Automated Early-Stage Glaucoma Detection Using a Robust Concatenated AI Model
Abstract
1. Introduction
- V-MD values between 0 dB and −6 dB indicate mild (early) glaucoma;
- V-MD values between −6 dB and −12 dB indicate moderate glaucoma;
- V-MD values below −12 dB indicate advanced glaucoma.
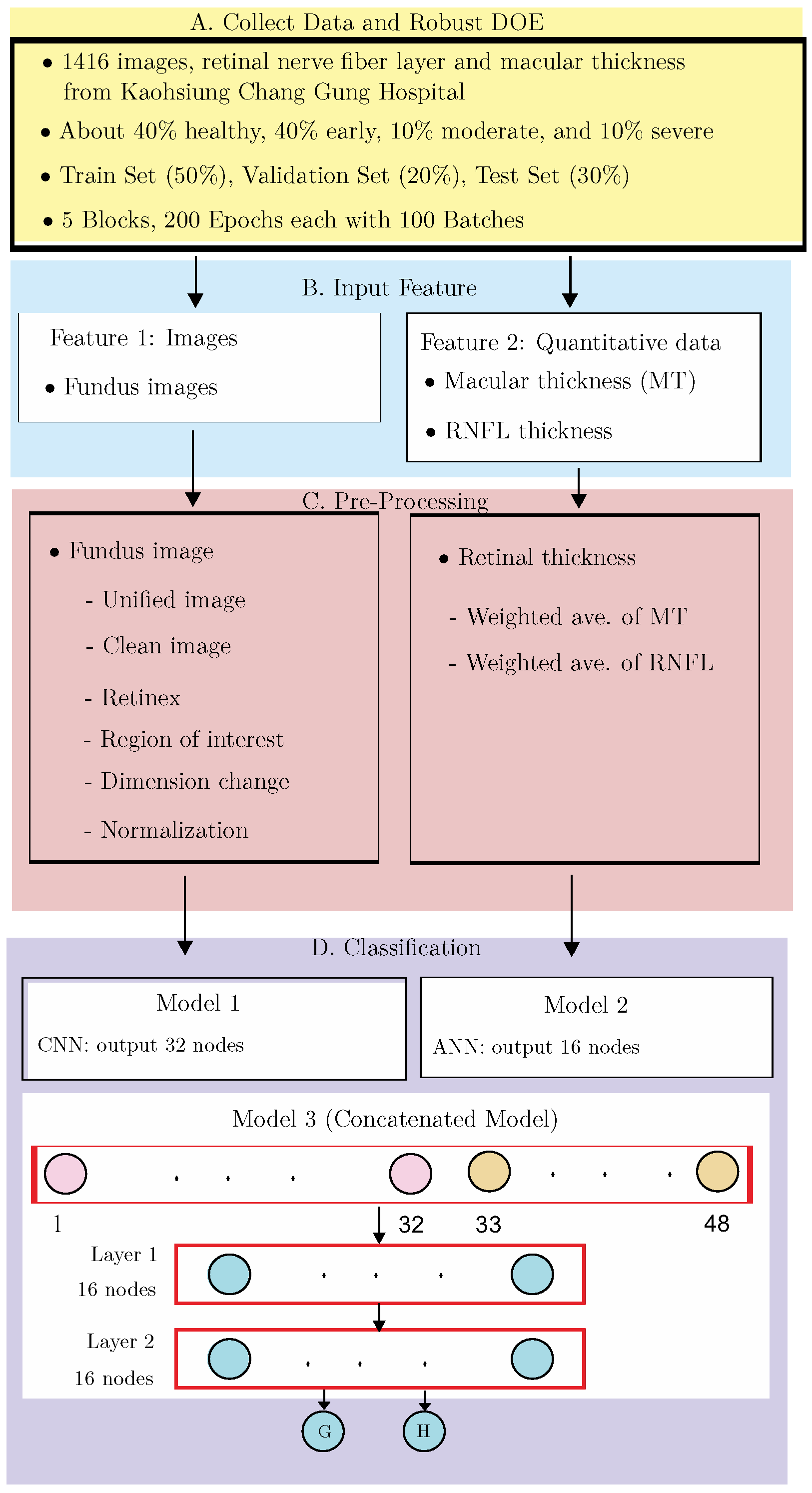
2. Materials and Methods
2.1. Data Collection and Design of Experiments
2.1.1. Blocks and Epochs
2.1.2. Epochs and Batches
2.1.3. Robust DOE
- : image size used;
- : convolutional layers;
- : number of kernels in each convolutional layer;
- : treatment for the area outside the ROI;
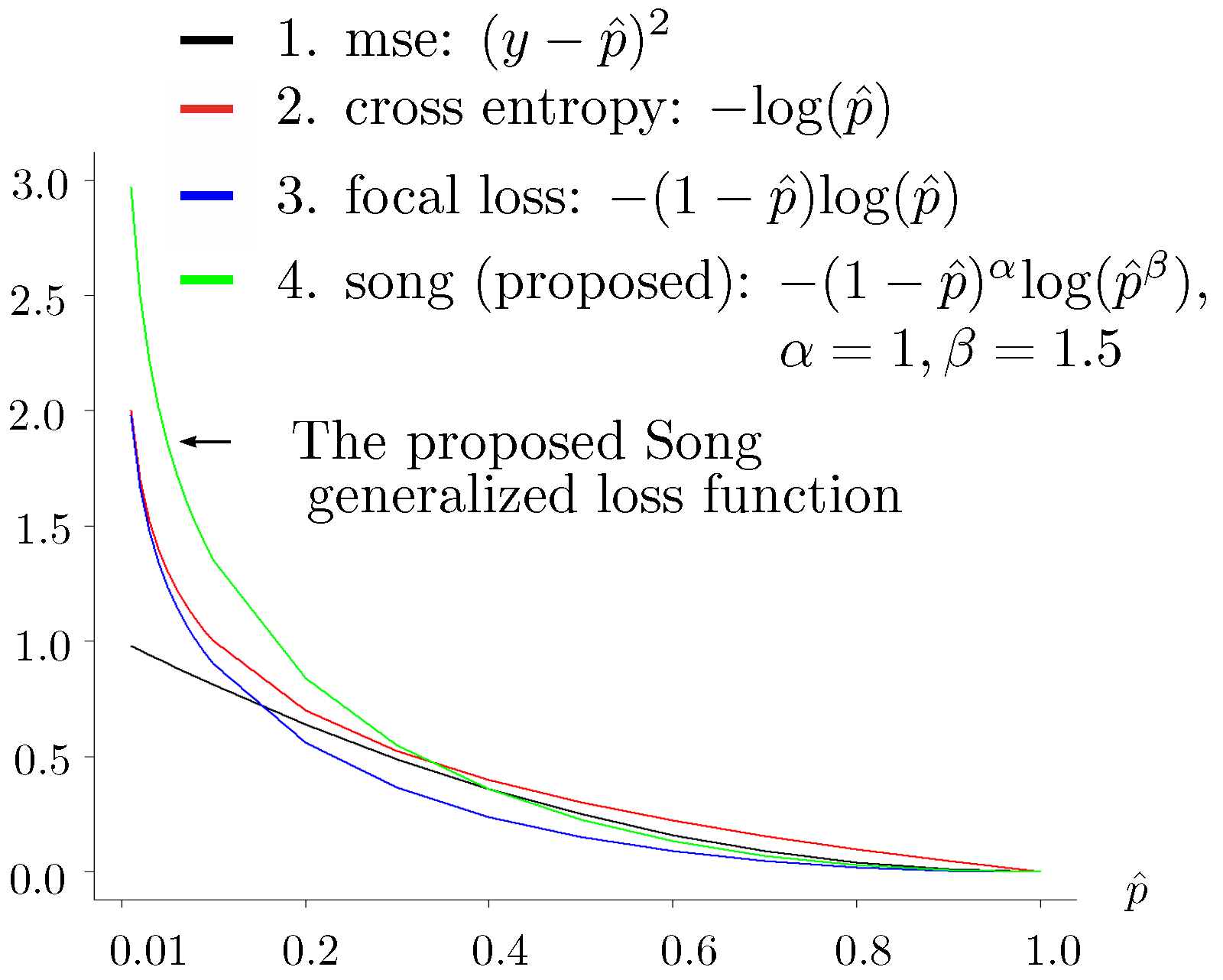
- : parameters and used in the proposed loss function (see Figure 2) for the training data:where the terms are defined as follows:
- -
- The response or indicates an early-stage glaucoma sample or a normal sample, respectively.
- -
- , a sigmoid function, gives the estimate of the true value y. Regarding the test set, the estimate of y for the test set data is if ; otherwise, the estimate is .
- Factor : No. of layers in the ANN;
- Factor : No. of nodes in the ANN.
- Factor : No. of layers in the concatenated model;
- Factor : No. of nodes in the concatenated model.
- Factor (formerly , number of layers in Model 2) with two levels: (2 layers), 1 (4 layers)
- Factor (formerly , number of layers in Model 2) with two levels: (4 nodes), 1 (16 nodes)
- Factor (formerly , number of layers in Model 3) with two levels: (2 layers), 1 (3 layers)
- Factor (formerly , number of layers in Model 3) with two levels: (4 nodes), 1 (16 nodes)
2.2. Input Features
Fundus Image Feature
- Retina: the light-sensitive tissue that detects visual information and sends signals to the brain;
- Optic disc: the region where the optic nerve exits the eye, appearing as a bright circular area;
- Macula: the central part of the retina responsible for detailed vision and color vision;
- Blood vessels: the network of veins and arteries that supply blood to the retina.
2.3. Smart Pre-Processing
2.3.1. Fundus Images
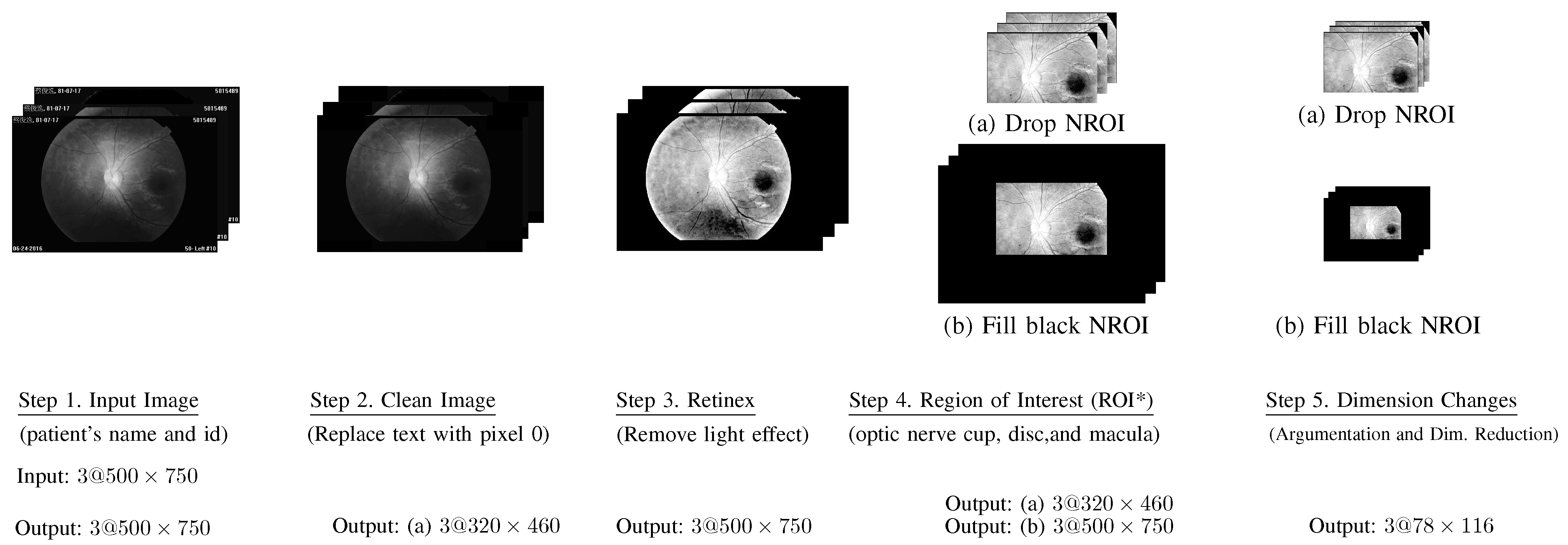
- Step 1. Input Images: All input fundus images include text about patient information and the date that the associated images were collected.
- Step 2. Clean Images: All text on each image was replaced by pixels with a value of 0, which is the color black.
- Step 3. Retinex: The word “Retinex” combines “retina” and “cortex”, where the retina is the part of the eye that detects color, while the visual cortex is the part of a brain that processes the information it receives from the retina. This step was used to remove the light effect from flash images.
- Step 4. Region of Interest (ROI*):Based on our prior knowledge, the region of interest should be near the optic nerve cup, and macular portion.Accordingly, we selected the (ROI*) as follows.where ROI* covers the optic nerve cup, optic disc, and macula.
- Step 5. Image augmentation and dimension reduction:Each input image was randomly rotated 30, 60, 90, 120, or 180 counterclockwise. Thus, each image was augmented into two images. Finally, we reduced each image, shifting the row and column sizes of into smaller sizes . Let denote the image processed at the end of this step, where regarding RGB.
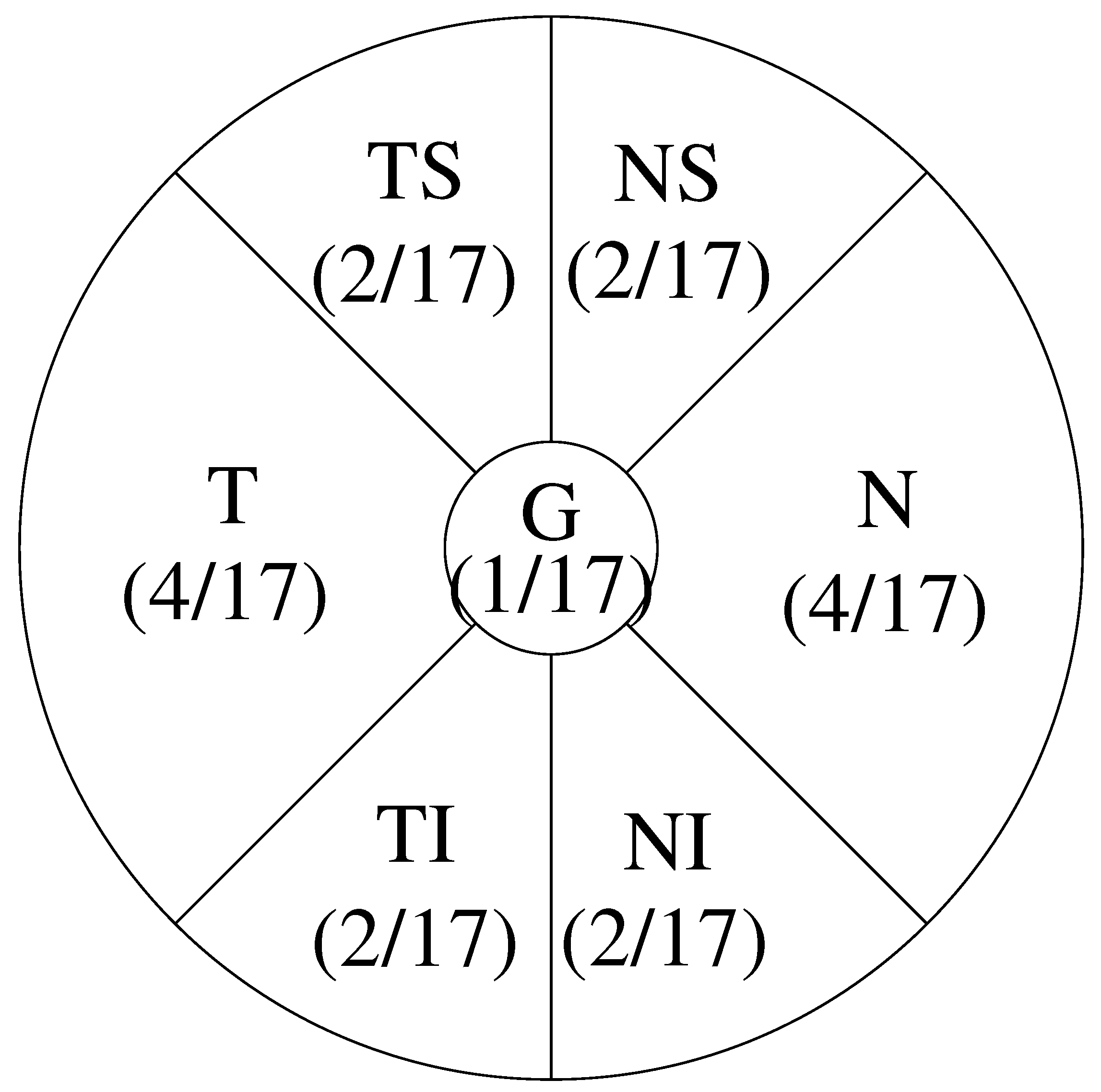
2.3.2. Quantitative Measure
- UO: denotes Upper Outer;
- UI: denotes Upper Inner;
- LO: denotes Lower Outer’
- LI: denotes Lower Inner.
- G: denotes Global, whose area is 1;
- T: denotes Temporal, which has area 4;
- TS: denotes Temporal Superior, which has area 2;
- TI: denotes Temporal Inferior, which has area 2;
- N: denotes Nasal, which has area 4;
- NS: denotes Nasal Superior, which has area 2;
- NI: denotes Nasal Inferior, which has area 2.
2.4. Classification
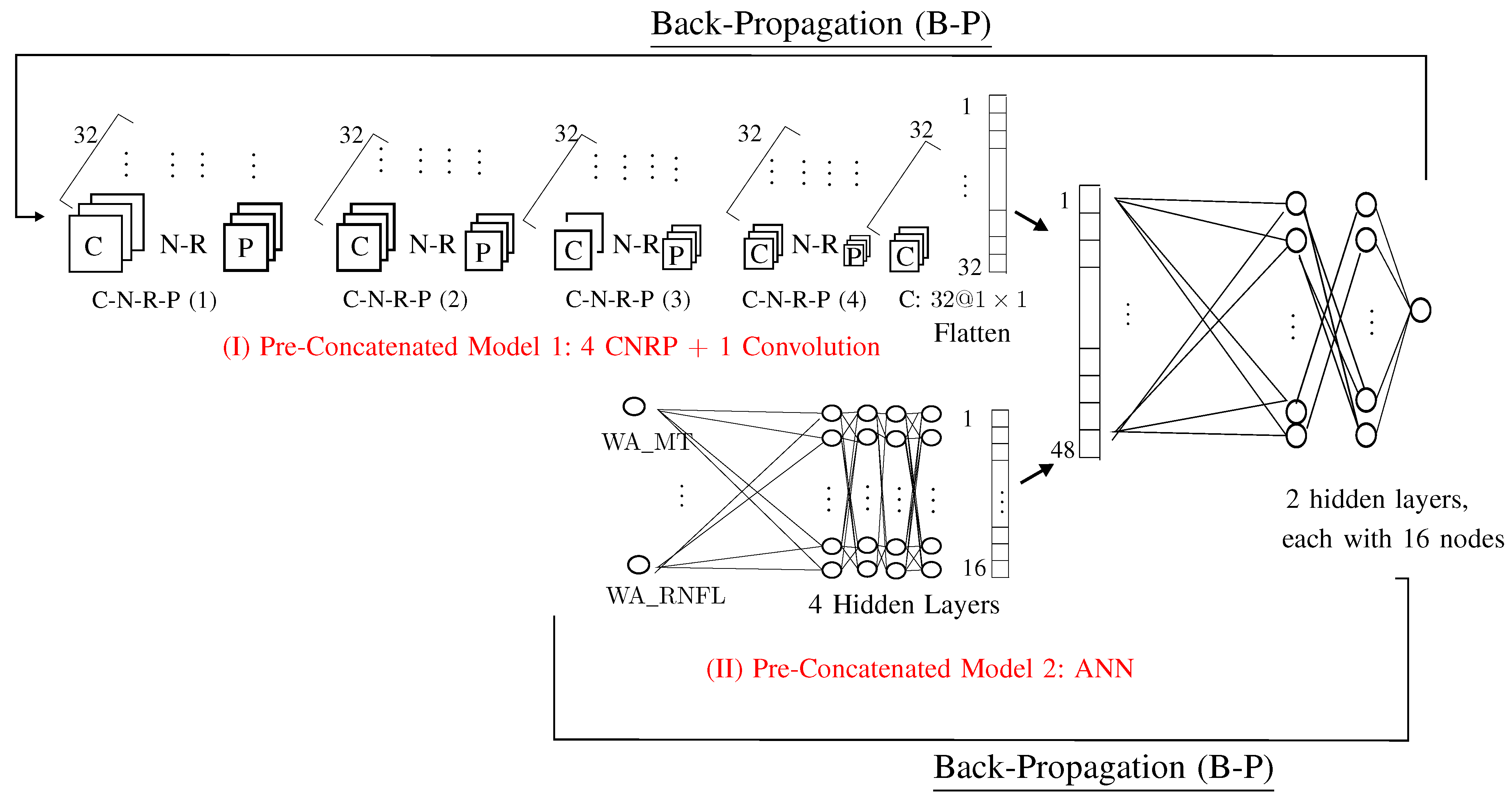
2.4.1. Pre-Concatenated Model 1
- Step 5.1. C-N-R-P(1):C: ;N: ;R: ;P: .
- Step 5.2. C-N-R-P(2):C: ;N: ;R: ;P: .
- Step 5.3. C-N-R-P(3):C: ;N: ;R: ;P: .
- Step 5.4. C-N-R-P(3):C: ;N: ;R: ;P: .
- Step 5.5. C: one convolution with kernel matrix , which yields one vector with 32 values.
- (1)
- : sizes used in images: ;
- (2)
- : convolutional layers: ;
- (3)
- : number of kernels in each convolutional layer: 32 (which yields 32 images);
- (4)
- : treatment for the area outside the ROI: delete (rather than fill with black);
- (5)
- : parameter used in the proposed loss function: parameter , used in the loss function defined in Equation (1).
2.4.2. Pre-Concatenated 2: ANN
- Factor (No. of layers in ANN): 4 layers;
- Factor (No. of nodes in ANN): 16 nodes.
- macular thickness
- (1)
- UO: Upper Outer
- (2)
- UI: Upper Inner;
- (3)
- LO: Lower Outer;
- (4)
- LI: Lower Inner;
- (5)
- Weighted average for the Upper region: average of UO and UI;
- (6)
- Weighted average for the Lower region: average of LO and LI;
- (7)
- Weighted average for the Inner region: average of UI and LI;
- (8)
- Weighted average for the Outer region: average of UO and LO;
- (9)
- Overall weighted average of macular thickness: , as defined in Equation (3).
- RNFL thickness
- (1)
- G: Global;
- (2)
- T: Temporal;
- (3)
- TS: Temporal Superior;
- (4)
- TI: Temporal Inferior;
- (5)
- N: Nasal;
- (6)
- NS: Nasal Superior;
- (7)
- NI: Nasal Inferior;
- (8)
- Overall weighted average of RNFL thickness: , as defined in Equation (4).
2.4.3. Proposed AI Model 3: Concatenated Model
- –
- Factor (No. of layers in Concatenate): 2 layers;
- –
- Factor (No. of nodes in Concatenate): 16 nodes.
3. Results
3.1. Commonly Used Performance Metrics in Glaucoma Detecting
- –
- TP: correctly identifying a positive test result, where “positive” refers to the result of the test rather than the factual condition;
- –
- TN: correctly detecting a negative case;
- –
- FP: incorrectly detecting a negative case as positive;
- –
- FN: incorrectly detecting a positive case as negative.
- (1)
- Accuracy: (TP+TN)/(TP+FP+FN+TN).
- (2)
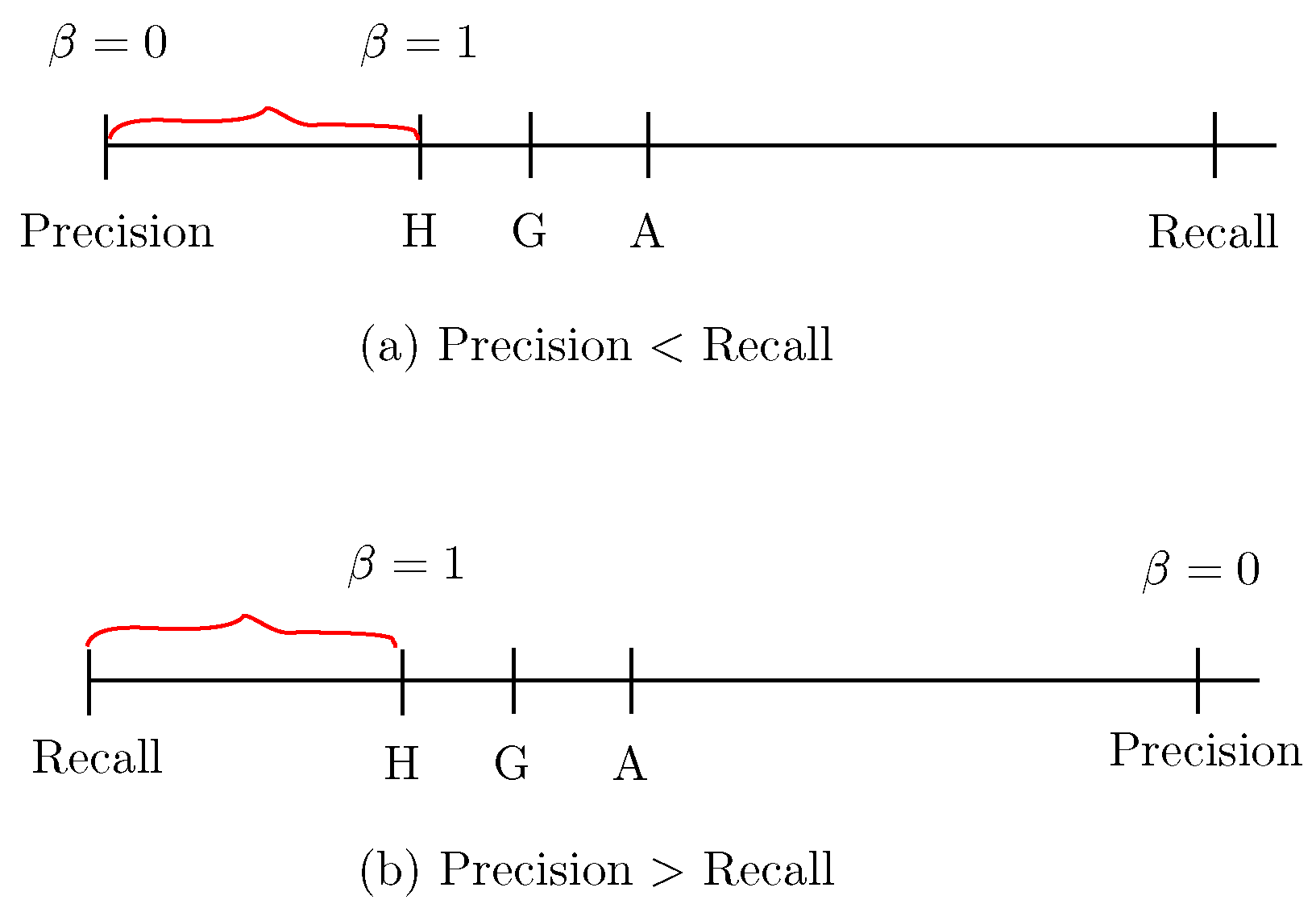
- Sensitivity, also referred to as recall: TP/(TP+FN),shown in Figure 8a,b. The insight of using recall is reflected in the name itself: it reflects how much the model recalls of the TP (true positive) results.
- (3)
- Specificity: TN/(FP+TN), shown in Figure 8a.
- (4)
- Precision: TP/(TP+FP), shown in Figure 8b.
- (5)
- -Score:
- (6)
- -Score (also referred to as the letter H):The F1-Score is a special case of the -Score with , which is referred to as the letter H because it is the harmonic mean of recall and precision, as shown in Figure 9.
- –
- A: The letter A represents the average mean of recall and precision, which is calculated as .
- –
- G: The letter G is referred to as the geometric mean of recall and precision, which is calculated as .
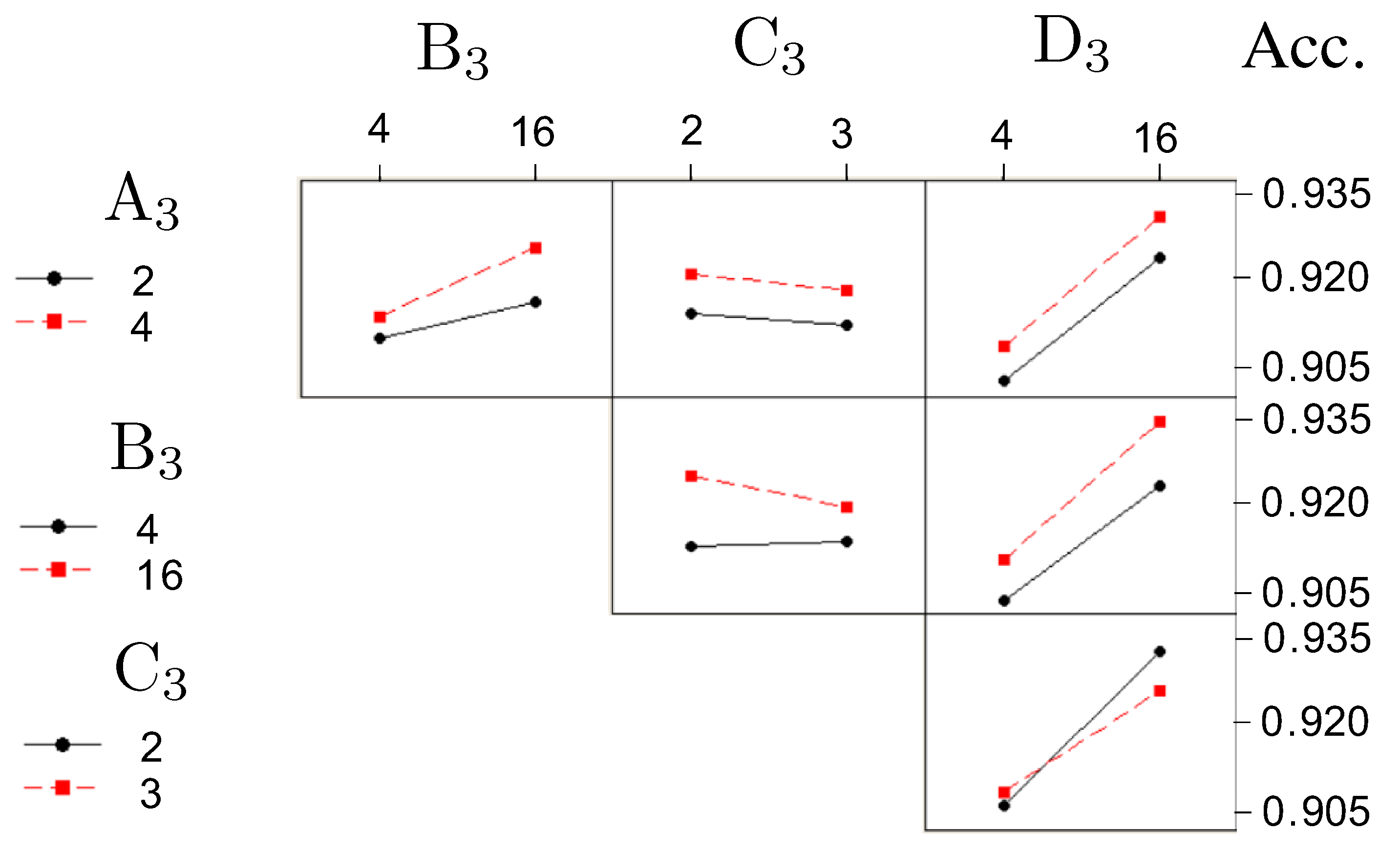
3.2. Determine the Optimal Values of the Four Factors Used in the Concatenated Model
- –
- Factor (No. of layers in ANN): four layers;
- –
- Factor (No. of nodes in ANN): sixteen nodes;
- –
- Factor (No. of layers in Concatenate): two layers;
- –
- Factor (No. of nodes in Concatenate): sixteen nodes.
3.3. Performance of the Concatenated Model
4. Discussion
- –
- Effectiveness: The proposed model achieves an accuracy of 0.93 for early-stage glaucoma detection, which is significantly higher than the 0.85 accuracy reported in [17]. Additionally, Ref. [17] did not report metrics such as sensitivity, specificity, precision, or -Score. In contrast, the concatenated model demonstrates superior performance, with all metrics exceeding 0.9: accuracy of 0.93, sensitivity of 0.91, specificity of 0.94, precision of 0.92, and -Score of 0.90.
- –
- Robustness: The standard errors for the estimated performance are all below 0.04. To the best of our knowledge, none of the previous studies appears to have used a statistically robust design of experiments (DOE) to optimize hyperparameters.
- –
- Expert Knowledge Integration: For any problem, such as detecting glaucoma, experts can always provide valuable insights. For example, they know that the thickness of the macular or peripapillary retinal nerve fiber layer is smaller in glaucoma cases than in normal cases. We believe that a concatenated model that incorporates expert knowledge should outperform one that relies solely on image data. Specifically, research from journal papers published after 2018 (including [9,10,11,12,13,14,15,16,17,18,19,20,21]) suggests that incorporating expert knowledge will certainly lead to better performance compared to using image data alone.
- –
- practicality for clinical use: The model requires approximately 1 min to process a single block (blocks are discussed in Section 2.1), demonstrating its practicality for clinical use. The computational system used in our study consisted of the following hardware:
- (1)
- Motherboard: ASUS, ROG STRIX Z590-E GAMING WIFI;
- (2)
- CPU: 11th Gen Intel(R) Core(TM) i9-11900KF @ 3.50 GHz/eight cores;
- (3)
- GPU: NVIDIA [GeForce RTX 3090], NVIDIA [GeForce RTX 2080 Ti];
- (4)
- Graphics card: NVIDIA [GeForce RTX 3090], NVIDIA [GeForce RTX 2080 Ti];
- (5)
- RAM: 64 GB DDR4 (16 GB X 4);
- (6)
- Operating system: Debian Linux 12;
- (7)
- Others: HDDs: 1 TB X 1, 6 TB X 4.
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Correction Statement
References
- Allison, K.; Patel, D.; Alabi, O. Epidemiology of glaucoma: The past, present, and predictions for the future. Cureus 2020, 12. [Google Scholar] [CrossRef] [PubMed]
- Bizios, D.; Heijl, A.; Hougaard, J.L.; Bengtsson, B. Machine learning classifiers for glaucoma diagnosis based on classification of retinal nerve fibre layer thickness parameters measured by Stratus OCT. Acta Ophthalmol. 2010, 88, 44–52. [Google Scholar] [CrossRef] [PubMed]
- Acharya, U.R.; Dua, S.; Du, X.; Chua, C.K. Automated diagnosis of glaucoma using texture and higher order spectra features. IEEE Trans. Inf. Technol. Biomed. 2011, 15, 449–455. [Google Scholar] [CrossRef] [PubMed]
- Khan, F.; Khan, S.A.; Yasin, U.U.; ul Haq, I.; Qamar, U. Detection of glaucoma using retinal fundus images. In Proceedings of the 6th 2013 Biomedical Engineering International Conference, Amphur Muang, Thailand, 23–25 October 2013; IEEE: Piscataway, NJ, USA, 2013; pp. 1–5. [Google Scholar]
- Samanta, S.; Ahmed, S.S.; Salem, M.A.M.M.; Nath, S.S.; Dey, N.; Chowdhury, S.S. Haralick features based automated glaucoma classification using back propagation neural network. In Proceedings of the 3rd International Conference on Frontiers of Intelligent Computing: Theory and Applications (FICTA), Bhubaneswar, India, 14–15 November 2014; Springer: Berlin/Heidelberg, Germany, 2015; pp. 351–358. [Google Scholar]
- Issac, A.; Sarathi, M.P.; Dutta, M.K. An adaptive threshold based image processing technique for improved glaucoma detection and classification. Comput. Methods Programs Biomed. 2015, 122, 229–244. [Google Scholar] [CrossRef] [PubMed]
- Maheshwari, S.; Pachori, R.B.; Acharya, U.R. Automated diagnosis of glaucoma using empirical wavelet transform and correntropy features extracted from fundus images. IEEE J. Biomed. Health Inform. 2016, 21, 803–813. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.J.; Cho, K.J.; Oh, S. Development of machine learning models for diagnosis of glaucoma. PLoS ONE 2017, 12, e0177726. [Google Scholar] [CrossRef]
- Hasan, M.M.; Phu, J.; Sowmya, A.; Meijering, E.; Kalloniatis, M. Artificial Intelligence in the diagnosis of glaucomq qnd neurodegenerative diseases. Clin. Exp. Optom. 2024, 107, 130–146. [Google Scholar] [CrossRef] [PubMed]
- Fu, H.; Cheng, J.; Xu, Y.; Zhang, C.; Wong, D.W.K.; Liu, J.; Cao, X. Disc-aware ensemble network for glaucoma screening from fundus image. IEEE Trans. Med. Imaging 2018, 37, 2493–2501. [Google Scholar] [CrossRef] [PubMed]
- Serte, S.; Serener, A. A generalized deep learning model for glaucoma detection. In Proceedings of the 2019 3rd International Symposium on Multidisciplinary Studies and Innovative Technologies (ISMSIT), Ankara, Turkey, 11–13 October 2019; IEEE: Piscataway, NJ, USA, 2019; pp. 1–5. [Google Scholar]
- Al Ghamdi, M.; Li, M.; Abdel-Mottaleb, M.; Abou Shousha, M. Semi-supervised transfer learning for convolutional neural networks for glaucoma detection. In Proceedings of the ICASSP 2019—2019 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Brighton, UK, 12–17 May 2019; IEEE: Piscataway, NJ, USA, 2019; pp. 3812–3816. [Google Scholar]
- Sreng, S.; Maneerat, N.; Hamamoto, K.; Win, K.Y. Deep learning for optic disc segmentation and glaucoma diagnosis on retinal images. Appl. Sci. 2020, 10, 4916. [Google Scholar] [CrossRef]
- Serte, S.; Serener, A. Graph-based saliency and ensembles of convolutional neural networks for glaucoma detection. IET Image Process. 2021, 15, 797–804. [Google Scholar] [CrossRef]
- Gheisari, S.; Shariflou, S.; Phu, J.; Kennedy, P.J.; Agar, A.; Kalloniatis, M.; Golzan, S.M. A combined convolutional and recurrent neural network for enhanced glaucoma detection. Sci. Rep. 2021, 11, 1945. [Google Scholar] [CrossRef] [PubMed]
- de Sales Carvalho, N.R.; Rodrigues, M.d.C.L.C.; de Carvalho Filho, A.O.; Mathew, M.J. Automatic method for glaucoma diagnosis using a three-dimensional convoluted neural network. Neurocomputing 2021, 438, 72–83. [Google Scholar] [CrossRef]
- Sun, S.; Ha, A.; Kim, Y.K.; Yoo, B.W.; Kim, H.C.; Park, K.H. Dual-input convolutional neural network for glaucoma diagnosis using spectral-domain optical coherence tomography. Br. J. Ophthalmol. 2021, 105, 1555–1560. [Google Scholar] [CrossRef] [PubMed]
- Song, W.T.; Lai, C.; Su, Y.Z. A Statistical Robust Glaucoma Detection Framework Combining Retinex, CNN, and DOE Using Fundus Images. IEEE Access 2021, 9, 103772–103783. [Google Scholar] [CrossRef]
- Basha, S.H.; Sudharsanam; Sriram, S.; Pasupuleti, S.C. Glaucoma screening using CNN classification. In Proceedings of the AIP Conference Proceedings: 12th National Conference on Recent Advancements in Biomedical Engineering, Chennai, India, 3 December 2020; AIP Publishing LLC: College Park, MD, USA, 2022; Volume 2405, p. 020018. [Google Scholar]
- Sunanthini, V.; Deny, J.; Govinda Kumar, E.; Vairaprakash, S.; Govindan, P.; Sudha, S.; Muneeswaran, V.; Thilagaraj, M. Comparison of CNN Algorithms for Feature Extraction on Fundus Images to Detect Glaucoma. J. Healthc. Eng. 2022, 2022. [Google Scholar] [CrossRef] [PubMed]
- Shyamalee, T.; Meedeniya, D. CNN based fundus images classification for glaucoma identification. In Proceedings of the 2022 2nd International Conference on Advanced Research in Computing (ICARC), Belihuloya, Sri Lanka, 23–24 February 2022; IEEE: Piscataway, NJ, USA, 2022; pp. 200–205. [Google Scholar]




| Comb. (Run) | , , , | Ori. Data | Mean Accuracy | Stand. Error |
|---|---|---|---|---|
| 1 | − − − − | |||
| 2 | + − − − | |||
| 3 | − + − − | |||
| 4 | + + − − | |||
| 5 | − − + − | |||
| 6 | + − + − | |||
| 7 | − + + − | |||
| 8 | + + + − | |||
| 9 | − − − + | |||
| 10 | + − − + | |||
| 11 | − + − + | |||
| 12 | + + − + | |||
| 13 | − − + + | |||
| 14 | + − + + | |||
| 15 | − + + + | |||
| 16 | + + + + |
| Term | Effect | Coef | T-Value | p-Value |
|---|---|---|---|---|
| 0.006 | 0.003 | 2.09 | 0.041 | |
| 0.009 | 0.004 | 2.92 | 0.005 | |
| −0.002 | −0.001 | −0.75 | 0.456 | |
| 0.021 | 0.011 | 7.04 | 0.000 |
| Acc. | Sen. | Spe. | Pre. | F1-S. | ||||
|---|---|---|---|---|---|---|---|---|
| 4 | 16 | 2 | 16 | 0.93 | 0.91 | 0.94 | 0.92 | 0.90 |
| 4 | 16 | 3 | 16 | 0.92 | 0.91 | 0.92 | 0.91 | 0.90 |
| 2 | 16 | 2 | 16 | 0.91 | 0.90 | 0.91 | 0.90 | 0.90 |
| 4 | 4 | 2 | 16 | 0.90 | 0.90 | 0.90 | 0.90 | 0.90 |
| 2 | 16 | 3 | 16 | 0.89 | 0.88 | 0.90 | 0.90 | 0.90 |
| Model | Acc. (s.e.) | Sen. (s.e.) | Spe. (s.e.) | Pre. (s.e.) | -S (s.e.) |
|---|---|---|---|---|---|
| Model 1 | 0.88 (0.03) | 0.88 (0.03) | 0.89 (0.02) | 0.88 (0.03) | 0.87 (0.03) |
| Model 2 | 0.68 (0.05) | 0.67 (0.06) | 0.69 (0.05) | 0.69 (0.05) | 0.68 (0.05) |
| Concatenated | 0.93 | 0.91 | 0.94 | 0.92 | 0.90 |
| Model | (0.04) | (0.04) | (0.04) | (0.03) | (0.04) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Song, W.; Lai, I.-C. Automated Early-Stage Glaucoma Detection Using a Robust Concatenated AI Model. Bioengineering 2025, 12, 516. https://doi.org/10.3390/bioengineering12050516
Song W, Lai I-C. Automated Early-Stage Glaucoma Detection Using a Robust Concatenated AI Model. Bioengineering. 2025; 12(5):516. https://doi.org/10.3390/bioengineering12050516
Chicago/Turabian StyleSong, Wheyming, and Ing-Chou Lai. 2025. "Automated Early-Stage Glaucoma Detection Using a Robust Concatenated AI Model" Bioengineering 12, no. 5: 516. https://doi.org/10.3390/bioengineering12050516
APA StyleSong, W., & Lai, I.-C. (2025). Automated Early-Stage Glaucoma Detection Using a Robust Concatenated AI Model. Bioengineering, 12(5), 516. https://doi.org/10.3390/bioengineering12050516







