An Effective and Interpretable Sleep Stage Classification Approach Using Multi-Domain Electroencephalogram and Electrooculogram Features
Abstract
1. Introduction
- We extracted multi-domain features from single-channel EEG signals that capture well the spectral and temporal characteristics of different sleep stages. We also proposed two novel EOG features that significantly improve the classification accuracy of the N1 and REM stages.
- We designed a novel two-step feature selection algorithm combining F-score prefiltering and XGBoost feature ranking that effectively identifies a small subset of discriminating features for sleep stage classification. This lays the foundation for the continued incorporation of new features in future works. The feature analysis results also provided quantifiable information for understanding the differences between sleep stages.
- We validated the proposed scheme on the popular Sleep-EDF database containing PSG data from 150 subjects following strict double cross-validation procedures and compared the results with state-of-the-art deep learning models. We showed that competitive performance can be achieved with a small number of representative features using an interpretable machine learning model.
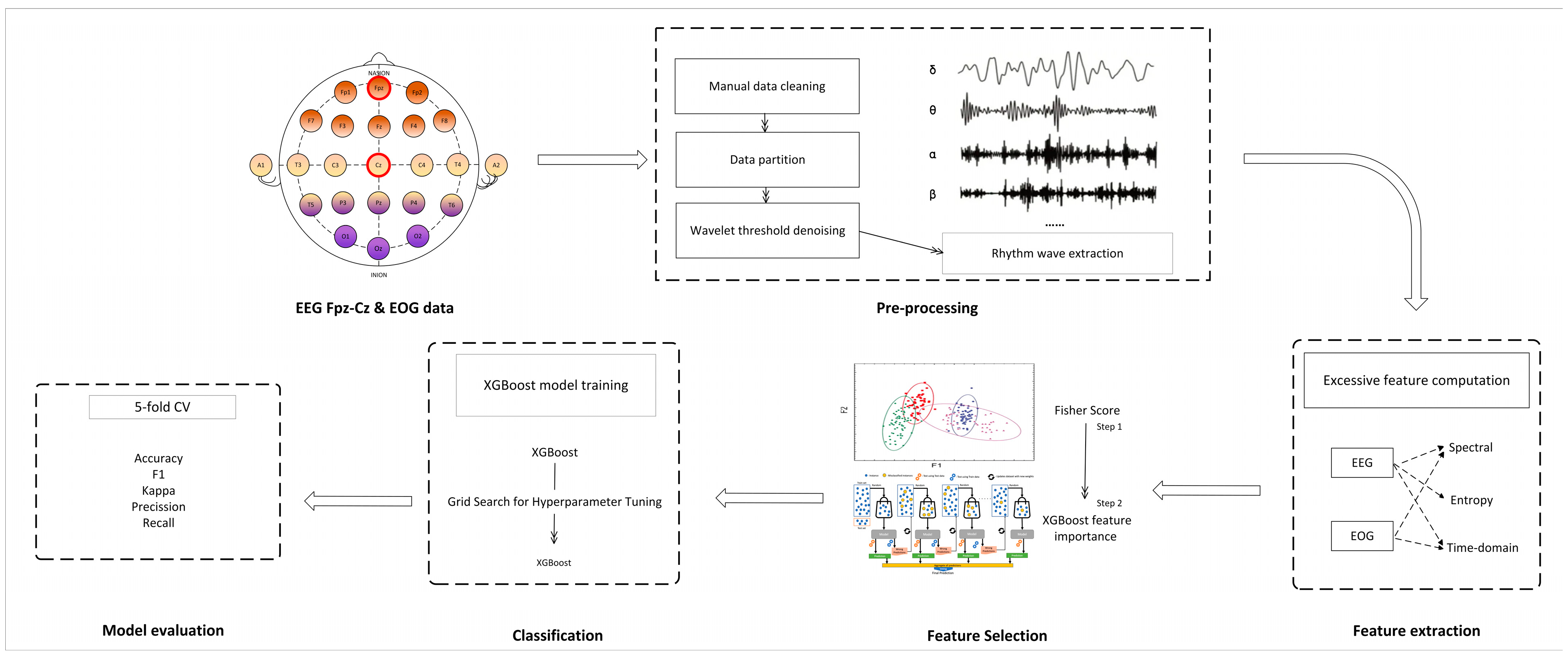
2. Proposed Method
2.1. Dataset
2.2. Preprocessing
2.3. Feature Extraction
2.3.1. Time Domain Features
2.3.2. Power Spectrum Density Features
2.3.3. Multiscale Entropy
2.4. Feature Selection
2.5. Classification Model
3. Experiments and Results
3.1. Evaluation Methods
3.2. Classification Results
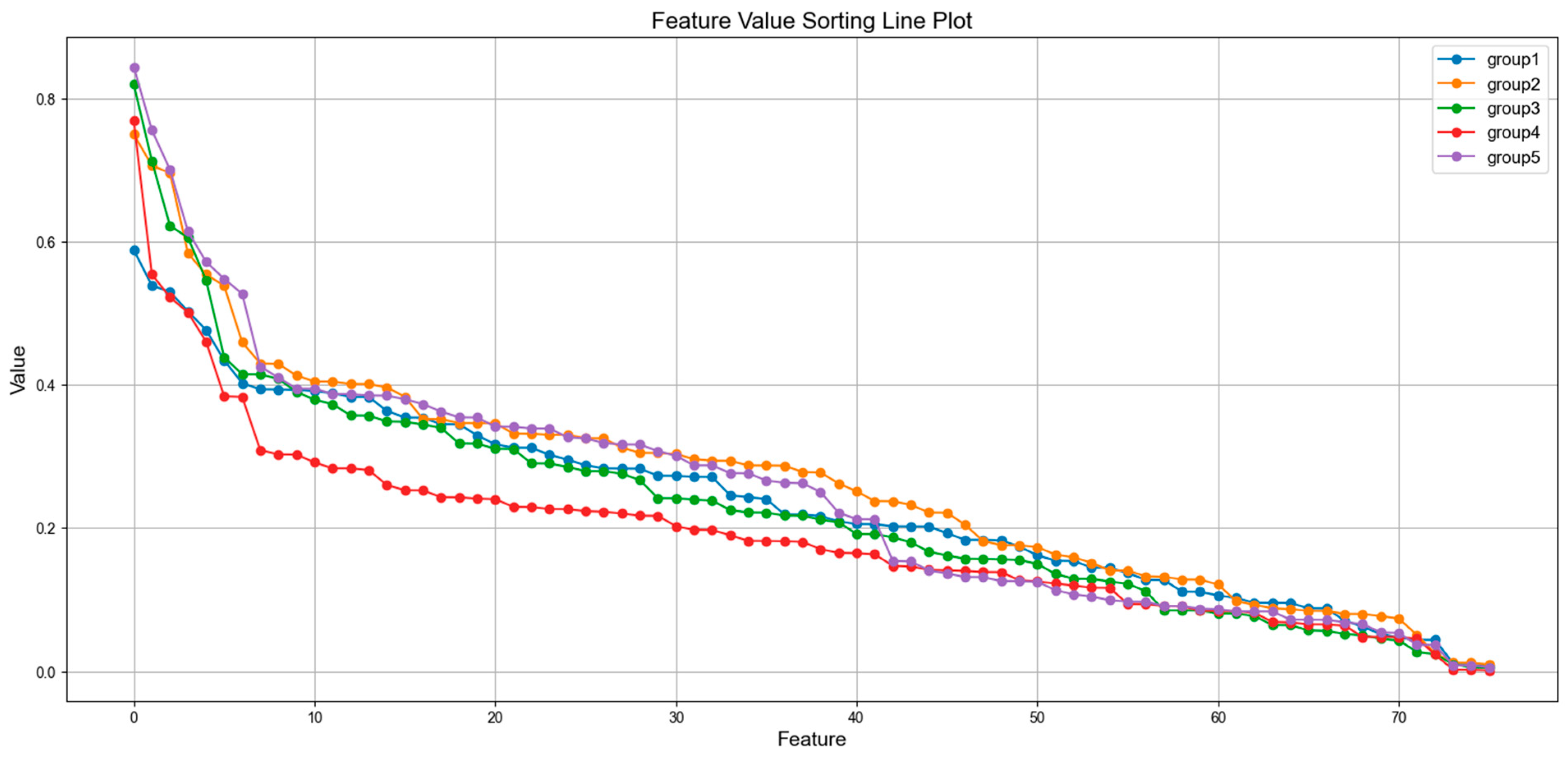
3.3. Feature Analysis
4. Discussions
5. Conclusions
- Developing more effective feature sets, particularly for the N1 stage, to further improve classification accuracy.
- Exploring more advanced feature selection algorithms to enhance the accuracy and adaptability of feature selection.
- Designing low-complexity, high-accuracy, and interpretable classification models to optimize N1 stage classification.
- Testing the models on more diverse datasets to validate their stability and adaptability across different populations and environments.
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ramar, K.; Malhotra, R.K.; Carden, K.A.; Martin, J.L.; Abbasi-Feinberg, F.; Aurora, R.N.; Kapur, V.K.; Olson, E.J.; Rosen, C.L.; Rowley, J.A.; et al. Sleep is essential to health: An American Academy of Sleep Medicine position statement. J. Clin. Sleep Med. JCSM Off. Publ. Am. Acad. Sleep Med. 2021, 17, 2115–2119. [Google Scholar] [CrossRef] [PubMed]
- Berry, R.B.; Brooks, R.; Gamaldo, C.E.; Harding, S.M.; Marcus, C.; Vaughn, B.V. The AASM Manual for the Scoring of Sleep and Associated Events: Rules, Terminology and Technical Specifications; American Academy of Sleep Medicine: Darien, IL, USA, 2012; Volume 176, p. 7. [Google Scholar]
- Hori, T.; Sugita, Y.; Koga, E.; Shirakawa, S.; Inoue, K.; Uchida, S.; Kuwahara, H.; Kousaka, M.; Kobayashi, T.; Tsuji, Y.; et al. Proposed supplements and amendments to ’A Manual of Standardized Terminology, Techniques and Scoring System for Sleep Stages of Human Subjects’, the Rechtschaffen & Kales (1968) standard. Psychiatry Clin. Neurosci. 2001, 55, 305–310. [Google Scholar] [CrossRef]
- Alickovic, E.; Subasi, A. Ensemble SVM Method for Automatic Sleep Stage Classification. IEEE Trans. Instrum. Meas. 2018, 67, 1258–1265. [Google Scholar] [CrossRef]
- Rahman, M.M.; Bhuiyan, M.I.H.; Hassan, A.R. Sleep stage classification using single-channel EOG. Comput. Biol. Med. 2018, 102, 211–220. [Google Scholar] [CrossRef] [PubMed]
- Supratak, A.; Dong, H.; Wu, C.; Guo, Y. DeepSleepNet: A Model for Automatic Sleep Stage Scoring Based on Raw Single-Channel EEG. IEEE Trans. Neural Syst. Rehabil. Eng. 2017, 25, 1998–2008. [Google Scholar] [CrossRef]
- Eldele, E.; Chen, Z.; Liu, C.; Wu, M.; Kwoh, C.; Li, X.; Guan, C. An Attention-Based Deep Learning Approach for Sleep Stage Classification With Single-Channel EEG. IEEE Trans. Neural Syst. Rehabil. Eng. 2021, 29, 809–818. [Google Scholar] [CrossRef]
- Supratak, A.; Guo, Y. TinySleepNet: An Efficient Deep Learning Model for Sleep Stage Scoring based on Raw Single-Channel EEG. In Proceedings of the 2020 42nd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), Montreal, QC, Canada, 20–24 July 2020; pp. 641–644. [Google Scholar]
- Van Der Donckt, J.; Van Der Donckt, J.; Deprost, E.; Vandenbussche, N.; Rademaker, M.; Vandewiele, G.; Van Hoecke, S. Do not sleep on traditional machine learning: Simple and interpretable techniques are competitive to deep learning for sleep scoring. Biomed. Signal Process. Control 2023, 81, 104429. [Google Scholar] [CrossRef]
- Pradeepkumar, J.; Anandakumar, M.; Kugathasan, V.; Suntharalingham, D.; Kappel, S.L.; Silva, A.C.D.; Edussooriya, C.U.S. Toward Interpretable Sleep Stage Classification Using Cross-Modal Transformers. IEEE Trans. Neural Syst. Rehabil. Eng. 2022, 32, 2893–2904. [Google Scholar] [CrossRef]
- Kemp, B.; Zwinderman, A.H.; Tuk, B.; Kamphuisen, H.A.; Oberyé, J.J. Analysis of a sleep-dependent neuronal feedback loop: The slow-wave microcontinuity of the EEG. IEEE Trans. Bio-Med. Eng. 2000, 47, 1185–1194. [Google Scholar] [CrossRef]
- Mikkelsen, K.B.; Tabar, Y.R.; Kappel, S.L.; Christensen, C.B.; Toft, H.O.; Hemmsen, M.C.; Rank, M.L.; Otto, M.; Kidmose, P. Accurate whole-night sleep monitoring with dry-contact ear-EEG. Sci. Rep. 2019, 9, 16824. [Google Scholar] [CrossRef]
- Young, L.R.; Sheena, D. Survey of eye movement recording methods. Behav. Res. Methods Instrum. 1975, 7, 397–429. [Google Scholar] [CrossRef]
- Sparks, D.L. The brainstem control of saccadic eye movements. Nat. Rev. Neurosci. 2002, 3, 952–964. [Google Scholar] [CrossRef]
- Collewijn, H.; Erkelens, C.J.; Steinman, R.M. Binocular co-ordination of human horizontal saccadic eye movements. J. Physiol. 1988, 404, 157–182. [Google Scholar] [CrossRef]
- Osadchiy, A.; Kamenev, A.; Saharov, V.; Chernyi, S. Signal Processing Algorithm Based on Discrete Wavelet Transform. Designs 2021, 5, 41. [Google Scholar] [CrossRef]
- Chen, W.; Anderson, B.D.O.; Deistler, M.; Filler, A. Solutions of Yule-Walker equations for singular AR processes. J. Time Ser. Anal. 2011, 32, 531–538. [Google Scholar] [CrossRef]
- Phan, H.; Mikkelsen, K. Automatic sleep staging of EEG signals: Recent development, challenges, and future directions. Physiol. Meas. 2022, 43, 04TR01. [Google Scholar] [CrossRef] [PubMed]
- Humeau-Heurtier, A. The Multiscale Entropy Algorithm and Its Variants: A Review. Entropy 2015, 17, 3110–3123. [Google Scholar] [CrossRef]
- Pincus, S.M. Approximate entropy as a measure of system complexity. Proc. Natl. Acad. Sci. USA 1991, 88, 2297–2301. [Google Scholar] [CrossRef]
- Liang, S.-F.; Kuo, C.-E.; Hu, Y.-H.; Pan, Y.-H.; Wang, Y.-H. Automatic Stage Scoring of Single-Channel Sleep EEG by Using Multiscale Entropy and Autoregressive Models. IEEE Trans. Instrum. Meas. 2012, 61, 1649–1657. [Google Scholar] [CrossRef]
- Luo, Y.; Mu, W.; Wang, L.; Wang, J.; Wang, P.; Gan, Z.; Zhang, L.; Kang, X. An EEG channel selection method for motor imagery based on Fisher score and local optimization. J. Neural Eng. 2024, 21, 036030. [Google Scholar] [CrossRef]
- Chen, T.; Guestrin, C. XGBoost: A Scalable Tree Boosting System. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016. [Google Scholar] [CrossRef]
- Tan, H. Machine Learning Algorithm for Classification. J. Phys. Conf. Ser. 2021, 1994, 012016. [Google Scholar] [CrossRef]
- Chicco, D.; Jurman, G. The advantages of the Matthews correlation coefficient (MCC) over F1 score and accuracy in binary classification evaluation. BMC Genom. 2020, 21, 6. [Google Scholar] [CrossRef] [PubMed]
- Davis, J.; Goadrich, M.H. The relationship between Precision-Recall and ROC curves. In Proceedings of the 23rd International Conference on Machine Learning, Pittsburgh, Pennsylvania, 25–29 June 2006. [Google Scholar] [CrossRef]
- Hsu, Y.-L.; Yang, Y.-T.C.; Wang, J.-S.; Hsu, C.-Y. Automatic sleep stage recurrent neural classifier using energy features of EEG signals. Neurocomputing 2013, 104, 105–114. [Google Scholar] [CrossRef]
- Sharma, R.; Pachori, R.B.; Upadhyay, A.B. Automatic sleep stages classification based on iterative filtering of electroencephalogram signals. Neural Comput. Appl. 2017, 28, 2959–2978. [Google Scholar] [CrossRef]
- Hassan, A.R.; Subasi, A. A decision support system for automated identification of sleep stages from single-channel EEG signals. Knowl. Based Syst. 2017, 128, 115–124. [Google Scholar] [CrossRef]
- Tsinalis, O.; Matthews, P.M.; Guo, Y. Automatic Sleep Stage Scoring Using Time-Frequency Analysis and Stacked Sparse Autoencoders. Ann. Biomed. Eng. 2016, 44, 1587–1597. [Google Scholar] [CrossRef]
- Tsinalis, O.; Matthews, P.M.; Guo, Y.; Zafeiriou, S. Automatic Sleep Stage Scoring with Single-Channel EEG Using Convolutional Neural Networks. arXiv 2016, arXiv:1610.01683. [Google Scholar] [CrossRef]
- Casciola, A.A.; Carlucci, S.K.; Kent, B.A.; Punch, A.M.; Muszynski, M.A.; Zhou, D.; Kazemi, A.; Mirian, M.S.; Valerio, J.; McKeown, M.J.; et al. A Deep Learning Strategy for Automatic Sleep Staging Based on Two-Channel EEG Headband Data. Sensors 2021, 21, 3316. [Google Scholar] [CrossRef]



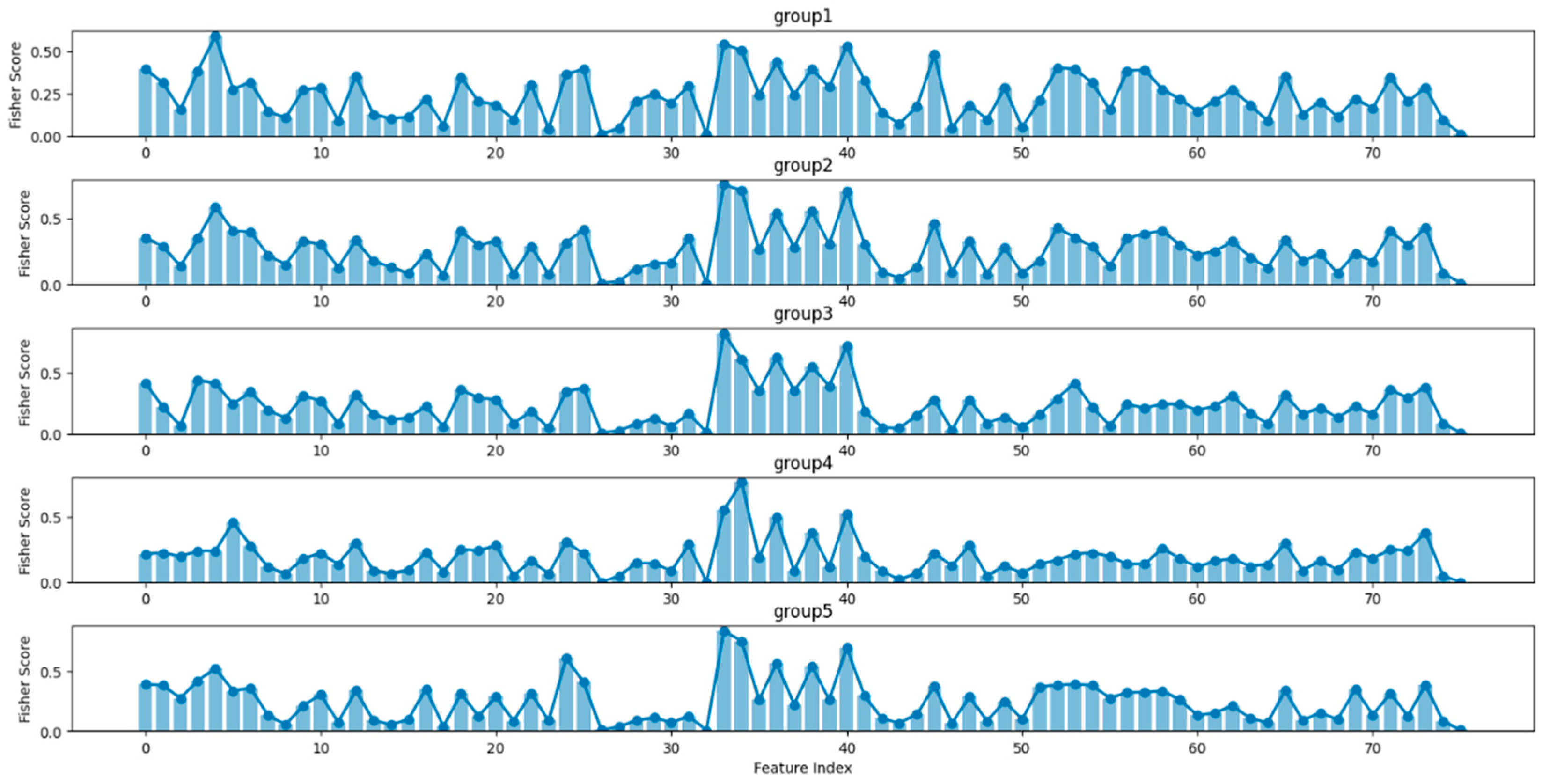
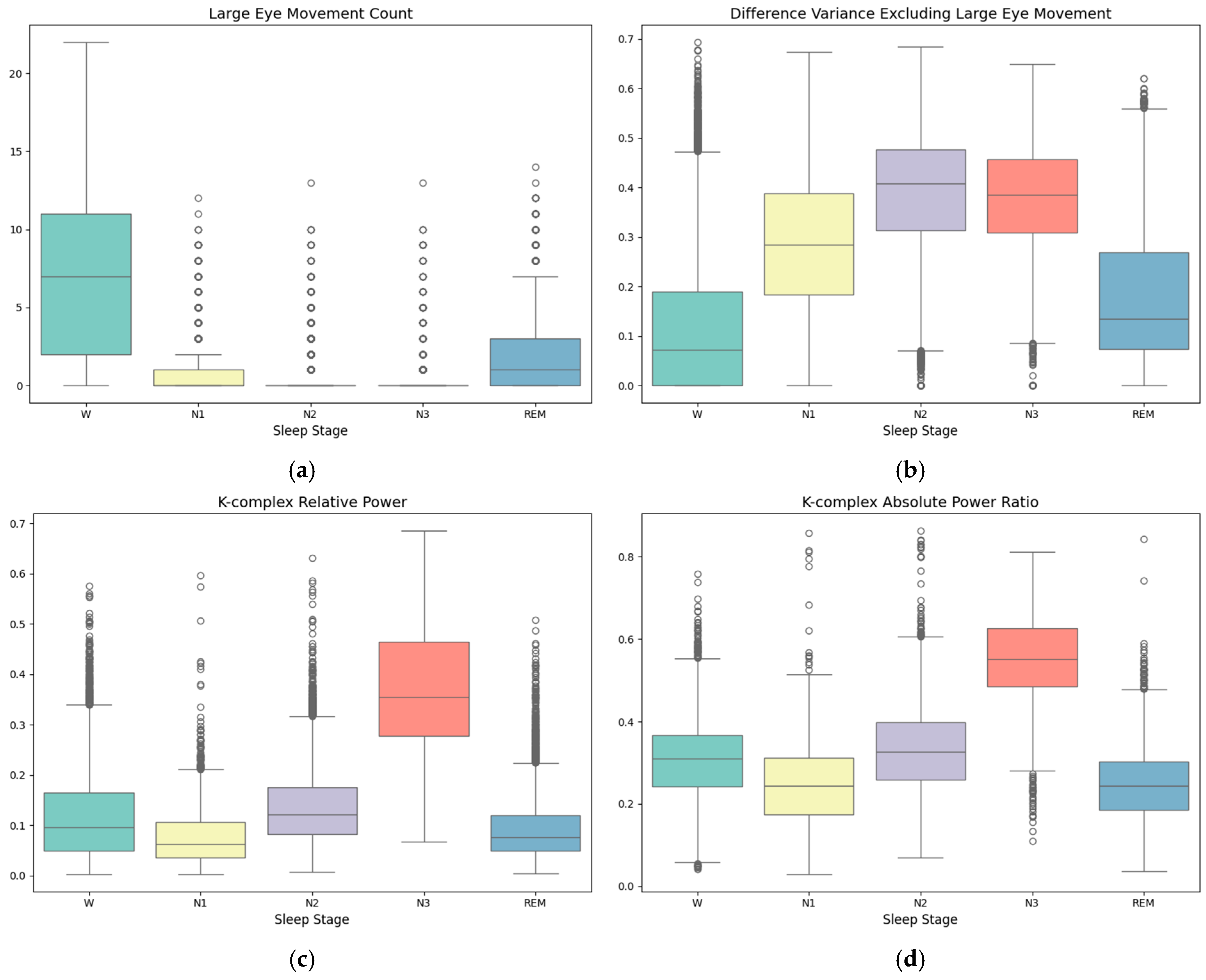
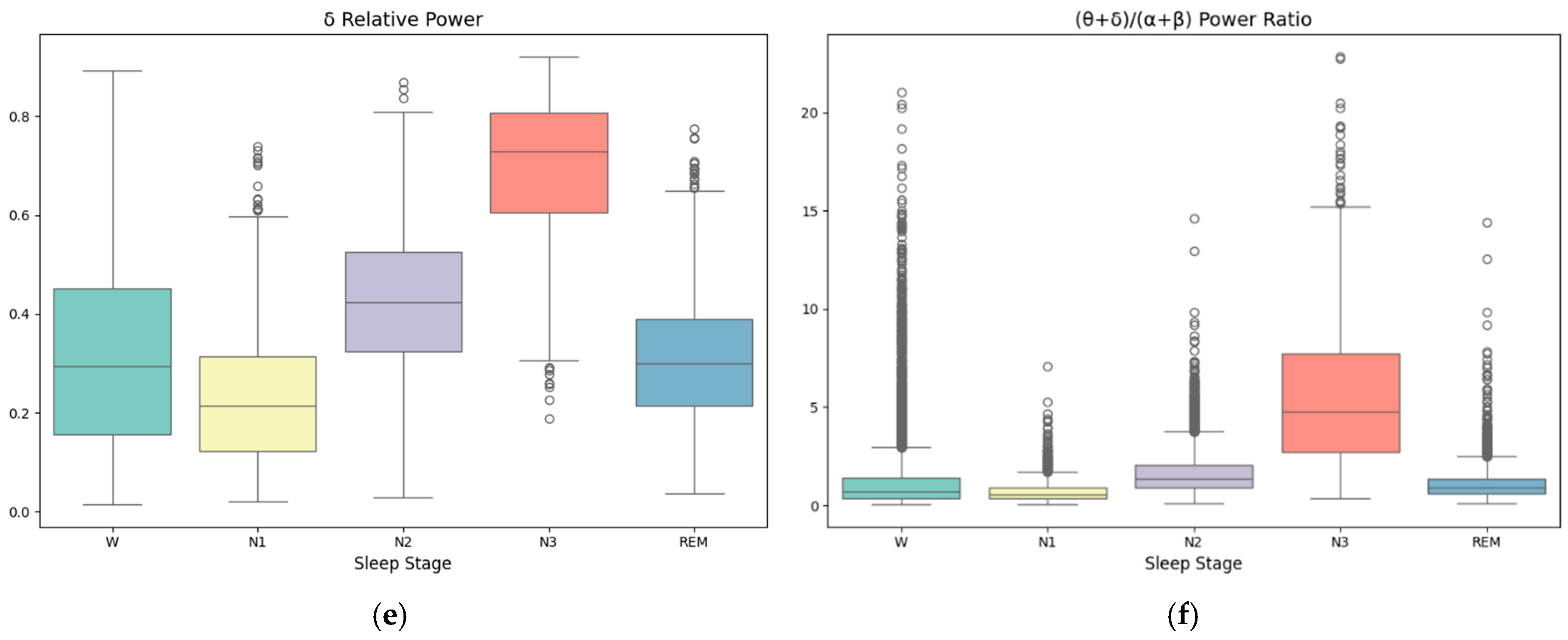
| Label | W | N1 | N2 | N3 | REM |
|---|---|---|---|---|---|
| Number of Frames | 52,492 | 15,064 | 60,473 | 8203 | 23,364 |
| Label | W | N1 | N2 | N3 | REM | Total |
|---|---|---|---|---|---|---|
| Part 1 | 9752 | 2808 | 12,430 | 1837 | 4648 | 31,475 |
| Part 2 | 10,900 | 3712 | 11,390 | 2128 | 4640 | 32,770 |
| Part 3 | 9652 | 3192 | 11,483 | 1488 | 4582 | 30,397 |
| Part 4 | 12,447 | 2561 | 12,525 | 1376 | 4699 | 33,608 |
| Part 5 | 9741 | 2791 | 12,645 | 1374 | 4795 | 31,346 |
| Total | 52,492 | 15,064 | 60,473 | 8203 | 23,364 | 159,596 |
| Function | EEG | #Features | EOG | #Features |
|---|---|---|---|---|
| Time-domain | ||||
| Range, Mean, Variance, Standard Deviation, Peak Count, Zero-crossing Count, Difference Variance | √ | 49 | √ | 7 |
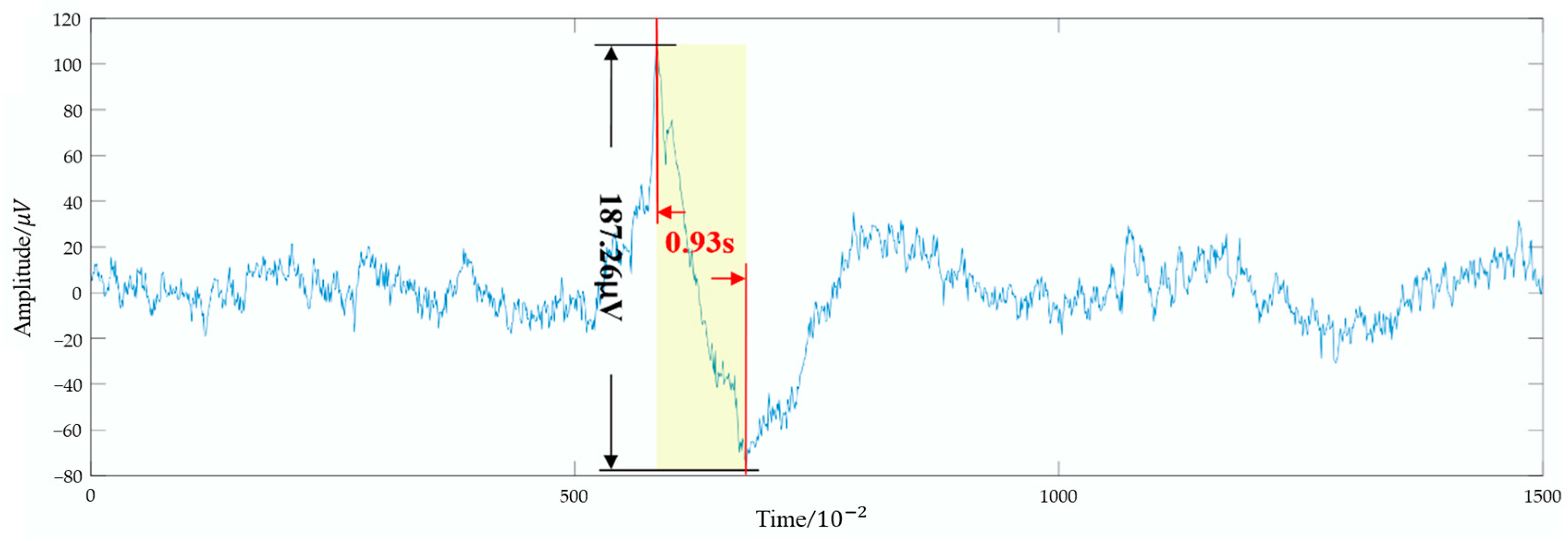
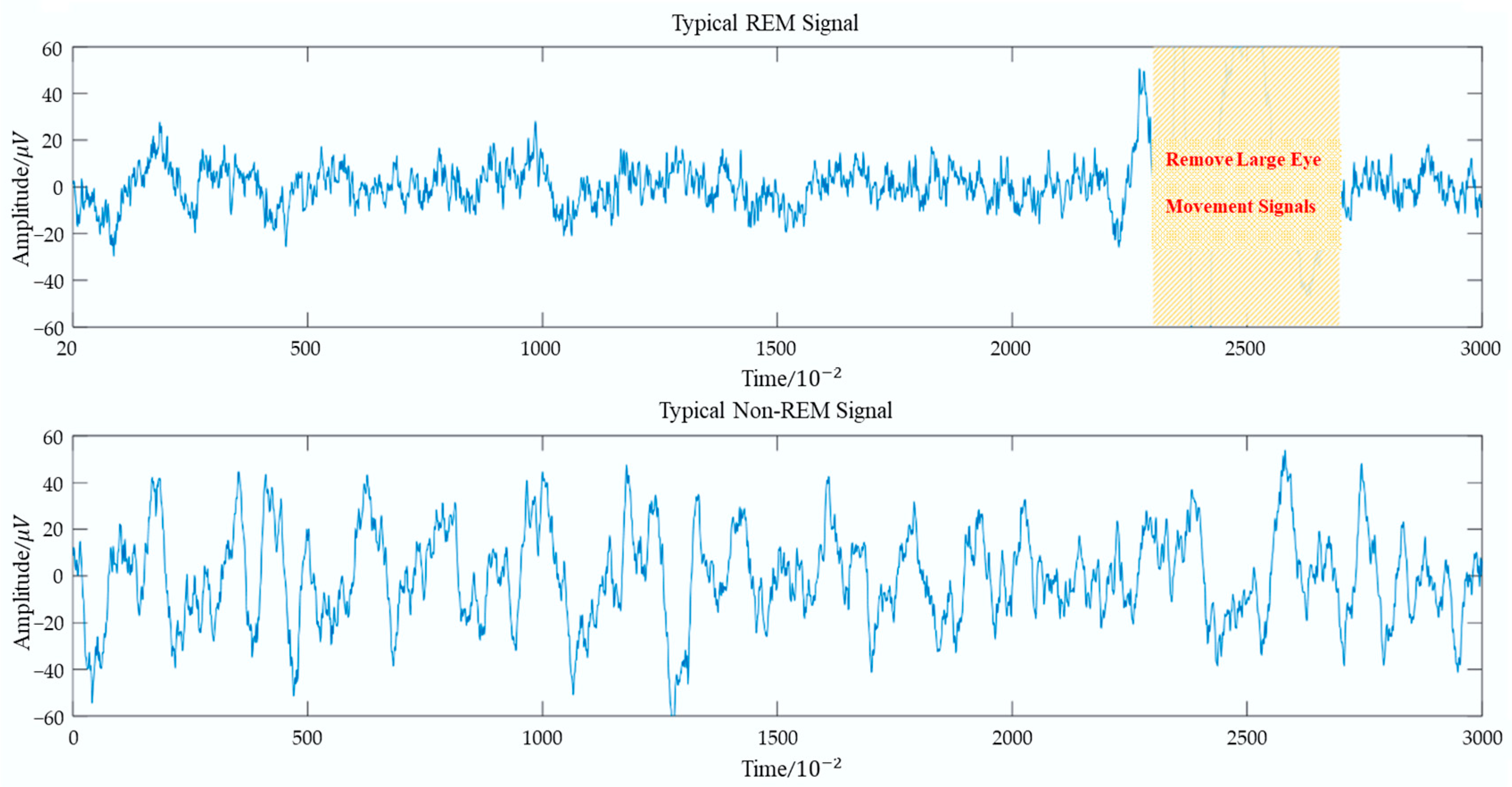
| Large Eye Movement Detection Difference Variance Excluding Large Eye Movement | - | - | √ | 2 |
| Power Spectrum Density | ||||
| Absolute power ratios of different frequency bands (Delta, Theta, Alpha, Beta, K-complex, Spindle and Sawtooth) | √ | 7 | - | - |
| Spectral Power Ratio: | √ | 4 | - | - |
| Eye movement power ratio: | - | - | √ | 2 |
| Multiscale Entropy | ||||
| Sample Entropy | √ | 5 | - | - |
| Predicted | Per-Class Metrics | |||||||
|---|---|---|---|---|---|---|---|---|
| W | N1 | N2 | N3 | REM | PR | RE | F1 | |
| W | 49,878 | 1371 | 629 | 12 | 781 | 92.68 | 94.63 | 93.64 |
| N1 | 1865 | 6178 | 4688 | 14 | 1575 | 57.30 | 42.37 | 48.54 |
| N2 | 884 | 1998 | 56,742 | 750 | 1768 | 86.62 | 91.28 | 88.85 |
| N3 | 120 | 3 | 1182 | 6803 | 6 | 88.88 | 83.33 | 85.98 |
| REM | 1070 | 1198 | 2223 | 17 | 17,840 | 81.22 | 79.87 | 80.54 |
| Method | Per-Class F1-Score | Overall Metrics | ||||||
|---|---|---|---|---|---|---|---|---|
| W | N1 | N2 | N3 | REM | Accuracy | MFI | κ | |
| 67 features (EEG only) | 92.0 | 43.3 | 88.2 | 85.9 | 77.5 | 84.4 | 83.8 | 0.78 |
| 76 features (EEG + EOG) | 94.8 | 53.3 | 89.9 | 87.4 | 84.1 | 87.5 | 87.1 | 0.82 |
| 25 features (EEG + EOG) | 93.6 | 48.5 | 88.9 | 86.0 | 80.5 | 87.0 | 86.6 | 0.81 |
| 10 features (EEG + EOG) | 90.4 | 36.1 | 86.1 | 80.3 | 69.3 | 83.0 | 82.3 | 0.76 |
| Methods | EEG Channel | Test Epochs | Feature Count | Overall Metrics | Per-Class F1-Score(F1) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| ACC | MFI | κ | W | N1 | N2 | N3 | REM | ||||
| Non-independent Training and Test Sets | |||||||||||
| Ref. [27] | Fpz-Cz | 960 | - | 90.3 | 76.5 | - | 77.3 | 46.5 | 94.9 | 72.2 | 91.8 |
| Ref. [28] | Pz-Oz | 15,136 | 50 | 91.3 | 77 | 0.86 | 97.8 | 30.4 | 89 | 85.5 | 82.5 |
| Ref. [29] | Pz-Oz | 7596 | - | 90.8 | 80 | 0.85 | 96.9 | 49.1 | 89 | 84.2 | 81.2 |
| Independent Training and Test Sets | |||||||||||
| This paper | Fpz-Cz EOG | 159,596 | 25 | 87.0 | 86.6 | 0.81 | 93.6 | 48.5 | 88.9 | 86.0 | 80.5 |
| Ref. [30] | Fpz-Cz | 37,022 | 35 | 78.9 | 73.7 | - | 71.6 | 47.0 | 84.6 | 84.0 | 81.4 |
| Ref. [31] | Fpz-Cz | 37,022 | 35 | 74.8 | 69.8 | - | 65.4 | 43.7 | 80.6 | 84.9 | 74.5 |
| Ref. [32] | F3-M2 F4-M1 | - | 62 | 77.0 | - | - | 84.6 | 31.1 | 77.8 | 85.3 | 75.4 |
| Ref. [7] | Fpz-Cz | 32,485 | - | 84.2 | 75.3 | 0.78 | 86.7 | 33.2 | 87.1 | 87.1 | 82.1 |
| Ref. [6] | Fpz-Cz C4-A1 | 41,950 | - | 82.0 | 76.9 | 0.76 | 84.7 | 46.6 | 85.9 | 84.8 | 82.4 |
| Ref. [6] | Pz-Oz | 41,950 | - | 79.8 | 73.1 | 0.72 | 88.1 | 37 | 82.7 | 77.3 | 80.3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, X.; Zhang, B.; Xu, T.; Tang, J. An Effective and Interpretable Sleep Stage Classification Approach Using Multi-Domain Electroencephalogram and Electrooculogram Features. Bioengineering 2025, 12, 286. https://doi.org/10.3390/bioengineering12030286
Xu X, Zhang B, Xu T, Tang J. An Effective and Interpretable Sleep Stage Classification Approach Using Multi-Domain Electroencephalogram and Electrooculogram Features. Bioengineering. 2025; 12(3):286. https://doi.org/10.3390/bioengineering12030286
Chicago/Turabian StyleXu, Xin, Bei Zhang, Tingting Xu, and Junyi Tang. 2025. "An Effective and Interpretable Sleep Stage Classification Approach Using Multi-Domain Electroencephalogram and Electrooculogram Features" Bioengineering 12, no. 3: 286. https://doi.org/10.3390/bioengineering12030286
APA StyleXu, X., Zhang, B., Xu, T., & Tang, J. (2025). An Effective and Interpretable Sleep Stage Classification Approach Using Multi-Domain Electroencephalogram and Electrooculogram Features. Bioengineering, 12(3), 286. https://doi.org/10.3390/bioengineering12030286






