Regional Brain Aging Disparity Index: Region-Specific Brain Aging State Index for Neurodegenerative Diseases and Chronic Disease Specificity
Abstract
1. Introduction
2. Materials and Methods
2.1. Database
2.2. Image Preprocessing
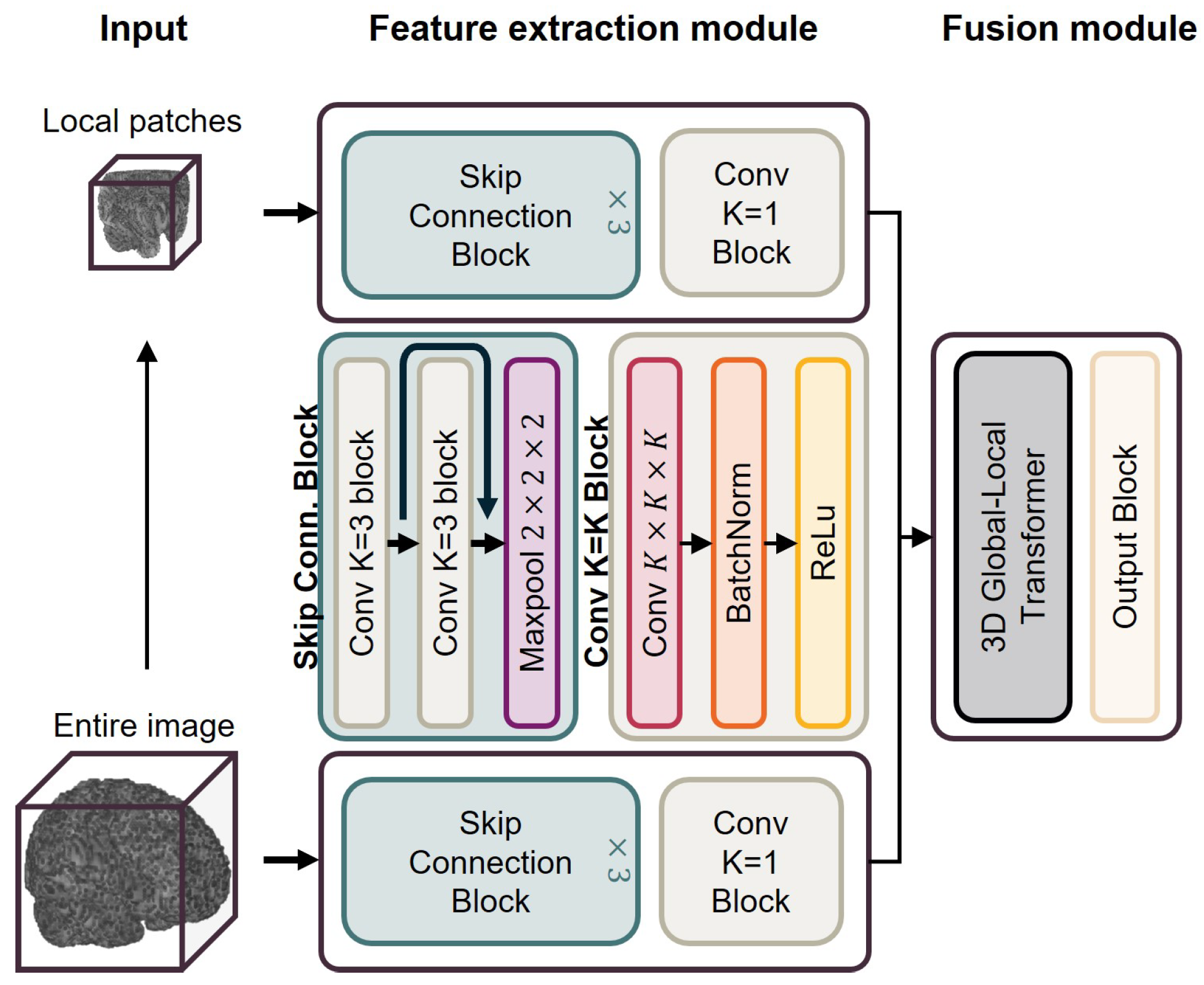
2.3. Brain Age Prediction Model
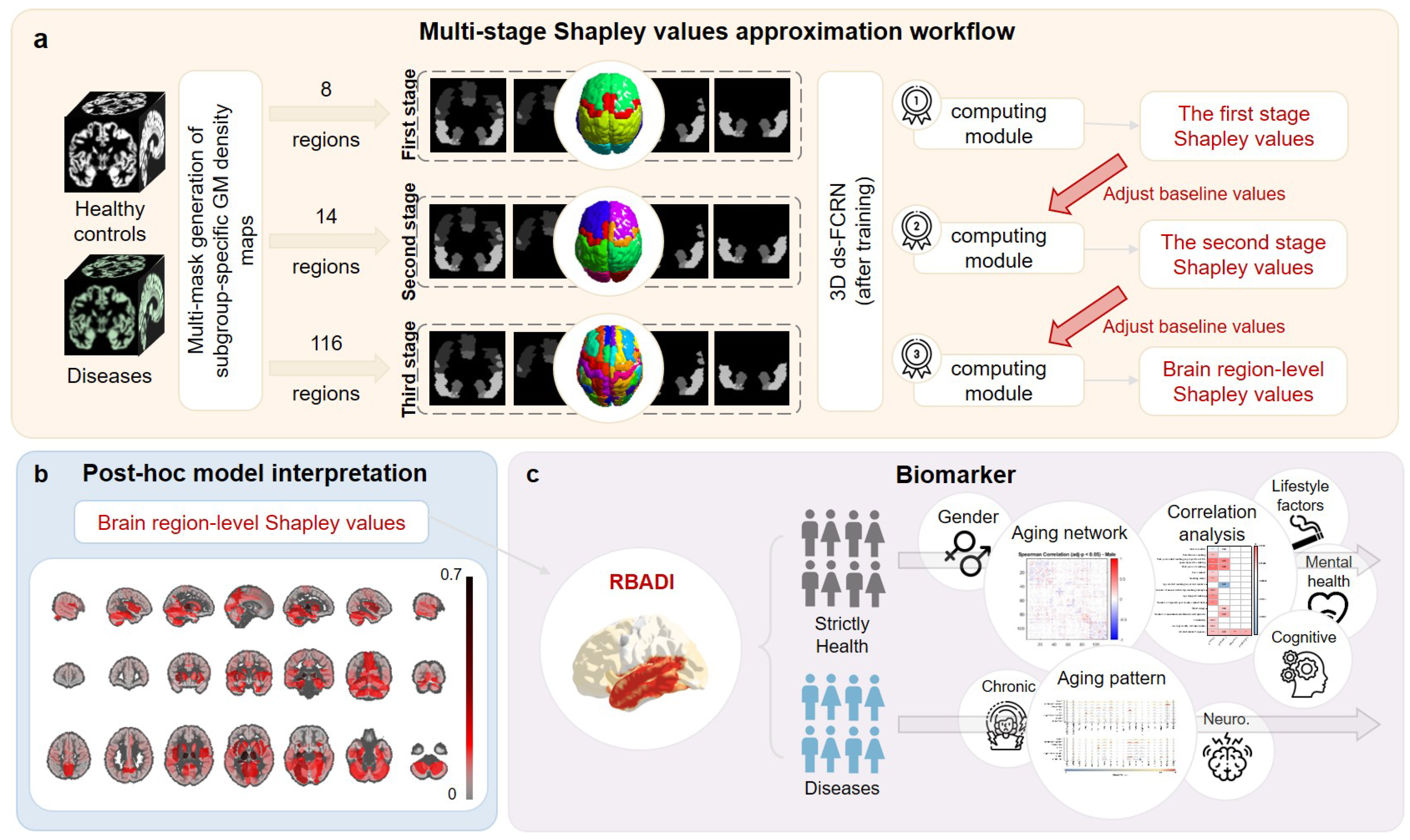
2.4. Multi-Stage Shapley Values Approximation Workflow
2.5. Model Interpretation and Regional Brain Aging Disparity Index (RBADI)
2.6. Identification of Regional Brain Aging Patterns
2.7. Classification Experiment for Stage Recognition of Neurodegenerative Diseases
3. Results
3.1. Performance and Prediction of the Brain Age Model
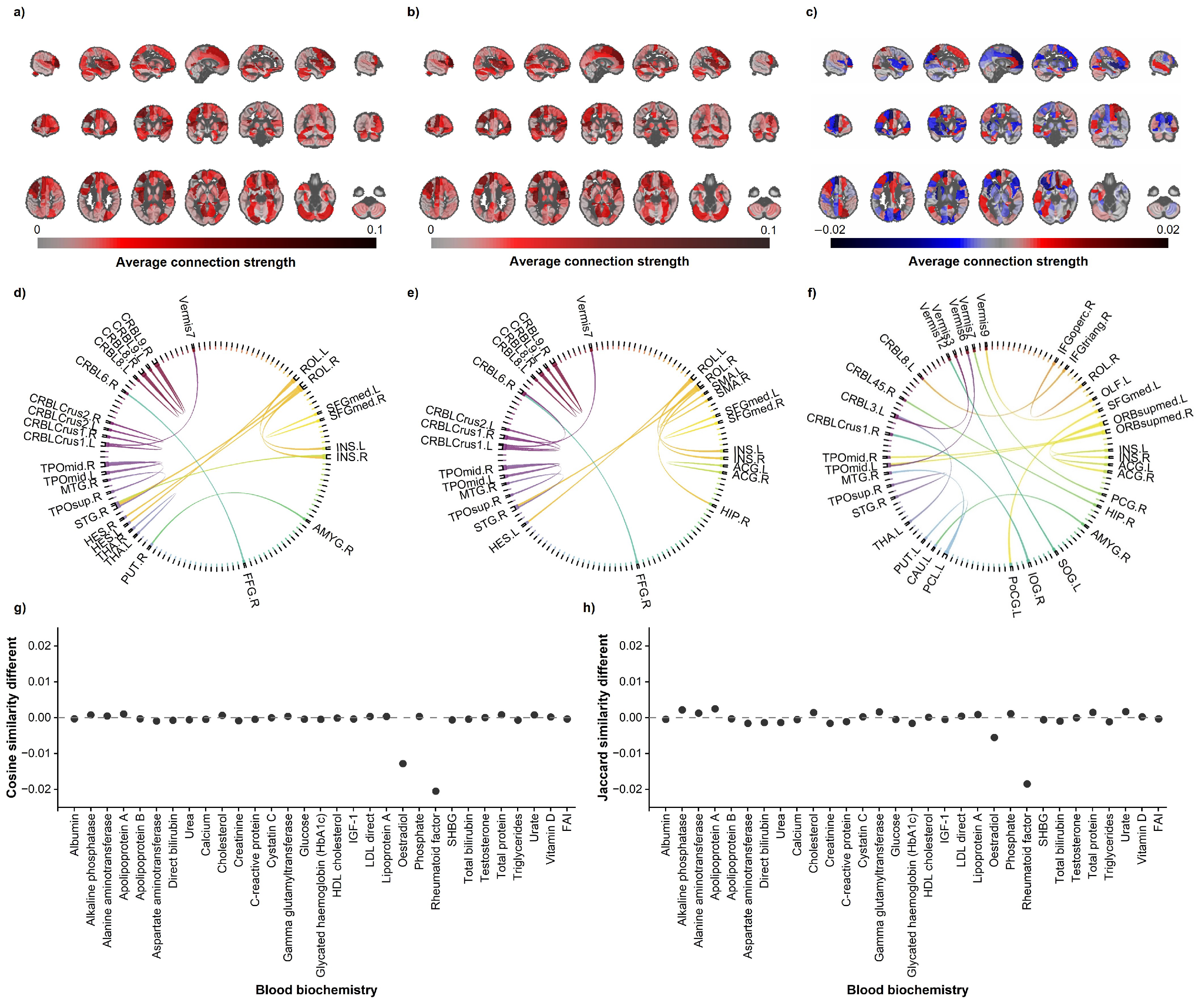
3.2. Regional-Level Contributions
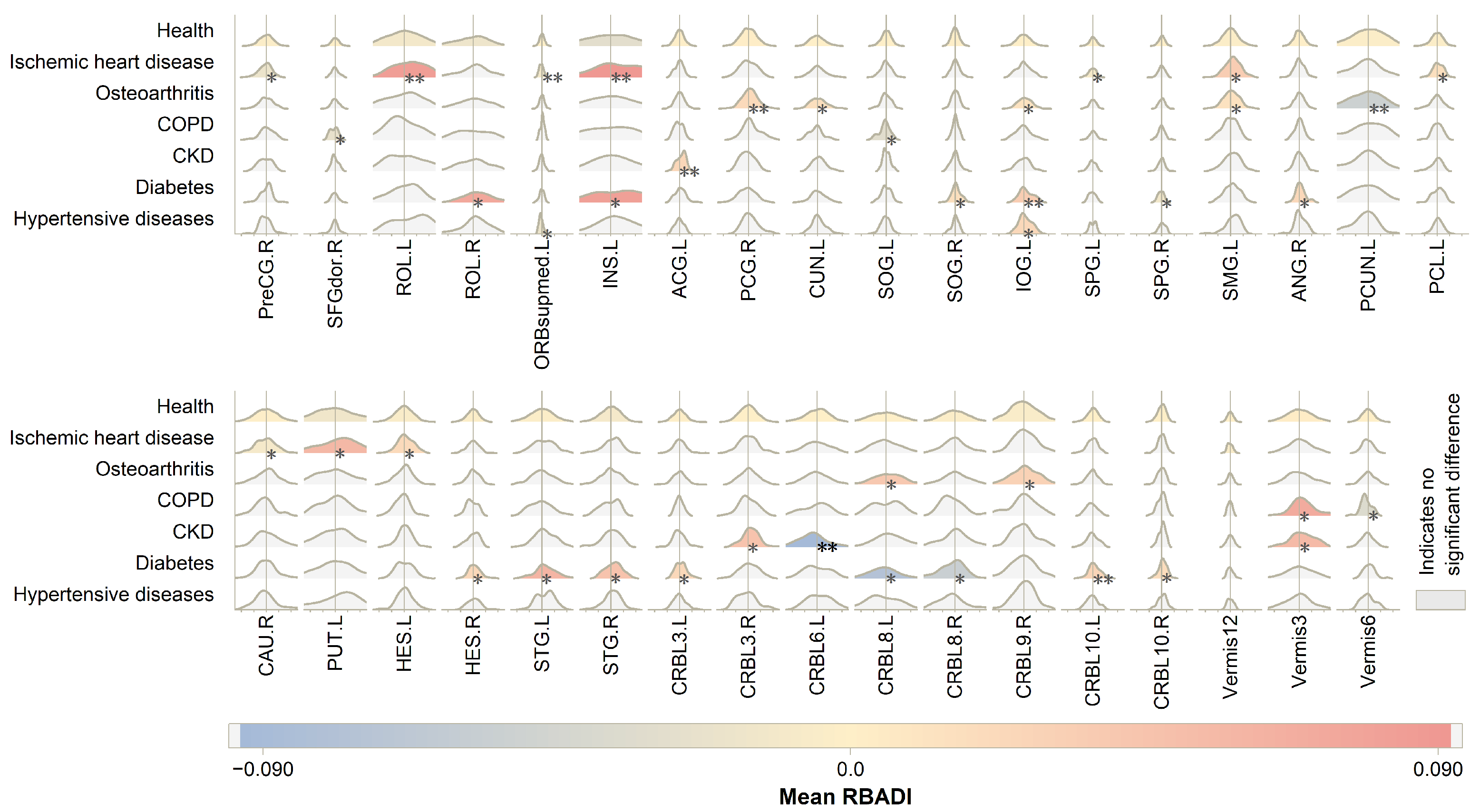
3.3. Regional Brain Aging Patterns
3.4. Classification Experiment for Stage Recognition of Neurodegenerative Diseases
4. Discussion
- (1)
- The SHAP method is not the sole interpretability approach applied in brain age prediction models. In fact, a diverse array of interpretability techniques has been successfully implemented in these frameworks. In addition to conventional methods such as CAM and saliency map techniques, other advanced approaches including Layer-wise Relevance Propagation (LRP) algorithms [32,33], occlusion sensitivity analysis [34,35], and attention-score-based approaches [36,37] have been systematically employed and incorporated into model interpretation architectures. However, the strength of the SHAP method lies in its role as a model-agnostic post hoc explanatory approach. This means SHAP demonstrates strong versatility in interpretation, applicable regardless of the model architecture or data type employed.
- (2)
- The multi-stage Shapley values computation workflow offers an effective approach for calculating Shapley values, as it constitutes a model-agnostic post hoc explanation method that enables modular application to any deep learning model. Compared with the original Shapley values algorithm’s complexity, this algorithm significantly reduces computational complexity, enabling the computation of Shapley values at the brain regional level. Taking the AAL116 template as an example, the original algorithm would require (equal to ) operations to obtain Shapley values for each brain region, making such computation completely infeasible for relatively complex deep learning models. Through our proposed approximation algorithm, the computational complexity is reduced to three sequential stages: (equal to ) operations in the first stage, (equal to ) operations in the second stage, and (equal to ) operations in the third stage. This achieves a remarkable reduction of approximately orders of magnitude in computational complexity compared to the original algorithm. Taking the 3D ds-FCRN architecture employed in this study as an example, performing a single age prediction for 3276 subjects in the test set requires approximately 15 s during the post hoc explanation process. The approximate computational workflow necessitates operations with a total execution time of 52 h, while the standard Shapley value computation method would require operations, amounting to h of computational time. This efficient approximation algorithm has enabled the application of Shapley value interpretation methods in deep learning, thereby facilitating the construction of the RBADI. The framework achieves region-level aging interpretation and reveals significant correlations between anomalous aging and factors including lifestyle patterns, mental health status, cognitive performance, and chronic disease profiles.
- (3)
- The prominent contributions of the THA and HIP in brain age prediction models highlight the central role of these regions in cerebral aging processes, identifying them as a key predictor of brain age in HCs. Notably, among all brain regions, THA demonstrates the highest contribution value and with left-lateralized characteristics. These findings align with previous research. For instance, Wang et al. reported that THA exhibits high predictive weights in structural MRI models, where reduced GM volume and increased WM volume serve as critical features for brain age prediction [38]. Similarly, Zhang et al. identified THA and the CAU as pivotal regions for brain age prediction across the adult lifespan through an occlusion analysis of brain network atlases, with the left hemisphere (particularly THA.L) showing more pronounced contributions [39]. A plausible explanation for this lateralization involves THA’s asymmetric structural development and aging patterns [40], coupled with the heightened sensitivity of THA.L-mediated language information processing to age-related changes [41,42]. Furthermore, HIP emerges as an exceptionally critical region within the TL. Responsible for memory formation and spatial navigation, HIP plays a vital role in cognitive processes vulnerable to aging [43]. This study further reveals the significant contribution of HIP, especially HIP.L, in brain age prediction frameworks.
- (4)
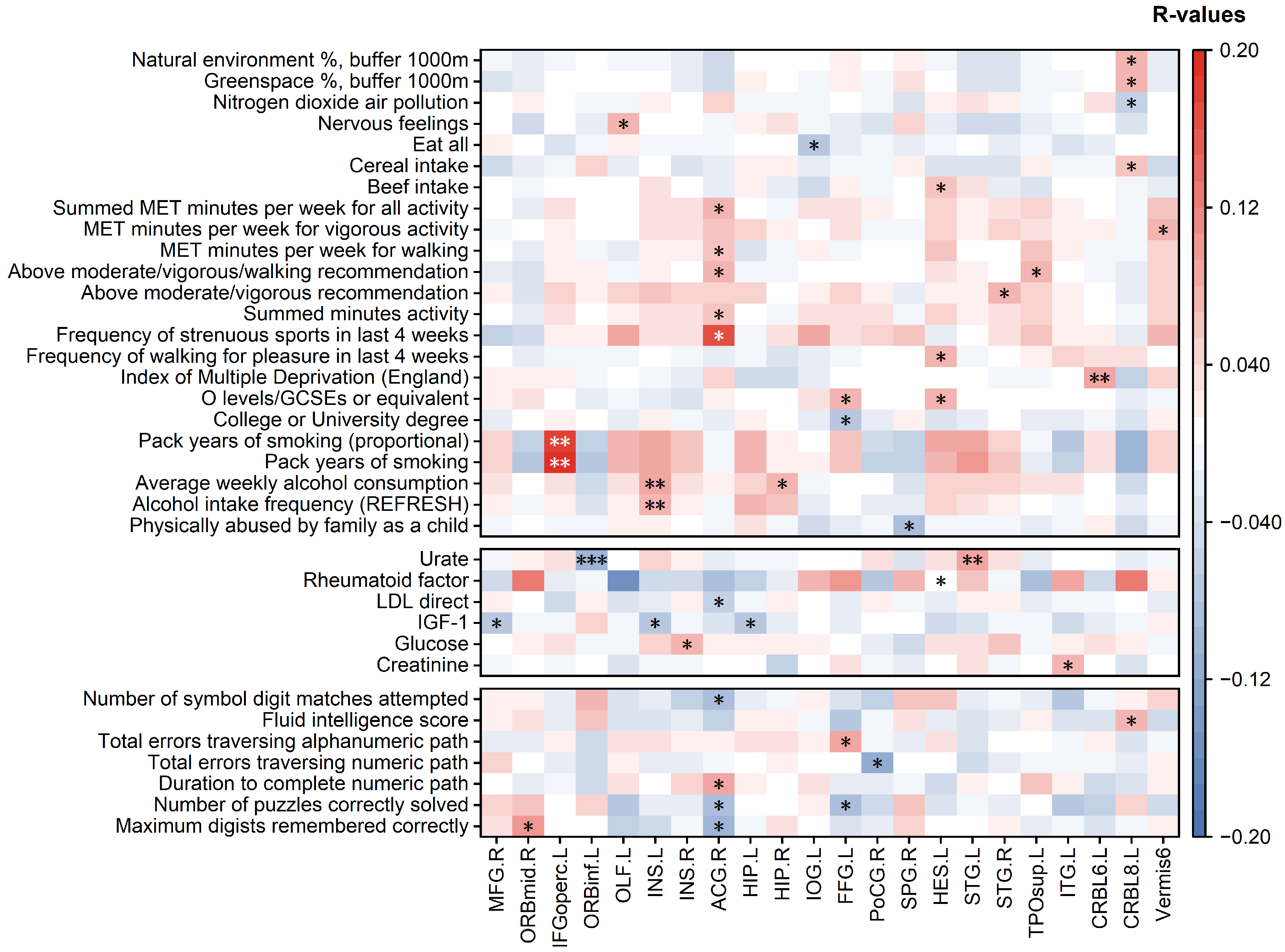
- The RBADI model provides an analytical framework with higher spatial resolution for deciphering the intricate relationships between lifestyle factors, mental health, cognitive abilities, and accelerated brain aging. Our findings reveal a significant association between abnormal aging in the IFGoperc.L and smoking duration. Previous studies have demonstrated that smoking behavior affects pain regulation, sleep quality, and depressive/anxiety symptoms through inferior frontal gyrus-mediated neural mechanisms [44,45]. Specifically, Addicott et al. found that smokers exhibited significantly enhanced task-based functional connectivity between the left inferior frontal gyrus (including the opercular part) and auditory cortex during cognitive-affective distress tasks, suggesting that smokers may require stronger IFGoperc activation to cope with distress-related stimuli [44]. Furthermore, Chen et al. demonstrated that enhanced functional connectivity between the hypothalamus and right inferior frontal gyrus in smokers was associated with poorer PSQI sleep quality scores, potentially reflecting neural compensatory mechanisms to maintain alertness and attentional control [45]. Additionally, the strong correlation between INS and alcohol consumption may stem from alcohol-induced alterations in insular functional connectivity patterns that impair cognitive control and reinforce positive alcohol expectancy [46]. Notably, the insula also plays a pivotal regulatory role in the stress–alcohol use pathway [47]. In conclusion, compared with BAG, the RBADI demonstrates superior regional specificity in brain aging assessments.
- (5)
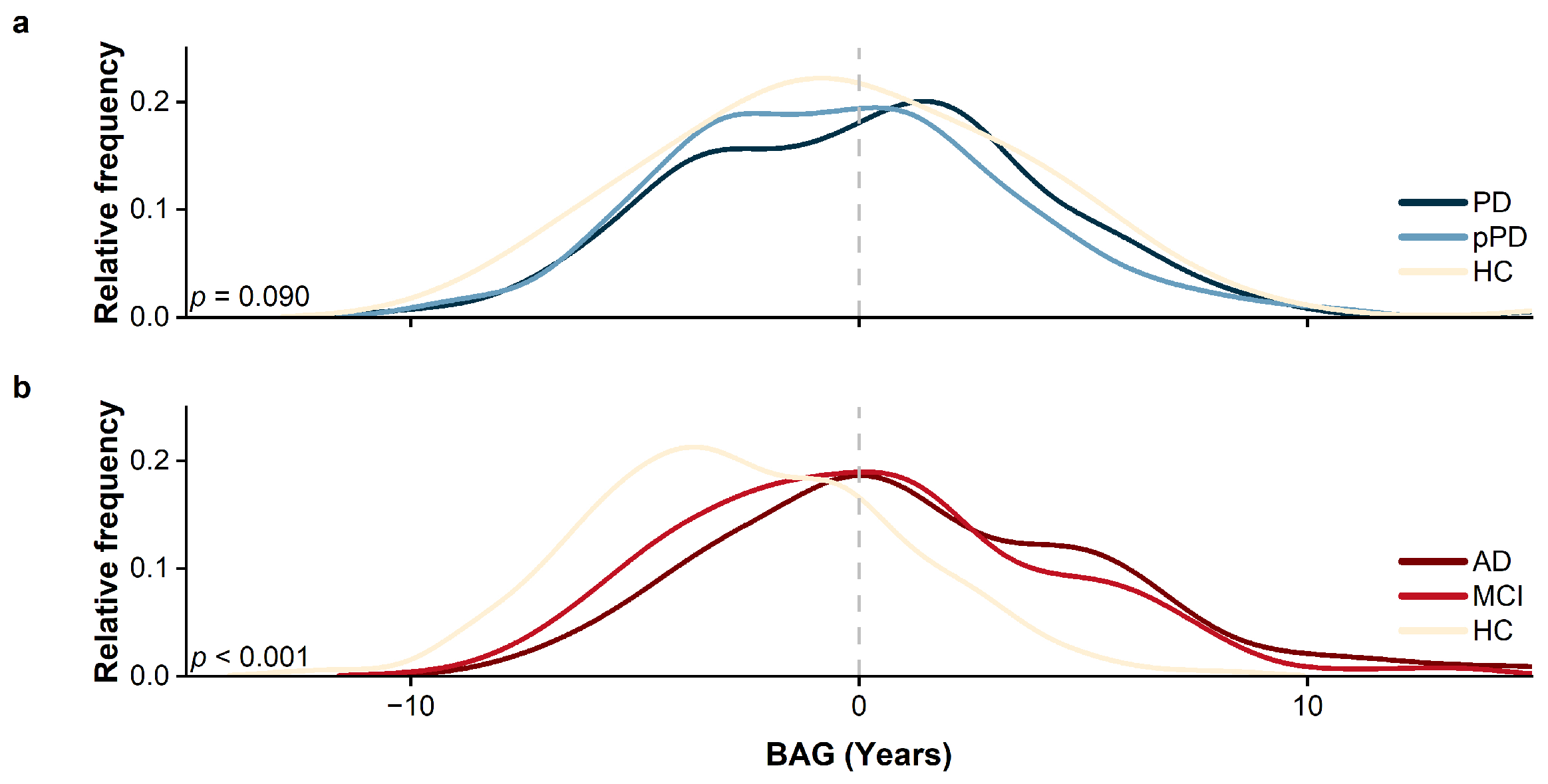
- When employing BAG for identifying prodromal neurodegenerative disorders (AD and PD), although significant differences were observed in the HC vs. MCI vs. AD groups, its discriminative capability remained unsatisfactory in distinguishing the HC vs. pPD vs. PD cohorts. The classification model trained with BAG, gender, and age as input features demonstrated a poor performance, only marginally better than random guessing. Compared with the previously well-performing BAV, our proposed RBADI provides age-correlated metrics that not only exhibit stronger competitiveness in neurodegenerative disease screening but also substantiate the pivotal role of brain aging in the progression of neurodegenerative disorders.
- (1)
- The dataset employed in this study is characterized by racial representativeness imbalance [48,49]. Although this phenomenon is prevalent across research domains, it may still compromise the representativeness of the RBADI analytical outcomes. Future investigations should prioritize the incorporation of datasets encompassing diverse ethnic populations to validate the generalizability of the RBADI. Additionally, consideration should be given to implementing multi-center data integration strategies to address potential representativeness limitations [50,51,52].
- (2)
- The current study solely utilized single-modality T1 MRI data, which is inherently limited to providing structural information. The integration of multimodal imaging data not only further enhances the predictive performance of models but also offers additional perspectives for post hoc model interpretability. Therefore, the primary focus of subsequent research will be to incorporate multimodal data to enable a more comprehensive investigation.
- (3)
- Although the rigorous data preprocessing protocols and stringent quality control criteria excluded low-quality T1 MRI data, this approach may limit the model’s generalizability to noise-corrupted data. Considering that adversarial examples containing noise can easily interfere with deep neural networks and compromise their performance [53], future developments in model design and interpretation modules should incorporate adversarial noise considerations to enhance the robustness of both the predictive models and explanatory methodologies.
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Franke, K.; Gaser, C. Ten years of BrainAGE as a neuroimaging biomarker of brain aging: What insights have we gained? Front. Neurol. 2019, 10, 789. [Google Scholar] [CrossRef] [PubMed]
- Cole, J.H.; Poudel, R.P.; Tsagkrasoulis, D.; Caan, M.W.; Steves, C.; Spector, T.D.; Montana, G. Predicting brain age with deep learning from raw imaging data results in a reliable and heritable biomarker. NeuroImage 2017, 163, 115–124. [Google Scholar] [CrossRef]
- Franke, K.; Ziegler, G.; Klöppel, S.; Gaser, C.; Alzheimer’s Disease Neuroimaging Initiative. Estimating the age of healthy subjects from T1-weighted MRI scans using kernel methods: Exploring the influence of various parameters. NeuroImage 2010, 50, 883–892. [Google Scholar] [CrossRef] [PubMed]
- Lemaitre, H.; Goldman, A.L.; Sambataro, F.; Verchinski, B.A.; Meyer-Lindenberg, A.; Weinberger, D.R.; Mattay, V.S. Normal age-related brain morphometric changes: Nonuniformity across cortical thickness, surface area and gray matter volume? Neurobiol. Aging 2012, 33, 617.e1–617.e9. [Google Scholar] [CrossRef] [PubMed]
- Long, X.; Liao, W.; Jiang, C.; Liang, D.; Qiu, B.; Zhang, L. Healthy aging: An automatic analysis of global and regional morphological alterations of human brain. Acad. Radiol. 2012, 19, 785–793. [Google Scholar] [CrossRef]
- Coupé, P.; Manjón, J.V.; Mansencal, B.; Tourdias, T.; Catheline, G.; Planche, V. Hippocampal-amygdalo-ventricular atrophy score: Alzheimer disease detection using normative and pathological lifespan models. Hum. Brain Mapp. 2022, 43, 3270–3282. [Google Scholar] [CrossRef]
- Coupé, P.; Manjón, J.V.; Lanuza, E.; Catheline, G. Lifespan changes of the human brain in Alzheimer’s disease. Sci. Rep. 2019, 9, 3998. [Google Scholar] [CrossRef]
- Cole, J.H.; Franke, K. Predicting age using neuroimaging: Innovative brain ageing biomarkers. Trends Neurosci. 2017, 40, 681–690. [Google Scholar] [CrossRef]
- Franke, K.; Gaser, C. Longitudinal changes in individual BrainAGE in healthy aging, mild cognitive impairment, and Alzheimer’s disease. GeroPsych 2012, 25, 235–245. [Google Scholar] [CrossRef]
- Gaser, C.; Franke, K.; Klöppel, S.; Koutsouleris, N.; Sauer, H.; Alzheimer’s Disease Neuroimaging Initiative. BrainAGE in mild cognitive impaired patients: Predicting the conversion to Alzheimer’s disease. PLoS ONE 2013, 8, e67346. [Google Scholar] [CrossRef]
- Beheshti, I.; Mishra, S.; Sone, D.; Khanna, P.; Matsuda, H. T1-weighted MRI-driven brain age estimation in Alzheimer’s disease and Parkinson’s disease. Aging Dis. 2019, 11, 618. [Google Scholar] [CrossRef]
- Johnson, K.A.; Fox, N.C.; Sperling, R.A.; Klunk, W.E. Brain imaging in Alzheimer disease. Cold Spring Harb. Perspect. Med. 2012, 2, a006213. [Google Scholar] [CrossRef]
- Lewis, M.M.; Du, G.; Lee, E.Y.; Nasralah, Z.; Sterling, N.W.; Zhang, L.; Wagner, D.; Kong, L.; Tröster, A.I.; Styner, M.; et al. The pattern of gray matter atrophy in Parkinson’s disease differs in cortical and subcortical regions. J. Neurol. 2016, 263, 68–75. [Google Scholar] [CrossRef]
- Nguyen, H.D.; Clément, M.; Mansencal, B.; Coupé, P. Brain structure ages—A new biomarker for multi-disease classification. Hum. Brain Mapp. 2024, 45, e26558. [Google Scholar] [CrossRef]
- Kaufmann, T.; van der Meer, D.; Doan, N.T.; Schwarz, E.; Lund, M.J.; Agartz, I.; Alnæs, D.; Barch, D.M.; Baur-Streubel, R.; Bertolino, A.; et al. Common brain disorders are associated with heritable patterns of apparent aging of the brain. Nat. Neurosci. 2019, 22, 1617–1623. [Google Scholar] [CrossRef]
- Bloch, L.; Friedrich, C.M. Developing a Machine Learning Workflow to Explain Black-box Models for Alzheimer’s Disease Classification. In Proceedings of the 14th International Joint Conference on Biomedical Engineering Systems and Technologies, SCITEPRESS—Science and Technology Publications, Vienna, Austria, 11–13 February 2021; pp. 87–99. [Google Scholar]
- Li, X.; Zhou, Y.; Dvornek, N.C.; Gu, Y.; Ventola, P.; Duncan, J.S. Efficient Shapley explanation for features importance estimation under uncertainty. In Proceedings of the Medical Image Computing and Computer Assisted Intervention—MICCAI 2020: 23rd International Conference, Lima, Peru, 4–8 October 2020; pp. 792–801. [Google Scholar] [CrossRef]
- Besson, P.; Parrish, T.; Katsaggelos, A.K.; Bandt, S.K. Geometric deep learning on brain shape predicts sex and age. Comput. Med. Imaging Graph. 2021, 91, 101939. [Google Scholar] [CrossRef]
- Khosla, M.; Jamison, K.; Kuceyeski, A.; Sabuncu, M.R. Ensemble learning with 3D convolutional neural networks for functional connectome-based prediction. NeuroImage 2019, 199, 651–662. [Google Scholar] [CrossRef]
- Hong, J.; Yun, H.J.; Park, G.; Kim, S.; Ou, Y.; Vasung, L.; Rollins, C.K.; Ortinau, C.M.; Takeoka, E.; Akiyama, S.; et al. Optimal method for fetal brain age prediction using multiplanar slices from structural magnetic resonance imaging. Front. Neurosci. 2021, 15, 714252. [Google Scholar] [CrossRef]
- Zhou, B.; Khosla, A.; Lapedriza, A.; Oliva, A.; Torralba, A. Learning deep features for discriminative localization. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 2921–2929. [Google Scholar]
- Simonyan, K.; Vedaldi, A.; Zisserman, A. Deep inside convolutional networks: Visualising image classification models and saliency maps. arXiv 2013, arXiv:1312.6034. [Google Scholar] [CrossRef]
- Salih, A.; Galazzo, I.B.; Raisi-Estabragh, Z.; Petersen, S.E.; Gkontra, P.; Lekadir, K.; Menegaz, G.; Radeva, P. A new scheme for the assessment of the robustness of explainable methods applied to brain age estimation. In Proceedings of the 2021 IEEE 34th International Symposium on Computer-Based Medical Systems (CBMS), Aveiro, Portugal, 7–9 June 2021; pp. 492–497. [Google Scholar] [CrossRef]
- Ballester, P.L.; Suh, J.S.; Ho, N.C.; Liang, L.; Hassel, S.; Strother, S.C.; Arnott, S.R.; Minuzzi, L.; Sassi, R.B.; Lam, R.W.; et al. Gray matter volume drives the brain age gap in schizophrenia: A SHAP study. Schizophrenia 2023, 9, 3. [Google Scholar] [CrossRef]
- Ran, C.; Yang, Y.; Ye, C.; Lv, H.; Ma, T. Brain age vector: A measure of brain aging with enhanced neurodegenerative disorder specificity. Hum. Brain Mapp. 2022, 43, 5017–5031. [Google Scholar] [CrossRef]
- Wu, Y.; Gao, H.; Zhang, C.; Ma, X.; Zhu, X.; Wu, S.; Lin, L. Machine learning and deep learning approaches in lifespan brain age prediction: A comprehensive review. Tomography 2024, 10, 1238–1262. [Google Scholar] [CrossRef]
- Sajedi, H.; Pardakhti, N. Age prediction based on brain MRI image: A survey. J. Med. Syst. 2019, 43, 279. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Knol, M.J.; Tiulpin, A.; Dubost, F.; de Bruijne, M.; Vernooij, M.W.; Adams, H.H.; Ikram, M.A.; Niessen, W.J.; Roshchupkin, G.V. Gray matter age prediction as a biomarker for risk of dementia. Proc. Natl. Acad. Sci. USA 2019, 116, 21213–21218. [Google Scholar] [CrossRef]
- Dufumier, B.; Gori, P.; Battaglia, I.; Victor, J.; Grigis, A.; Duchesnay, E. Benchmarking cnn on 3d anatomical brain mri: Architectures, data augmentation and deep ensemble learning. arXiv 2021, arXiv:2106.01132. [Google Scholar] [CrossRef]
- Wu, Y.; Zhang, C.; Ma, X.; Zhu, X.; Lin, L.; Tian, M. ds-FCRN: Three-dimensional dual-stream fully convolutional residual networks and transformer-based global–local feature learning for brain age prediction. Brain Struct. Funct. 2025, 230, 1–18. [Google Scholar] [CrossRef]
- Wang, L.; Li, J. The value of serum-free androgen index in the diagnosis of polycystic ovary syndrome: A systematic review and meta-analysis. J. Obstet. Gynaecol. Res. 2021, 47, 1221–1231. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.V.; Chaudhari, G.; Hess, C.P.; Glenn, O.A.; Sugrue, L.P.; Rauschecker, A.M.; Li, Y. Deep Learning to Predict Neonatal and Infant Brain Age from Myelination on Brain MRI Scans. Radiology 2022, 305, 678–687. [Google Scholar] [CrossRef]
- Hofmann, S.M.; Beyer, F.; Lapuschkin, S.; Goltermann, O.; Loeffler, M.; Müller, K.R.; Villringer, A.; Samek, W.; Witte, A.V. Towards the interpretability of deep learning models for multi-modal neuroimaging: Finding structural changes of the ageing brain. NeuroImage 2022, 261, 119504. [Google Scholar] [CrossRef]
- Jiang, H.; Lu, N.; Chen, K.; Yao, L.; Li, K.; Zhang, J.; Guo, X. Predicting brain age of healthy adults based on structural MRI parcellation using convolutional neural networks. Front. Neurol. 2020, 10, 1346. [Google Scholar] [CrossRef]
- Kuchcinski, G.; Rumetshofer, T.; Zervides, K.A.; Lopes, R.; Gautherot, M.; Pruvo, J.P.; Bengtsson, A.A.; Hansson, O.; Jönsen, A.; Sundgren, P.C.M. MRI BrainAGE demonstrates increased brain aging in systemic lupus erythematosus patients. Front. Aging Neurosci. 2023, 15, 1274061. [Google Scholar] [CrossRef]
- Shi, W.; Yan, G.; Li, Y.; Li, H.; Liu, T.; Sun, C.; Wang, G.; Zhang, Y.; Zou, Y.; Wu, D. Fetal brain age estimation and anomaly detection using attention-based deep ensembles with uncertainty. NeuroImage 2020, 223, 117316. [Google Scholar] [CrossRef] [PubMed]
- Cai, H.; Gao, Y.; Liu, M. Graph transformer geometric learning of brain networks using multimodal MR images for brain age estimation. IEEE Trans. Med. Imaging 2022, 42, 456–466. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Hu, K.; Wang, M.; Zhao, Y.; Liu, Y.; Fan, L.; Liu, B. Predicting brain age during typical and atypical development based on structural and functional neuroimaging. Hum. Brain Mapp. 2021, 42, 5943–5955. [Google Scholar] [CrossRef]
- Zhang, X.; Pan, Y.; Wu, T.; Zhao, W.; Zhang, H.; Ding, J.; Ji, Q.; Jia, X.; Li, X.; Lee, Z.; et al. Brain age prediction using interpretable multi-feature-based convolutional neural network in mild traumatic brain injury. NeuroImage 2024, 297, 120751. [Google Scholar] [CrossRef]
- Ahsan, R.L.; Allom, R.; Gousias, I.S.; Habib, H.; Turkheimer, F.E.; Free, S.; Lemieux, L.; Myers, R.; Duncan, J.S.; Brooks, D.J.; et al. Volumes, spatial extents and a probabilistic atlas of the human basal ganglia and thalamus. NeuroImage 2007, 38, 261–270. [Google Scholar] [CrossRef] [PubMed]
- Llano, D.A. Functional imaging of the thalamus in language. Brain Lang. 2013, 126, 62–72. [Google Scholar] [CrossRef]
- Federmeier, K.D.; Kutas, M. Aging in context: Age-related changes in context use during language comprehension. Psychophysiology 2005, 42, 133–141. [Google Scholar] [CrossRef]
- Bird, C.M.; Burgess, N. The hippocampus and memory: Insights from spatial processing. Nat. Rev. Neurosci. 2008, 9, 182–194. [Google Scholar] [CrossRef]
- Addicott, M.A.; Oliveto, A.H.; Daughters, S.B. Smoking status affects cognitive, emotional and neural-connectivity response to distress-inducing auditory feedback. Drug Alcohol Depend. 2023, 246, 109855. [Google Scholar] [CrossRef]
- Chen, Y.; Chaudhary, S.; Li, G.; Fucito, L.M.; Bi, J.; Li, C.S.R. Deficient sleep, altered hypothalamic functional connectivity, depression and anxiety in cigarette smokers. NeuroImage Rep. 2024, 4, 100200. [Google Scholar] [CrossRef]
- Le, T.M.; Malone, T.; Li, C.S.R. Positive alcohol expectancy and resting-state functional connectivity of the insula in problem drinking. Drug Alcohol Depend. 2022, 231, 109248. [Google Scholar] [CrossRef] [PubMed]
- Bach, P.; Zaiser, J.; Zimmermann, S.; Gessner, T.; Hoffmann, S.; Gerhardt, S.; Berhe, O.; Bekier, N.K.; Abel, M.; Radler, P.; et al. Stress-Induced Sensitization of Insula Activation Predicts Alcohol Craving and Alcohol Use in Alcohol Use Disorder. Biol. Psychiatry 2024, 95, 245–255. [Google Scholar] [CrossRef]
- Sudlow, C.; Gallacher, J.; Allen, N.; Beral, V.; Burton, P.; Danesh, J.; Downey, P.; Elliott, P.; Green, J.; Landray, M.; et al. UK biobank: An open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015, 12, e1001779. [Google Scholar] [CrossRef] [PubMed]
- Petersen, R.C.; Aisen, P.S.; Beckett, L.A.; Donohue, M.C.; Gamst, A.C.; Harvey, D.J.; Jack, C.R., Jr.; Jagust, W.J.; Shaw, L.M.; Toga, A.W.; et al. Alzheimer’s Disease Neuroimaging Initiative (ADNI). Neurology 2010, 74, 201–209. [Google Scholar] [CrossRef] [PubMed]
- Leonardsen, E.H.; Peng, H.; Kaufmann, T.; Agartz, I.; Andreassen, O.A.; Celius, E.G.; Espeseth, T.; Harbo, H.F.; Høgestøl, E.A.; De Lange, A.M.; et al. Deep neural networks learn general and clinically relevant representations of the ageing brain. NeuroImage 2022, 256, 119210. [Google Scholar] [CrossRef]
- Feng, X.; Lipton, Z.C.; Yang, J.; Small, S.A.; Provenzano, F.A.; Alzheimer’s Disease Neuroimaging Initiative; Australian Imaging Biomarkers and Lifestyle Flagship Study of Ageing; Frontotemporal Lobar Degeneration Neuroimaging Initiative. Estimating brain age based on a uniform healthy population with deep learning and structural magnetic resonance imaging. Neurobiol. Aging 2020, 91, 15–25. [Google Scholar] [CrossRef]
- Levakov, G.; Rosenthal, G.; Shelef, I.; Raviv, T.R.; Avidan, G. From a deep learning model back to the brain—Identifying regional predictors and their relation to aging. Hum. Brain Mapp. 2020, 41, 3235–3252. [Google Scholar] [CrossRef]
- Kwon, H. AudioGuard: Speech Recognition System Robust against Optimized Audio Adversarial Examples. Multimed. Tools Appl. 2024, 83, 57943–57962. [Google Scholar] [CrossRef]

| Data Set | UKB | ADNI | PPMI | ||||
|---|---|---|---|---|---|---|---|
| Category | HCs | HCs | MCI | AD | HCs | pPD | PD |
| Number of subjects | 16,377 | 582 | 432 | 252 | 36 | 426 | 264 |
| Female/male | 8566/7811 | 346/236 | 188/244 | 125/127 | 16/20 | 219/217 | 109/155 |
| Age (years) | / | / | / | / | / | / | / |
| Data Set | UKB | ADNI | PPMI | ||||
|---|---|---|---|---|---|---|---|
| Category | HCs | HCs | MCI | AD | HCs | pPD | PD |
| MAE | |||||||
| 0.83 | 0.52 | 0.58 | 0.52 | 0.68 | 0.69 | 0.76 | |
| Task | HCs vs. MCI vs. AD | HCs vs. pPD vs. PD | |||||
|---|---|---|---|---|---|---|---|
| Model | Performance\Input | RBADI | BAV | BAG | RBADI | BAV | BAG |
| SVM | ACC | ||||||
| SEN | |||||||
| SPE | |||||||
| AUC | |||||||
| XGboost | ACC | ||||||
| SEN | |||||||
| SPE | |||||||
| AUC | |||||||
| Lasso | ACC | ||||||
| SEN | |||||||
| SPE | |||||||
| AUC | |||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, Y.; Sun, S.; Zhang, C.; Ma, X.; Zhu, X.; Li, Y.; Lin, L.; Fu, Z. Regional Brain Aging Disparity Index: Region-Specific Brain Aging State Index for Neurodegenerative Diseases and Chronic Disease Specificity. Bioengineering 2025, 12, 607. https://doi.org/10.3390/bioengineering12060607
Wu Y, Sun S, Zhang C, Ma X, Zhu X, Li Y, Lin L, Fu Z. Regional Brain Aging Disparity Index: Region-Specific Brain Aging State Index for Neurodegenerative Diseases and Chronic Disease Specificity. Bioengineering. 2025; 12(6):607. https://doi.org/10.3390/bioengineering12060607
Chicago/Turabian StyleWu, Yutong, Shen Sun, Chen Zhang, Xiangge Ma, Xinyu Zhu, Yanxue Li, Lan Lin, and Zhenrong Fu. 2025. "Regional Brain Aging Disparity Index: Region-Specific Brain Aging State Index for Neurodegenerative Diseases and Chronic Disease Specificity" Bioengineering 12, no. 6: 607. https://doi.org/10.3390/bioengineering12060607
APA StyleWu, Y., Sun, S., Zhang, C., Ma, X., Zhu, X., Li, Y., Lin, L., & Fu, Z. (2025). Regional Brain Aging Disparity Index: Region-Specific Brain Aging State Index for Neurodegenerative Diseases and Chronic Disease Specificity. Bioengineering, 12(6), 607. https://doi.org/10.3390/bioengineering12060607








