG | 0.042 | 0.037 | 0.229 | 0.006 | 0.176 | 0.419 | 0.883 | 0.644 | 0.519 | 0.289 | 0.688 | 0.513 | 0.007 | 0.229 |
| FT vs. FT+RAG | 0.235 | 0.034 | 0.345 | 0.190 | 0.316 | 0.089 | 0.891 | 0.210 | 0.601 | 0.916 | 0.235 | 0.669 | 0.194 | 0.945 | |
| RAG vs. FT+RAG | 0.941 | 0.677 | 0.665 | 0.720 | 0.724 | 0.438 | 0.958 | 0.496 | 0.329 | 0.163 | 0.170 | 0.078 | 0.022 | 0.245 | |
| PHI | FT vs. RAG | 3.4 × | 3.6 × | 2.3 × | 1.6 × | 8.6 × | 0.028 | 1.3 × | 0.150 | 0.154 | 3.9 × | 0.296 | 3.2 × | 0.050 | 2.6 × |
| FT vs. FT+RAG | 1.4 × | 6.3 × | 1.0 × | 2.0 × | 2.6 × | 0.004 | 1.4 × | 0.133 | 0.039 | 2.3 × | 0.217 | 9.9 × | 0.014 | 3.0 × | |
| RAG vs. FT+RAG | 0.289 | 0.043 | 0.170 | 0.054 | 0.139 | 0.284 | 0.088 | 0.985 | 0.203 | 0.009 | 0.005 | 0.002 | 0.598 | 0.122 | |
| QWEN | FT vs. RAG | 0.891 | 0.477 | 0.233 | 0.044 | 0.345 | 0.751 | 1.3 × | 0.026 | 0.207 | 0.005 | 0.306 | 0.296 | 0.135 | 0.983 |
| FT vs. FT+RAG | 0.523 | 0.170 | 0.196 | 0.003 | 0.231 | 0.404 | 4.2 × | 0.072 | 0.205 | 9.5 × | 0.572 | 0.035 | 0.688 | 0.434 | |
| RAG vs. FT+RAG | 0.672 | 0.414 | 0.893 | 0.832 | 0.924 | 0.559 | 0.966 | 0.280 | 0.347 | 0.452 | 0.268 | 0.275 | 0.220 | 0.186 | |
| LLAMA | FT vs. RAG | 1.2 × | 2.2 × | 8.6 × | 4.2 × | 8.4 × | 2.0 × | 2.0 × | 9.5 × | 7.5 × | 2.2 × | 5.2 × | 3.5 × | 1.8 × | 2.0 × |
| FT vs. FT+RAG | 2.9 × | 2.2 × | 7.2 × | 1.0 × | 6.6 × | 2.1 × | 5.6 × | 3.4 × | 3.2 × | 1.6 × | 3.8 × | 5.9 × | 1.0 × | 6.7 × | |
| RAG vs. FT+RAG | 0.512 | 0.748 | 0.657 | 0.117 | 0.591 | 0.141 | 0.069 | 0.117 | 0.336 | 0.052 | 0.482 | 0.097 | 0.846 | 0.167 | |
| MISTRAL | FT vs. RAG | 0.513 | 0.053 | 0.004 | 9.2 × | 0.004 | 0.788 | 1.2 × | 3.7 × | 0.387 | 2.9 × | 0.587 | 6.8 × | 0.037 | 1.7 × |
| FT vs. FT+RAG | 0.513 | 0.537 | 0.256 | 0.004 | 0.245 | 0.384 | 3.9 × | 0.001 | 0.051 | 5.1 × | 0.099 | 0.026 | 0.825 | 0.321 | |
| RAG vs. FT+RAG | 0.963 | 0.686 | 0.181 | 0.252 | 0.195 | 0.867 | 0.001 | 0.781 | 0.650 | 0.005 | 0.765 | 0.049 | 0.268 | 0.091 |
| Paper | Models Used | Most Effective Method | Notes |
|---|---|---|---|
| [10] | Llama2-13B, GPT-3.5, GPT-4 | FT+RAG | Fine-tuning improved accuracy by 6 percentage points and RAG added 5 more percentage points |
| [11] | Not explicitly named, generic LLMs | RAG | RAG consistently outperformed fine-tuning for both new and existing knowledge |
| [15] | Not specified | FT+RAG | Combined approach yielded best results in Portuguese QA tasks |
| [33] | openassistant-guanaco | FT | Achieved highest accuracy (87.8%) and BLEU (0.81); RAG and prompt engineering were competitive |
| [34] | Not specified | RAG | Fine-tuning degraded RAG performance across domains |
| Proposed | Llama-3.1-8B, Gemma-2-9B, Mistral-7B-Instruct, Qwen2.5-7B, Phi-3.5-Mini-Instruct | RAG | retrieval-augmented generation (RAG) emerged as most effective adaptation strategy. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pingua, B.; Sahoo, A.; Kandpal, M.; Murmu, D.; Rautaray, J.; Barik, R.K.; Saikia, M.J. Medical LLMs: Fine-Tuning vs. Retrieval-Augmented Generation. Bioengineering 2025, 12, 687. https://doi.org/10.3390/bioengineering12070687
Pingua B, Sahoo A, Kandpal M, Murmu D, Rautaray J, Barik RK, Saikia MJ. Medical LLMs: Fine-Tuning vs. Retrieval-Augmented Generation. Bioengineering. 2025; 12(7):687. https://doi.org/10.3390/bioengineering12070687
Chicago/Turabian StylePingua, Bhagyajit, Adyakanta Sahoo, Meenakshi Kandpal, Deepak Murmu, Jyotirmayee Rautaray, Rabindra Kumar Barik, and Manob Jyoti Saikia. 2025. "Medical LLMs: Fine-Tuning vs. Retrieval-Augmented Generation" Bioengineering 12, no. 7: 687. https://doi.org/10.3390/bioengineering12070687
APA StylePingua, B., Sahoo, A., Kandpal, M., Murmu, D., Rautaray, J., Barik, R. K., & Saikia, M. J. (2025). Medical LLMs: Fine-Tuning vs. Retrieval-Augmented Generation. Bioengineering, 12(7), 687. https://doi.org/10.3390/bioengineering12070687









