Research on Medical Image Segmentation Based on SAM and Its Future Prospects
Abstract
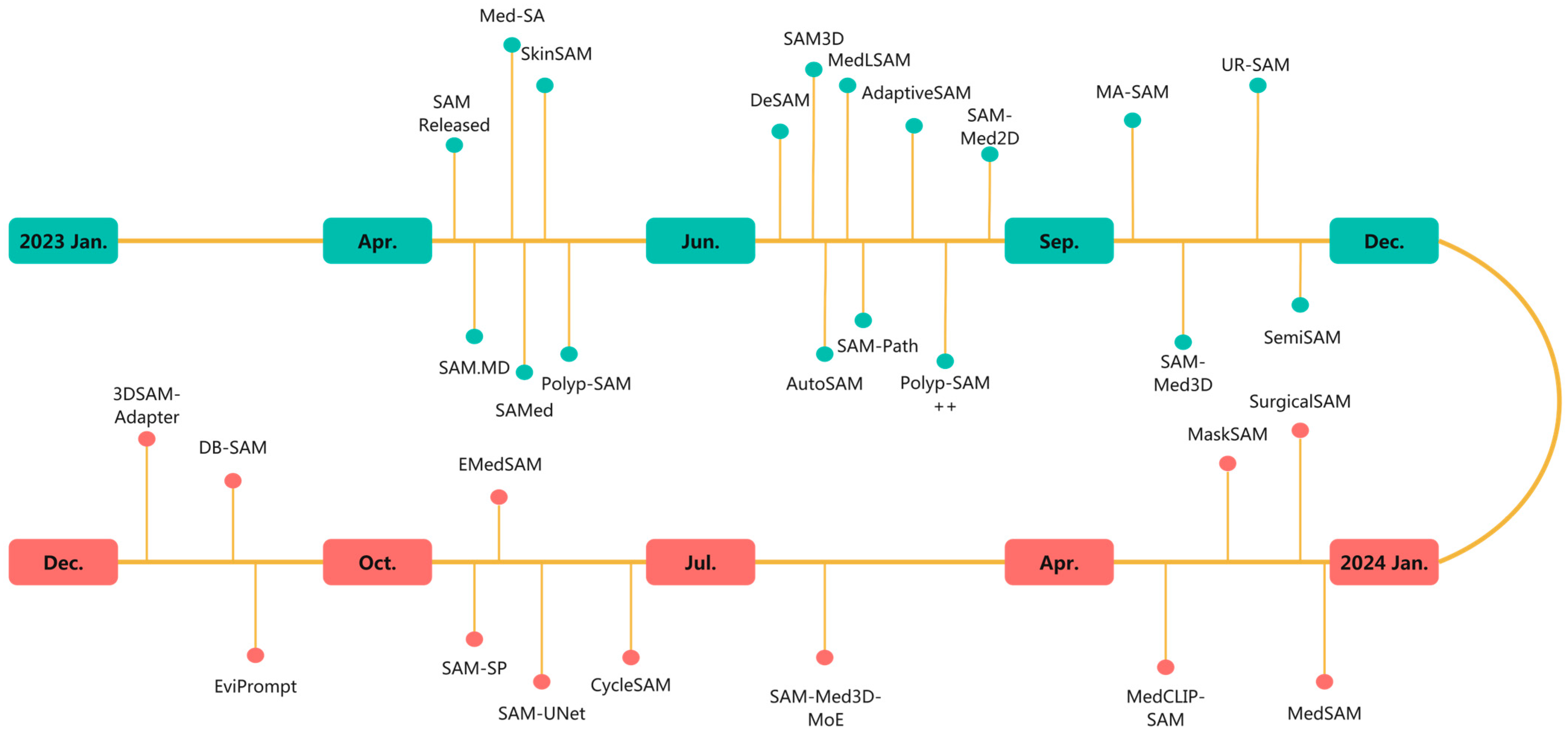
1. Introduction
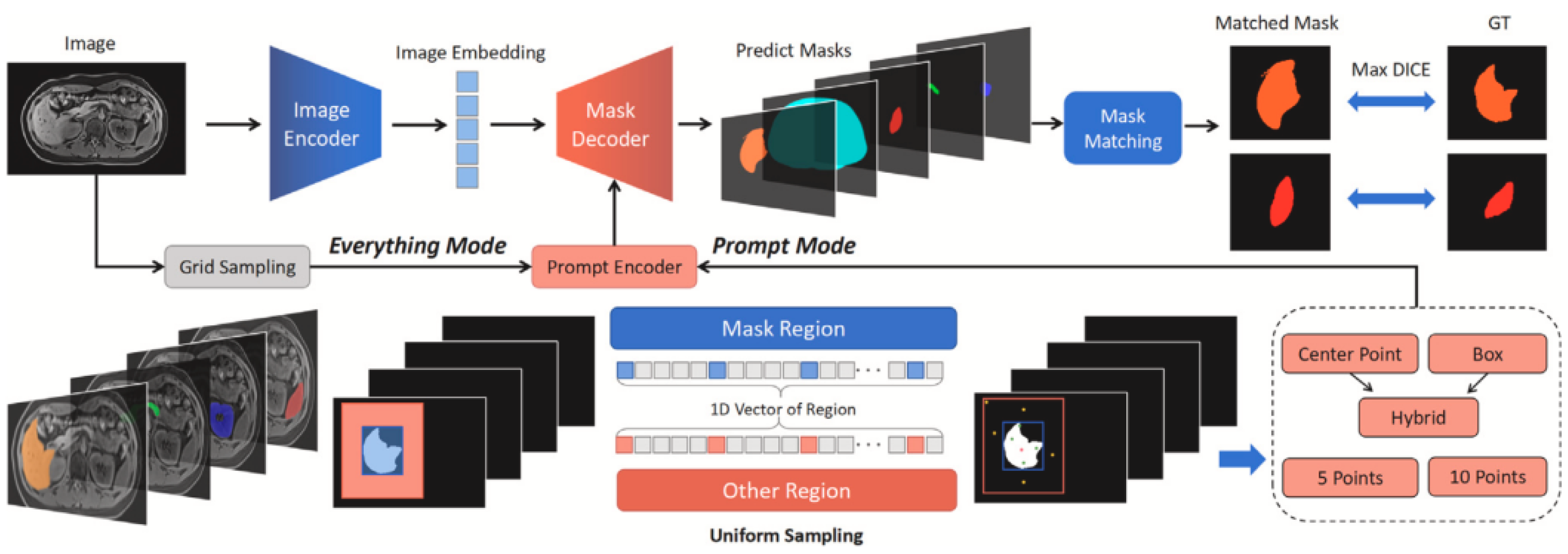
2. The Architecture of SAM
3. Evaluation of the Zero-Shot Capability of the Segment Anything Model (SAM) in the Field of Medical Image Segmentation
3.1. Evaluation on Specific Datasets
3.1.1. CT Image Segmentation
3.1.2. Colonoscopic Image Segmentation
3.2. Evaluation on Multimodal Datasets
3.3. Challenges in Low-Contrast Imaging Modalities
4. The Application of Segment Anything Model (SAM) in the Field of Medical Image Segmentation
4.1. Fine-Tuning on Medical Image Datasets
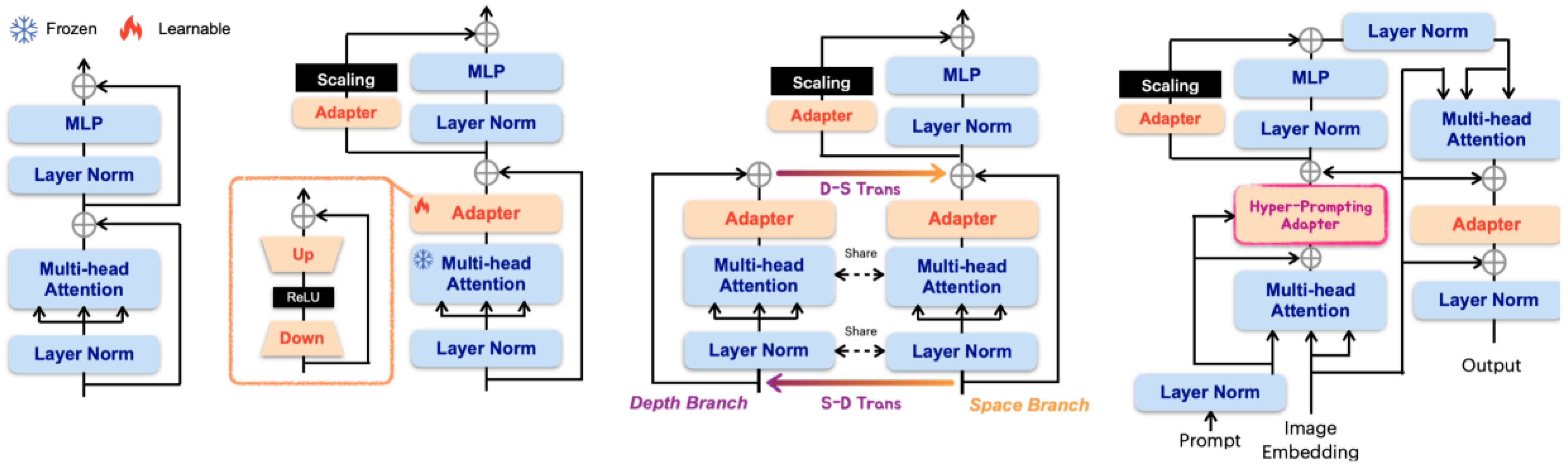
4.1.1. Full-Parameter Fine-Tuning
4.1.2. Parameter-Efficient Fine-Tuning (PEFT)
4.1.3. Trade-Offs Between Full Fine-Tuning and PEFT
4.2. Automated Prompt Mechanism
4.2.1. Automated Prompt Generation
4.2.2. Learnable Prompts
4.2.3. Enhance Robustness to Uncertain Prompts
4.3. Architecture Modification
4.3.1. Synergistic Effects of Training Segmentation Models
4.3.2. Promote Efficient Annotation Learning
4.4. Towards 3D Medical Images
4.4.1. Adaptation from 2D to 3D
4.4.2. Training from Scratch
4.5. Comparison Between SAM and Non-Prompt-Based Models
5. Discussion
5.1. Challenges in Achieving Robust Zero-Shot Segmentation
5.2. Domain-Specific Adaptation and Generalization Limitations
5.3. Persistent Challenges and Future Research Directions
- Computational Overhead: Unlike lightweight, task-specific models designed for deployment efficiency, the SAM’s generalized architecture imposes substantial inference latency and memory costs. These constraints limit real-time applicability, especially in resource-constrained hospital environments. While model pruning, quantization, and hardware acceleration offer possible solutions, these optimizations remain an active area of research.
- Prompt Sensitivity: The SAM’s segmentation results are highly sensitive to variations in prompt type, location, and quality. Even minor prompt changes can cause inconsistent outputs, raising concerns regarding diagnostic reliability. Although the SAM is envisioned as an assistive tool rather than a standalone system, robust prompt engineering protocols and consensus-driven oversight are necessary to ensure consistent, interpretable outputs across users and tasks.
- Dataset Scarcity and Annotation Costs: Foundational models like the SAM rely on vast, high-quality datasets for pretraining and fine-tuning. However, medical image datasets—especially 3D volumetric scans—require expert annotation, which is time-consuming, labor-intensive, and expensive. For comparison, the SAM was trained on SA-1B, which contains 11 million images and 1.1 billion masks [17], whereas constructing medical datasets of this scale is significantly more challenging.
- Dataset Development: Curate large-scale, domain-specific medical image datasets, particularly 3D volumetric datasets, to enable more effective fine-tuning and benchmarking;
- Hybrid Frameworks: Integrate SAM with lightweight, task-optimized backbones for improved performance on specialized medical tasks;
- Deployment Optimization: Enhance clinical usability through latency reduction, model compression, and inference acceleration to meet real-time processing requirements;
- Prompt Robustness: Establish reliable prompt engineering protocols and develop tools to ensure consistent, reproducible segmentation outputs across users and institutions.
5.4. Clinical Feasibility and Ethical Risks
5.5. Outlook
Author Contributions
Funding
Data Availability Statement
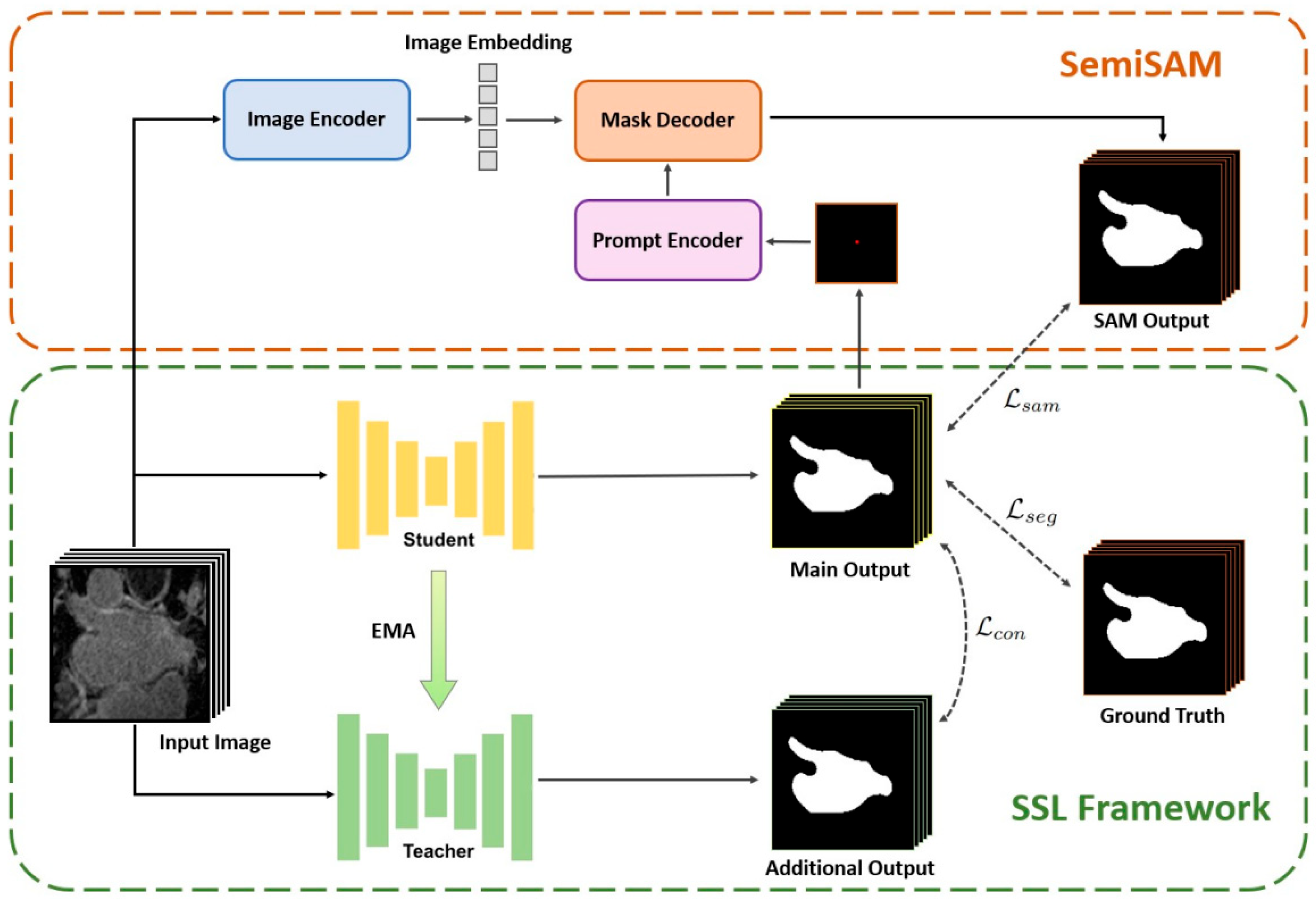
Acknowledgments
Conflicts of Interest
Appendix A
| No. | Method | Title | Code Repository | Paper Link | Description |
|---|---|---|---|---|---|
| 1 | Review | Deep Interactive Segmentation of Medical Images: A Systematic Review and Taxonomy | - | https://ieeexplore.ieee.org/document/10660300 (accessed on 1 May 2025) | Review and taxonomy of deep interactive segmentation in medical images. |
| 2 | Med-SA | Medical SAM Adapter: Adapting Segment Anything Model for Medical Image Segmentation | https://github.com/SuperMedIntel/Medical-SAM-Adapter (accessed on 27 March 2025) | https://www.sciencedirect.com/science/article/pii/S1361841525000945 (accessed on 1 May 2025) | Medical SAM adapter for 3D segmentation with minimal parameter updates. |
| 3 | SAM | Segment Anything | https://github.com/facebookresearch/segment-anything (accessed on 27 March 2025) | https://openaccess.thecvf.com/content/ICCV2023/papers/Kirillov_Segment_Anything_ICCV_2023_paper.pdf (accessed on 1 May 2025) | Segment Anything Model Origin Paper |
| 9 | SAM.MD | SAM.MD: Zero-shot medical image segmentation capabilities of the Segment Anything Model | - | https://openreview.net/forum?id=iilLHaINUW (accessed on 1 May 2025) | Evaluates SAM’s zero-shot medical image segmentation on abdominal CT. |
| 11 | Review | When sam meets medical images: An investigation of segment anything model (sam) on multi-phase liver tumor segmentation | - | https://arxiv.org/pdf/2304.08506 (accessed on 1 May 2025) | Evaluating the SAM for multi-phase liver tumor segmentation. |
| 13 | SAMPolyp | Can SAM Segment Polyps? | - | https://arxiv.org/pdf/2304.07583 (accessed on 1 May 2025) | Evaluating the SAM for polyp segmentation. |
| 14 | Segment-Anything-Model-for-Medical-Images | Segment anything model for medical images? | https://github.com/yuhoo0302/Segment-Anything-Model-for-Medical-Images (accessed on 27 March 2025) | https://www.sciencedirect.com/science/article/pii/S1361841523003213 (accessed on 1 May 2025) | Evaluating the SAM’s performance on medical image segmentation. |
| 17 | Review | Sam on medical images: A comprehensive study on three prompt modes | - | https://arxiv.org/pdf/2305.00035 (accessed on 1 May 2025) | Evaluating the SAM’s zero-shot segmentation on medical images with various prompts. |
| 19 | SkinSAM | SkinSAM: Empowering Skin Cancer Segmentation with Segment Anything Model | - | https://arxiv.org/pdf/2304.13973 (accessed on 1 May 2025) | Fine-tuned SAM for skin cancer segmentation. |
| 20 | Polyp-SAM | Polyp-SAM: Transfer SAM for Polyp Segmentation | https://github.com/ricklisz/Polyp-SAM (accessed on 27 March 2025) | https://www.spiedigitallibrary.org/conference-proceedings-of-spie/12927/1292735/Polyp-SAM-transfer-SAM-for-polyp-segmentation/10.1117/12.3006809.short (accessed on 1 May 2025) | Polyp-SAM adapts the SAM for polyp segmentation, achieving state-of-the-art performance. |
| 21 | MedSAM | Segment anything in medical images | - | https://www.nature.com/articles/s41467-024-44824-z (accessed on 1 May 2025) | MedSAM for universal medical image segmentation. |
| 23 | SAMed | Customized Segment Anything Model for Medical Image Segmentation | https://github.com/hitachinsk/SAMed (accessed on 27 March 2025) | https://arxiv.org/pdf/2304.13785 (accessed on 1 May 2025) | LoRA-finetuned SAM for medical image segmentation. |
| 24 | Fine-tuning SAM on Few Exemplars | Cheap lunch for medical image segmentation by fine-tuning sam on few exemplars | - | https://link.springer.com/chapter/10.1007/978-3-031-76160-7_2 (accessed on 1 May 2025) | Fine-tuning the SAM for medical images with few exemplars. |
| 25 | Adaptivesam | Adaptivesam: Towards efficient tuning of sam for surgical scene segmentation | https://github.com/JayParanjape/biastuning (accessed on 27 March 2025) | https://link.springer.com/chapter/10.1007/978-3-031-66958-3_14 (accessed on 1 May 2025) | AdaptiveSAM for efficient surgical image segmentation with text prompts. |
| 26 | SAM-Med2D | SAM-Med2D | - | https://arxiv.org/pdf/2308.16184 (accessed on 1 May 2025) | SAM fine-tuned for medical 2D image segmentation with comprehensive prompts. |
| 27 | YOLOv8+SAM | Comprehensive Multimodal Segmentation in Medical Imaging: Combining YOLOv8 with SAM and HQ-SAM Models | - | https://openaccess.thecvf.com/content/ICCV2023W/CVAMD/papers/Pandey_Comprehensive_Multimodal_Segmentation_in_Medical_Imaging_Combining_YOLOv8_with_SAM_ICCVW_2023_paper.pdf (accessed on 1 May 2025) | YOLOv8 with the SAM for medical image segmentation. |
| 29 | MedLSAM | MedLSAM: Localize and segment anything model for 3D CT images | https://github.com/openmedlab/MedLSAM (accessed on 27 March 2025) | https://www.sciencedirect.com/science/article/pii/S1361841524002950 (accessed on 1 May 2025) | MedLSAM for 3D CT localization and segmentation with minimal annotations. |
| 30 | One Shot Localization And Segmentation | One shot localization and segmentation of medical images with Foundation Models | - | https://arxiv.org/pdf/2310.18642 (accessed on 1 May 2025) | One-shot medical image segmentation using natural-image pretrained foundation models. |
| 31 | AutoSAM | AutoSAM: Adapting SAM to Medical Images by Overloading the Prompt Encoder | https://github.com/talshaharabany/AutoSAM (accessed on 27 March 2025) | https://arxiv.org/pdf/2306.06370 (accessed on 1 May 2025) | AutoSAM adapts the SAM to medical images via prompt encoder overloading. |
| 32 | All-in-sam | All-in-sam: from weak annotation to pixel-wise nuclei segmentation with prompt-based finetuning | - | https://iopscience.iop.org/article/10.1088/1742-6596/2722/1/012012/meta (accessed on 1 May 2025) | All-in-SAM automates nuclei segmentation via prompt-based finetuning from weak annotations. |
| 33 | DeSAM | DeSAM: Decoupled Segment Anything Model for Generalizable Medical Image Segmentation | https://github.com/yifangao112/DeSAM (accessed on 27 March 2025) | https://link.springer.com/chapter/10.1007/978-3-031-72390-2_48 (accessed on 1 May 2025) | DeSAM improves the SAM for generalizable medical image segmentation by decoupling prompts and mask generation. |
| 34 | SurgicalSAM | SurgicalSAM: Efficient class promptable surgical instrument segmentation | https://github.com/wenxi-yue/SurgicalSAM (accessed on 27 March 2025) | https://ojs.aaai.org/index.php/AAAI/article/view/28514 (accessed on 1 May 2025) | SurgicalSAM enables class-promptable instrument segmentation via prototype-based tuning. |
| 35 | EviPrompt | EviPrompt: A Training-Free Evidential Prompt Generation Method for Segment Anything Model in Medical Images | https://github.com/SPIresearch/EviPrompt (accessed on 27 March 2025) | http://ieeexplore.ieee.org/document/10729707/ (accessed on 1 May 2025) | EviPrompt generates training-free evidential prompts for SAM in medical images. |
| 36 | SAM-U | SAM-U: Multi-box prompts triggered uncertainty estimation for reliable SAM in medical image | - | https://link.springer.com/chapter/10.1007/978-3-031-47425-5_33 (accessed on 1 May 2025) | Multi-box prompts for an uncertainty-aware SAM in medical images. |
| 37 | Ur-sam | Ur-sam: Enhancing the reliability of segment anything model for auto-prompting medical image segmentation with uncertainty rectification | - | https://papers.ssrn.com/sol3/papers.cfm?abstract_id=4878606 (accessed on 1 May 2025) | Uncertainty-rectified SAM for auto-prompting medical segmentation. |
| 38 | nnSAM | Plug-and-play Segment Anything Model Improves nnUNet Performance | https://github.com/Kent0n-Li/nnSAM (accessed on 27 March 2025) | https://arxiv.org/pdf/2309.16967 (accessed on 1 May 2025) | nnSAM combines SAM and nnUNet for improved medical image segmentation with limited data. |
| 39 | SAMAug | Input Augmentation with SAM: Boosting Medical Image Segmentation with Segmentation Foundation Model | https://github.com/yizhezhang2000/SAMAug (accessed on 27 March 2025) | https://link.springer.com/chapter/10.1007/978-3-031-47401-9_13 (accessed on 1 May 2025) | SAMAug boosts medical image segmentation via input augmentation with the SAM. |
| 40 | DB-SAM | DB-SAM: Delving into High Quality Universal Medical Image Segmentation | https://github.com/AlfredQin/DB-SAM (accessed on 27 March 2025) | https://link.springer.com/chapter/10.1007/978-3-031-72390-2_47 (accessed on 1 May 2025) | Dual-branch SAM adapter for universal medical image segmentation. |
| 41 | SamDSK | SamDSK: Combining Segment Anything Model with Domain-Specific Knowledge for Semi-Supervised Learning in Medical Image Segmentation | https://github.com/yizhezhang2000/SamDSK (accessed on 27 March 2025) | https://link.springer.com/chapter/10.1007/978-981-97-8496-7_24 (accessed on 1 May 2025) | Combining the SAM with domain knowledge for semi-supervised medical image segmentation. |
| 42 | SAM for Semi-Supervised Medical Image Segmentation | Segment anything model for semi-supervised medical image segmentation via selecting reliable pseudo-labels | - | https://link.springer.com/chapter/10.1007/978-981-99-8141-0_11 (accessed on 1 May 2025) | An SAM for selecting reliable pseudo-labels in semi-supervised medical segmentation. |
| 43 | SemiSAM | SemiSAM: Enhancing Semi-Supervised Medical Image Segmentation via SAM-Assisted Consistency Regularization | - | https://ieeexplore.ieee.org/abstract/document/10821951 (accessed on 1 May 2025) | SAM-enhanced semi-supervised medical image segmentation. |
| 44 | 3DSAM-adapter | 3DSAM-adapter: Holistic adaptation of SAM from 2D to 3D for promptable tumor segmentation | - | https://www.sciencedirect.com/science/article/pii/S1361841524002494 (accessed on 1 May 2025) | Three-dimensional adapter for the SAM enabling promptable tumor segmentation in volumetric medical images. |
| 45 | MA-SAM | MA-SAM: Modality-agnostic SAM Adaptation for 3D Medical Image Segmentation | https://github.com/cchen-cc/MA-SAM (accessed on 27 March 2025) | https://www.sciencedirect.com/science/article/pii/S1361841524002354 (accessed on 1 May 2025) | Modality-agnostic SAM adaptation for 3D medical image segmentation. |
| 46 | Promise | Promise: Prompt-driven 3d medical image segmentation using pretrained image foundation models | https://github.com/MedICL-VU/ProMISe (accessed on 27 March 2025) | https://ieeexplore.ieee.org/document/10635207 (accessed on 1 May 2025) | Prompt-driven 3D medical segmentation using a pretrained SAM. |
| 47 | Sam3d | Sam3d: Segment anything model in volumetric medical images | https://github.com/UARK-AICV/SAM3D (accessed on 27 March 2025) | https://ieeexplore.ieee.org/document/10635844 (accessed on 1 May 2025) | SAM3D for 3D medical image segmentation. |
| 48 | SAM-Med3D | SAM-Med3D: Towards General-purpose Segmentation Models for Volumetric Medical Images | https://github.com/uni-medical/SAM-Med3D (accessed on 27 March 2025) | https://arxiv.org/pdf/2310.15161 (accessed on 1 May 2025) | General-purpose 3D medical image segmentation with promptable SAM-Med3D. |
| 49 | Segvol | Segvol: Universal and interactive volumetric medical image segmentation | https://github.com/BAAI-DCAI/SegVol (accessed on 27 March 2025) | https://proceedings.neurips.cc/paper_files/paper/2024/file/c7c7cf10082e454b9662a686ce6f1b6f-Paper-Conference.pdf (accessed on 1 May 2025) | SegVol: Universal 3D medical image segmentation with interactive prompts. |
| 50 | SAM+nnUNet | SAM+nnUNet: Deep-learning-based Head and Neck Tumor Segmentation on FDG PET | - | https://ieeexplore.ieee.org/document/10657090 (accessed on 1 May 2025) | SAM+nnUNet improves head and neck tumor segmentation on PET. |
| 51 | SAM-UNet | SAM-UNet: Enhancing Zero-Shot Segmentation of SAM for Universal Medical Images | https://github.com/Hhankyangg/sam-unet (accessed on 27 March 2025) | https://arxiv.org/pdf/2408.09886v1 (accessed on 1 May 2025) | SAM-UNet improves medical image segmentation via a U-Net-enhanced SAM. |
| 52 | SAM ViT-H D-50 PVTv2 fusion | Improving existing segmentators performance with zero-shot segmentators | https://github.com/LorisNanni/Improving-existing-segmentators-performance-with-zero-shot-segmentators (accessed on 27 March 2025) | https://www.mdpi.com/1099-4300/25/11/1502 (accessed on 1 May 2025) | The SAM enhances existing segmentation models via zero-shot fusion. |
| 54 | GazeSAM | GazeSAM: Interactive Image Segmentation with Eye Gaze and Segment Anything Model | https://github.com/ukaukaaaa/GazeSAM (accessed on 27 March 2025) | https://proceedings.mlr.press/v226/wang24a.html (accessed on 1 May 2025) | GazeSAM uses eye-tracking and SAM for automated medical image segmentation. |
| 56 | GlanceSeg | GlanceSeg: Real-time microaneurysm lesion segmentation with gaze-map-guided foundation model for early detection of diabetic retinopathy | - | https://ieeexplore.ieee.org/document/10472575/ (accessed on 1 May 2025) | Gaze-guided SAM for real-time diabetic retinopathy lesion segmentation. |
| 57 | Uni-COAL | Uni-COAL: A Unified Framework for Cross-Modality Synthesis and Super-Resolution of MR Images | - | https://www.sciencedirect.com/science/article/abs/pii/S0957417424031087 (accessed on 1 May 2025) | Unified framework for MRI cross-modality synthesis and super-resolution. |
| 58 | FastSAM-3DSlicer | FastSAM-3DSlicer: A 3D-Slicer Extension for 3D Volumetric Segment Anything Model with Uncertainty Quantification | https://github.com/arcadelab/FastSAM3D_slicer (accessed on 27 March 2025) | https://link.springer.com/chapter/10.1007/978-3-031-73471-7_1 (accessed on 1 May 2025) | 3D-Slicer extension for volumetric SAM with uncertainty quantification. |
References
- Marinov, Z.; Jäger, P.F.; Egger, J.; Kleesiek, J.; Stiefelhagen, R. Deep interactive segmentation of medical images: A systematic review and taxonomy. IEEE Trans. Pattern Anal. Mach. Intell. 2024, 46, 10998–11018. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Wang, Z.; Hong, M.; Ji, W.; Fu, H.; Xu, Y.; Xu, M.; Jin, Y. Medical sam adapter: Adapting segment anything model for medical image segmentation. Med. Image Anal. 2025, 102, 103547. [Google Scholar] [CrossRef]
- Zhang, C.; Liu, L.; Cui, Y.; Huang, G.; Lin, W.; Yang, Y.; Hu, Y. A Comprehensive Survey on Segment Anything Model for Vision and Beyond. arXiv 2023, arXiv:2305.08196. [Google Scholar]
- Kirillov, A.; Mintun, E.; Ravi, N.; Mao, H.; Rolland, C.; Gustafson, L.; Xiao, T.; Whitehead, S.; Berg, A.C.; Lo, P.D.; et al. Segment anything. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Paris, France, 4–6 October 2023; pp. 4015–4026. [Google Scholar]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An Image Is Worth 16X16 Words: Transformers for Image Recognition at Scale. In Proceedings of the ICLR 2021—9th International Conference on Learning Representations, Virtual, 3–7 May 2021. [Google Scholar]
- He, K.; Chen, X.; Xie, S.; Li, Y.; Dollár, P.; Girshick, R. Masked autoencoders are scalable vision learners. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, New Orleans, LA, USA, 21–24 June 2022; pp. 16000–16009. [Google Scholar]
- Vaswani, A. Attention is all you need. Adv. Neural Inf. Process. Syst. 2017, 30, 1. [Google Scholar]
- Ross, T.Y.; Dollár, G. Focal loss for dense object detection. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 2980–2988. [Google Scholar]
- Milletari, F.; Navab, N.; Ahmadi, S.A. V-net: Fully convolutional neural networks for volumetric medical image segmentation. In Proceedings of the 2016 Fourth International Conference on 3D Vision (3DV), Stanford, CA, USA, 25–28 October 2016; pp. 565–571. [Google Scholar]
- Roy, S.; Wald, T.; Koehler, G.; Rokuss, M.; Disch, N.; Holzschuh, J.C.; Zimmerer, D.; Maier-Hein, K.H. SAM.MD: Zero-shot medical image segmentation capabilities of the Segment Anything Model. arXiv 2023, arXiv:abs/2304.05396. [Google Scholar]
- Ji, Y.; Bai, H.; Ge, C.; Yang, J.; Zhu, Y.; Zhang, R.; Li, Z.; Zhang, L.; Ma, W.; Wan, X.; et al. Amos: A large-scale abdominal multi-organ benchmark for versatile medical image segmentation. Adv. Neural Inf. Process. Syst. 2022, 35, 36722–36732. [Google Scholar]
- Hu, C.; Li, X. When sam meets medical images: An investigation of segment anything model (sam) on multi-phase liver tumor segmentation. arXiv 2023, arXiv:2304.08506. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Medical Image Computing and Computer-Assisted Intervention–MICCAI 2015, Proceedings of the 18th International Conference, Munich, Germany, 5–9 October 2015; Proceedings, Part III 18; Springer International Publishing: Berlin/Heidelberg, Germany, 2015; pp. 234–241. [Google Scholar]
- Zhou, T.; Zhang, Y.; Zhou, Y.; Wu, Y.; Gong, C. Can sam segment polyps? arXiv 2023, arXiv:2304.07583. [Google Scholar]
- Huang, Y.; Yang, X.; Liu, L.; Zhou, H.; Chang, A.; Zhou, X.; Chen, R.; Yu, J.; Chen, J.; Chen, C.; et al. Segment anything model for medical images? Med. Image Anal. 2024, 92, 103061. [Google Scholar] [CrossRef]
- He, S.; Bao, R.; Li, J.; Grant, P.E.; Ou, Y. Accuracy of segment-anything model (sam) in medical image segmentation tasks. CoRR 2023, arXiv:2304.09324. [Google Scholar]
- Mazurowski, M.A.; Dong, H.; Gu, H.; Yang, J.; Konz, N.; Zhang, Y. Segment anything model for medical image analysis: An experimental study. Med. Image Anal. 2023, 89, 102918. [Google Scholar] [CrossRef]
- Cheng, D.; Qin, Z.; Jiang, Z.; Zhang, S.; Lao, Q.; Li, K. Sam on medical images: A comprehensive study on three prompt modes. arXiv 2023, arXiv:2305.00035. [Google Scholar]
- Zhang, L.; Liu, Z.; Zhang, L.; Wu, Z.; Yu, X.; Holmes, J.; Feng, H.; Dai, H.; Li, X.; Li, Q.; et al. Segment anything model (sam) for radiation oncology. arXiv 2023, arXiv:2306.11730. [Google Scholar]
- Hu, M.; Li, Y.; Yang, X. Skinsam: Empowering skin cancer segmentation with segment anything model. arXiv 2023, arXiv:2304.13973. [Google Scholar]
- Li, Y.; Hu, M.; Yang, X. Polyp-sam: Transfer sam for polyp segmentation. In Proceedings of the Medical Imaging 2024: Computer-Aided Diagnosis, San Diego, CA, USA, 18–22 February 2024; Volume 12927, pp. 759–765. [Google Scholar]
- Ma, J.; He, Y.; Li, F.; Han, L.; You, C.; Wang, B. Segment anything in medical images. Nat. Commun. 2024, 15, 654. [Google Scholar] [CrossRef]
- Hu, E.; Shen, Y.; Wallis, P.; Allen-Zhu, Z.; Li, Y.; Wang, S.; Wang, L.; Chen, W. Lora: Low-Rank Adaptation of Large Language Models. In Proceedings of the ICLR 2022—10th International Conference on Learning Representations, Virtually, 25–29 April 2022. [Google Scholar]
- Zhang, K.; Liu, D. Customized segment anything model for medical image segmentation. arXiv 2023, arXiv:2304.13785. [Google Scholar]
- Feng, W.; Zhu, L.; Yu, L. Cheap lunch for medical image segmentation by fine-tuning sam on few exemplars. In International MICCAI Brainlesion Workshop; Springer Nature: Cham, Switzerland, 2023; pp. 13–22. [Google Scholar]
- Paranjape, J.N.; Nair, N.G.; Sikder, S.; Vedula, S.S.; Patel, V.M. Adaptivesam: Towards efficient tuning of sam for surgical scene segmentation. In Annual Conference on Medical Image Understanding and Analysis; Springer Nature: Cham, Switzerland, 2024; pp. 187–201. [Google Scholar]
- Cheng, J.; Ye, J.; Deng, Z.; Chen, J.; Li, T.; Wang, H.; Su, Y.; Huang, Z.; Chen, J.; Jiang, L.; et al. Sam-med2d. arXiv 2023, arXiv:2308.16184. [Google Scholar]
- Xie, W.; Willems, N.; Patil, S.; Li, Y.; Kumar, M. Sam fewshot finetuning for anatomical segmentation in medical images. In Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision, Waikoloa, HI, USA, 3–8 January 2024; pp. 3253–3261. [Google Scholar]
- Paranjape, J.N.; Sikder, S.; Vedula, S.S.; Patel, V.M. S-sam: Svd-based fine-tuning of segment anything model for medical image segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Marrakesh, Morocco, 6–10 October 2024; Springer Nature: Cham, Switzerland, 2024; pp. 720–730. [Google Scholar]
- Pandey, S.; Chen, K.F.; Dam, E.B. Comprehensive multimodal segmentation in medical imaging: Combining yolov8 with sam and hq-sam models. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Paris, France, 4–6 October 2023; pp. 2592–2598. [Google Scholar]
- Reis, D.; Kupec, J.; Hong, J.; Daoudi, A. Real-time flying object detection with YOLOv8. arXiv 2023, arXiv:2305.09972. [Google Scholar]
- Lei, W.; Xu, W.; Li, K.; Zhang, X.; Zhang, S. MedLSAM: Localize and segment anything model for 3D CT images. Med. Image Anal. 2025, 99, 103370. [Google Scholar] [CrossRef]
- Anand, D.; Singhal, V.; Shanbhag, D.D.; Ks, S.; Patil, U.; Bhushan, C.; Manickam, K.; Gui, D.; Mullick, R.; Gopal, A.; et al. One-shot localization and segmentation of medical images with foundation models. arXiv 2023, arXiv:2310.18642. [Google Scholar]
- Shaharabany, T.; Dahan, A.; Giryes, R.; Wolf, L. AutoSAM: Adapting SAM to Medical Images by Overloading the Prompt Encoder. arXiv 2023, arXiv:2306.06370. [Google Scholar]
- Cui, C.; Deng, R.; Liu, Q.; Yao, T.; Bao, S.; Remedios, L.W.; Landman, B.A.; Tang, Y.; Hou, Y. All-in-sam: From weak annotation to pixel-wise nuclei segmentation with prompt-based finetuning. J. Phys. Conf. Ser. 2024, 2722, 012012. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Xia, W.; Hu, D.; Wang, W.; Gao, X. DeSAM: Decoupling Segment Anything Model for Generalizable Medical Image Segmentation. arXiv 2023, arXiv:2306.00499. [Google Scholar]
- Yue, W.; Zhang, J.; Hu, K.; Xia, Y.; Luo, J.; Wang, Z. Surgicalsam: Efficient class promptable surgical instrument segmentation. In Proceedings of the AAAI Conference on Artificial Intelligence, Vancouver, BC, Canada, 26–27 February 2024; Volume 38, pp. 6890–6898. [Google Scholar]
- Xu, Y.; Tang, J.; Men, A.; Chen, Q. EviPrompt: A Training-Free Evidential Prompt Generation Method for Adapting Segment Anything Model in Medical Images. IEEE Trans. Image Process. 2024, 33, 6204–6215. [Google Scholar] [CrossRef]
- Deng, G.; Zou, K.; Ren, K.; Wang, M.; Yuan, X.; Ying, S.; Fu, H. SAM-U: Multi-box prompts triggered uncertainty estimation for reliable SAM in medical image. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Vancouver, BC, Canada, 8–12 October 2023; Springer Nature: Cham, Switzerland, 2023; pp. 368–377. [Google Scholar]
- Zhang, Y.; Hu, S.; Ren, S.; Pan, T.; Jiang, C.; Cheng, Y.; Qi, Y. Ur-sam: Enhancing the reliability of segment anything model for auto-prompting medical image segmentation with uncertainty rectification. Available online: https://ssrn.com/abstract=4878606 (accessed on 1 May 2025).
- Zhang, J.; Ma, K.; Kapse, S.; Saltz, J.; Vakalopoulou, M.; Prasanna, P.; Samaras, D. Sam-path: A segment anything model for semantic segmentation in digital pathology. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Vancouver, BC, Canada, 8–12 October 2023; Springer Nature: Cham, Switzerland, 2023; pp. 161–170. [Google Scholar]
- Chai, S.; Jain, R.K.; Teng, S.; Liu, J.; Li, Y.; Tateyama, T.; Chen, Y.-W. Ladder fine-tuning approach for sam integrating complementary network. Procedia Comput. Sci. 2024, 246, 4951–4958. [Google Scholar] [CrossRef]
- Li, Y.; Jing, B.; Li, Z.; Wang, J.; Zhang, Y. Plug-and-play segment anything model improves nnUNet performance. Med. Phys. 2024, 52, 899–912. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhou, T.; Wang, S.; Liang, P.; Zhang, Y.; Chen, D.Z. Input augmentation with sam: Boosting medical image segmentation with segmentation foundation model. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Vancouver, BC, Canada, 8–12 October 2023; Springer Nature: Cham, Switzerland, 2023; pp. 129–139. [Google Scholar]
- Qin, C.; Cao, J.; Fu, H.; Khan, F.S.; Anwer, R.M. DB-SAM: Delving into High Quality Universal Medical Image Segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Marrakesh, Morocco, 6–10 October 2024; Springer Nature: Cham, Switzerland, 2024; pp. 498–508. [Google Scholar]
- Zhang, Y.; Zhou, T.; Wu, Y.; Gu, P.; Wang, S. Combining Segment Anything Model with Domain-Specific Knowledge for Semi-Supervised Learning in Medical Image Segmentation. In Proceedings of the Chinese Conference on Pattern Recognition and Computer Vision (PRCV), Urumqi, China, 18–20 October 2024; Springer Nature: Singapore, 2024; pp. 343–357. [Google Scholar]
- Li, N.; Xiong, L.; Qiu, W.; Pan, Y.; Luo, Y.; Zhang, Y. Segment anything model for semi-supervised medical image segmentation via selecting reliable pseudo-labels. In Proceedings of the International Conference on Neural Information Processing, Changsha, China, 20–23 November 2023; Springer Nature: Singapore, 2023; pp. 138–149. [Google Scholar]
- Zhang, Y.; Yang, J.; Liu, Y.; Cheng, Y.; Qi, Y. SemiSAM: Enhancing Semi-Supervised Medical Image Segmentation via SAM-Assisted Consistency Regularization. In Proceedings of the 2024 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Istanbul, Turkey, 5–8 December 2024; pp. 3982–3986. [Google Scholar]
- Gong, S.; Zhong, Y.; Ma, W.; Li, J.; Wang, Z.; Zhang, J.; Heng, P.-A.; Dou, Q. 3dsam-adapter: Holistic adaptation of sam from 2d to 3d for promptable tumor segmentation. Med. Image Anal. 2024, 98, 103324. [Google Scholar] [CrossRef]
- Chen, C.; Miao, J.; Wu, D.; Zhong, A.; Yan, Z.; Kim, S.; Hu, J.; Liu, Z.; Sun, L.; Li, X.; et al. Ma-sam: Modality-agnostic sam adaptation for 3d medical image segmentation. Med. Image Anal. 2024, 98, 103310. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Liu, H.; Hu, D.; Wang, J.; Oguz, I. Promise: Prompt-driven 3d medical image segmentation using pretrained image foundation models. In Proceedings of the 2024 IEEE International Symposium on Biomedical Imaging (ISBI), Athens, Greece, 27–30 May 2024; pp. 1–5. [Google Scholar]
- Bui, N.T.; Hoang, D.H.; Tran, M.T.; Doretto, G.; Adjeroh, D.; Patel, B. Sam3d: Segment anything model in volumetric medical images. In Proceedings of the 2024 IEEE International Symposium on Biomedical Imaging (ISBI), Athens, Greece, 27–30 May 2024; pp. 1–4. [Google Scholar]
- Wang, H.; Guo, S.; Ye, J.; Deng, Z.; Cheng, J.; Li, T.; Chen, J.; Su, Y.; Huang, Z.; Shen, Y.; et al. SAM-Med3D: Towards General-purpose Segmentation Models for Volumetric Medical Images. arXiv 2023, arXiv:2310.15161. [Google Scholar]
- Du, Y.; Bai, F.; Huang, T.; Zhao, B. Segvol: Universal and interactive volumetric medical image segmentation. Adv. Neural Inf. Process. Syst. 2024, 37, 110746–110783. [Google Scholar]
- Fang, Z.; Lu, Z.; Liu, H.; Liu, Y.; Mok, G.S.P. SAM+ nnUNet: Deep-learning-based Head and Neck Tumor Segmentation on FDG PET. In Proceedings of the 2024 IEEE Nuclear Science Symposium (NSS), Medical Imaging Conference (MIC) and Room Temperature Semiconductor Detector Conference (RTSD), Tampa, FL, USA, 26 October–2 November 2024; pp. 1–2. [Google Scholar]
- Yang, S.; Bi, H.; Zhang, H.; Sun, J. SAM-UNet: Enhancing Zero-Shot Segmentation of SAM for Universal Medical Images. arXiv 2024, arXiv:2408.09886. [Google Scholar]
- Nanni, L.; Fusaro, D.; Fantozzi, C.; Pretto, A. Improving existing segmentators performance with zero-shot segmentators. Entropy 2023, 25, 1502. [Google Scholar] [CrossRef]
- Fu, G.; Nichelli, L.; Herran, D.; Valabregue, R.; Alentom, A.; Hoang-Xuan, K.; Houiller, C.; Dormont, D.; Lehéricy, S.; Colliot, O. Comparing foundation models and nnU-Net for segmentation of primary brain lymphoma on clinical routine post-contrast T1-weighted MRI. In Medical Imaging 2025: Clinical and Biomedical Imaging; SPIE: Bellingham, WA, USA, 2025; p. 13410. [Google Scholar]
- Wang, B.; Aboah, A.; Zhang, Z.; Pan, H.; Bagci, U. GazeSAM: Interactive Image Segmentation with Eye Gaze and Segment Anything Model. In Proceedings of the Gaze Meets Machine Learning Workshop, New Orleans, LA, USA, 30 November 2024; Volume 226, pp. 254–265. [Google Scholar]
- Ning, G.; Liang, H.; Jiang, Z.; Zhang, H.; Liao, H. The potential of Segment Anything’ (SAM) for universal intelligent ultrasound image guidance. Biosci. Trends 2023, 17, 230–233. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Gao, M.; Liu, Z.; Tang, C.; Zhang, X.; Jiang, S.; Yuan, W.; Liu, J. GlanceSeg: Real-time microaneurysm lesion segmentation with gaze-map-guided foundation model for early detection of diabetic retinopathy. IEEE J. Biomed. Health Inform. 2024, 2024, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Song, Z.; Qi, Z.; Wang, X.; Zhao, X.; Shen, Z.; Wang, S.; Fei, M.; Wang, Z.; Zang, D.; Chen, D.; et al. Uni-COAL: A unified framework for cross-modality synthesis and super-resolution of MR images. Expert Syst. Appl. 2025, 270, 126241. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, J.; She, Z.; Kheradmand, A.; Armand, M. Samm (segment any medical model): A 3d slicer integration to sam. arXiv 2023, arXiv:2304.05622. [Google Scholar]
- Shen, C.; Li, W.; Zhang, Y.; Wang, Y.; Wang, X. Temporally-extended prompts optimization for sam in interactive medical image segmentation. In Proceedings of the 2023 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Istanbul, Turkiye, 5–8 December 2023; pp. 3550–3557. [Google Scholar]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fan, K.; Liang, L.; Li, H.; Situ, W.; Zhao, W.; Li, G. Research on Medical Image Segmentation Based on SAM and Its Future Prospects. Bioengineering 2025, 12, 608. https://doi.org/10.3390/bioengineering12060608
Fan K, Liang L, Li H, Situ W, Zhao W, Li G. Research on Medical Image Segmentation Based on SAM and Its Future Prospects. Bioengineering. 2025; 12(6):608. https://doi.org/10.3390/bioengineering12060608
Chicago/Turabian StyleFan, Kangxu, Liang Liang, Hao Li, Weijun Situ, Wei Zhao, and Ge Li. 2025. "Research on Medical Image Segmentation Based on SAM and Its Future Prospects" Bioengineering 12, no. 6: 608. https://doi.org/10.3390/bioengineering12060608
APA StyleFan, K., Liang, L., Li, H., Situ, W., Zhao, W., & Li, G. (2025). Research on Medical Image Segmentation Based on SAM and Its Future Prospects. Bioengineering, 12(6), 608. https://doi.org/10.3390/bioengineering12060608






