Real-Time Object Detector for Medical Diagnostics (RTMDet): A High-Performance Deep Learning Model for Brain Tumor Diagnosis
Abstract
1. Introduction
2. Related Works
3. Methodology
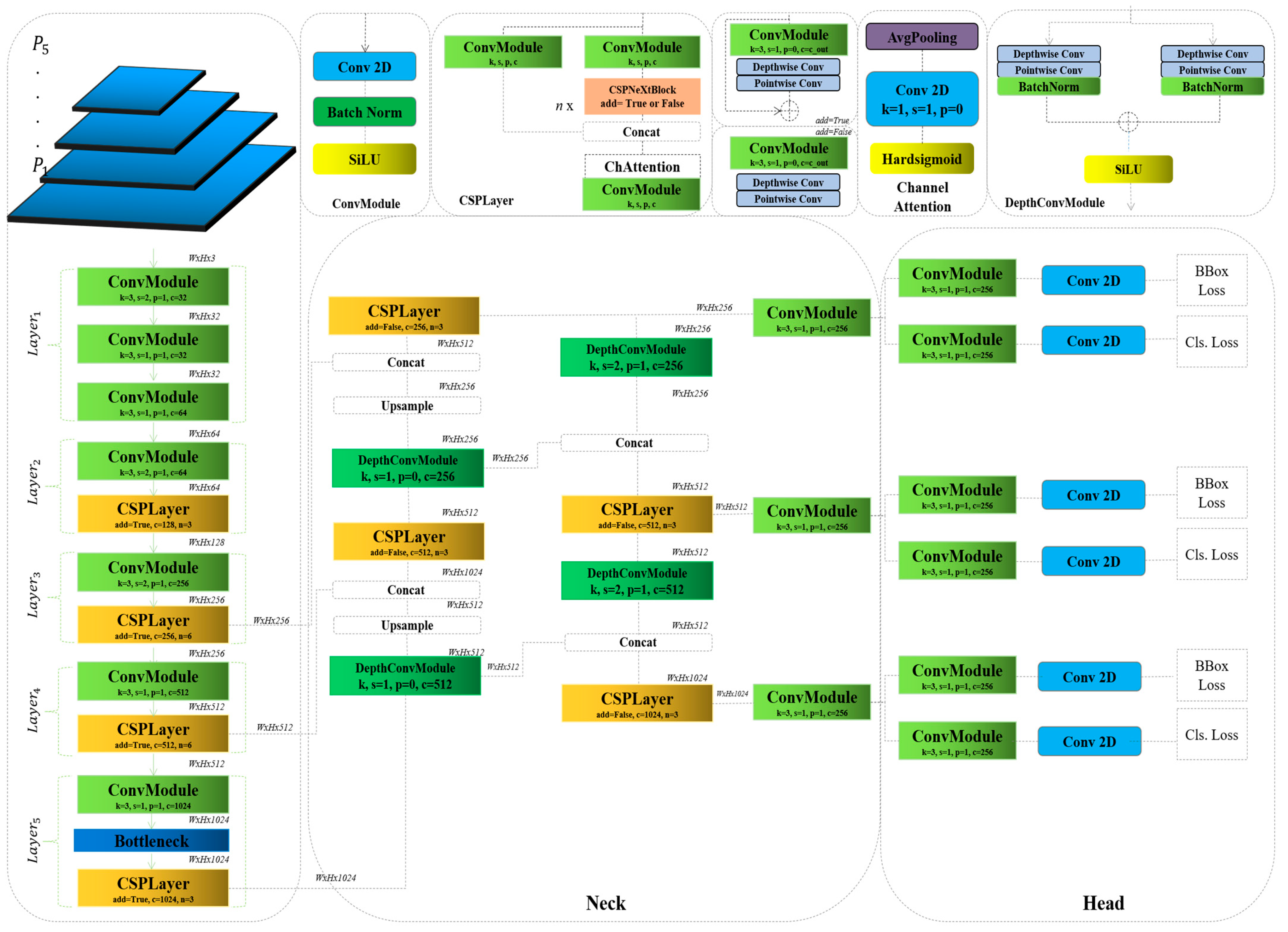
3.1. RTMDet
3.2. Rethinking the RTMDet
| Algorithm 1. Depthwise convolution module |
| 1: class DepthConvModule: |
| 2: #In this context, ‘dw1_conv’ and ‘pw1_conv’ refer to the depthwise and pointwise convolutions, |
| 3: respectively, for the first pathway. |
| 4: dw1_conv : Conv2d(in_channels, in_channels, kernel, padding, stride, groups) |
| 5: pw1_conv : Conv2d(in_channels, out_channels, kernel, padding, stride) |
| 6: #Here same structure for, ‘dw2_conv’ and ‘pw2_conv’ refer to the depthwise and pointwise |
| 7: convolutions, respectively, for the second pathway. |
| 8: dw2_conv : Conv2d(in_channels, in_channels, kernel, padding, stride, groups) |
| 9: pw2_conv : Conv2d(in_channels, out_channels, kernel, padding, stride) |
| 10: Connection: Residual |
| 11: Normalization: BatchNormalization |
| 12: Activation: SiLU() |
4. The Experiment and Analyses
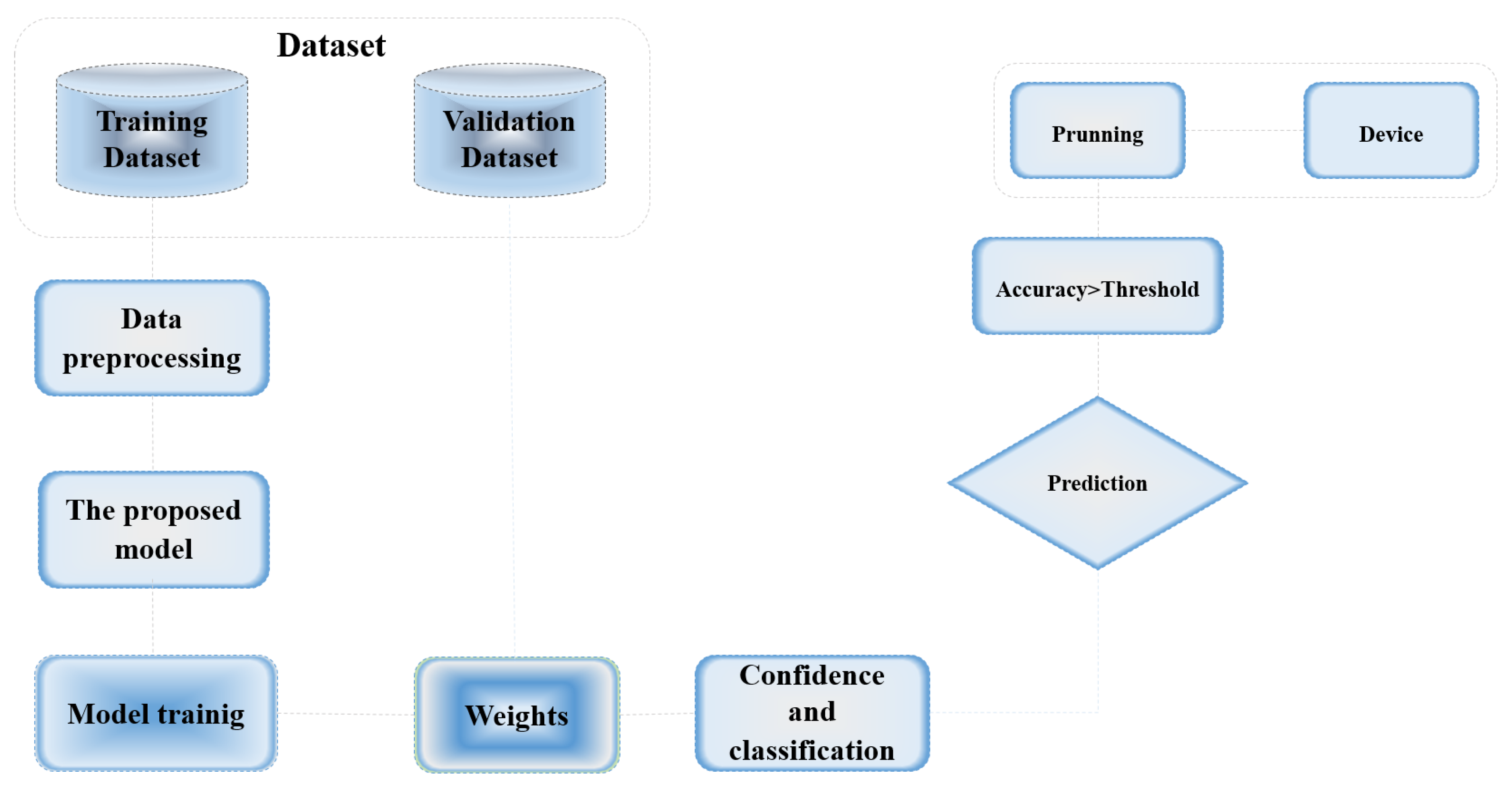
4.1. The Dataset
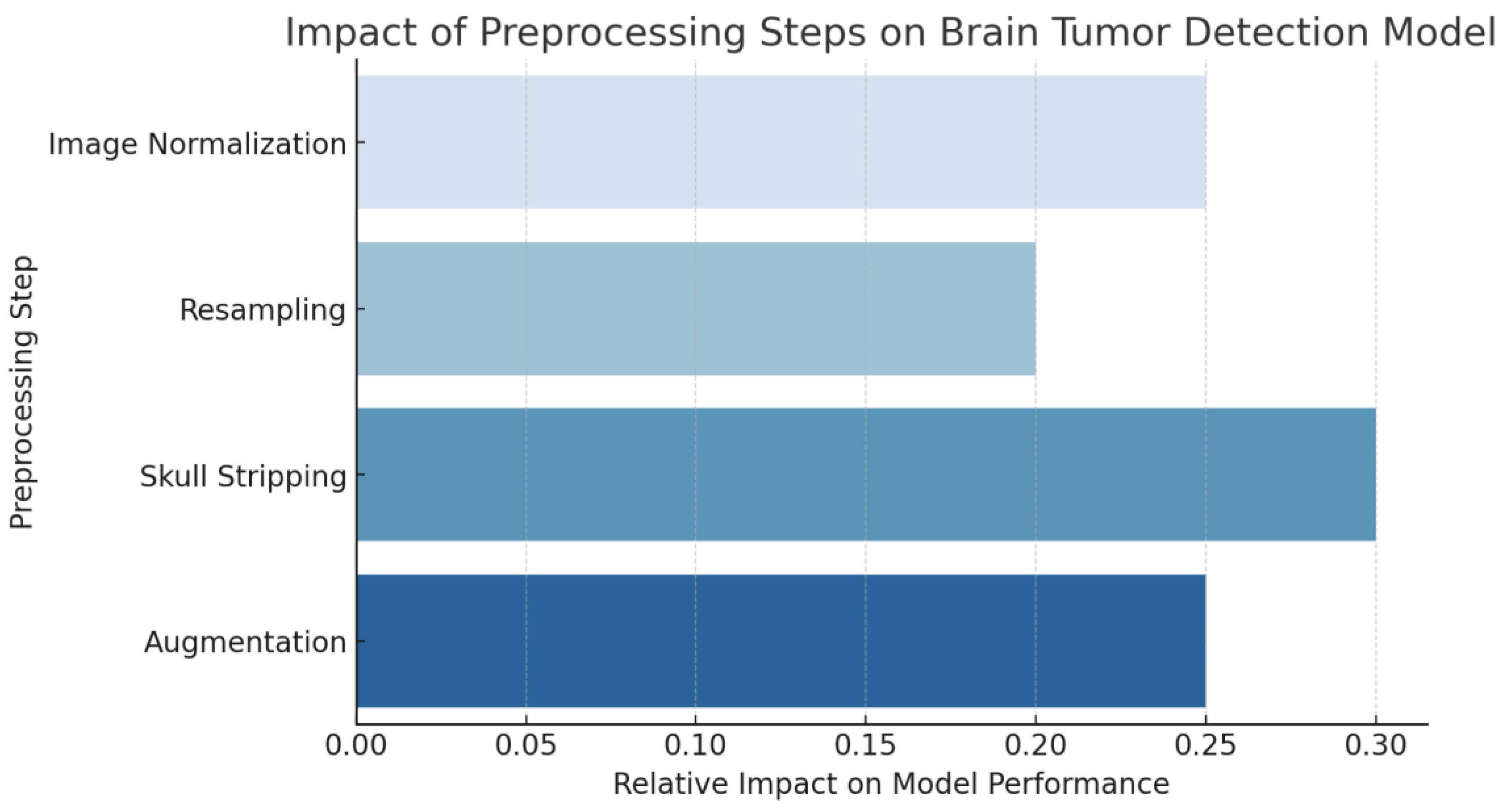
4.2. Data Preprocessing
4.3. The Analyses of the Results and Comparison of the Baseline Models
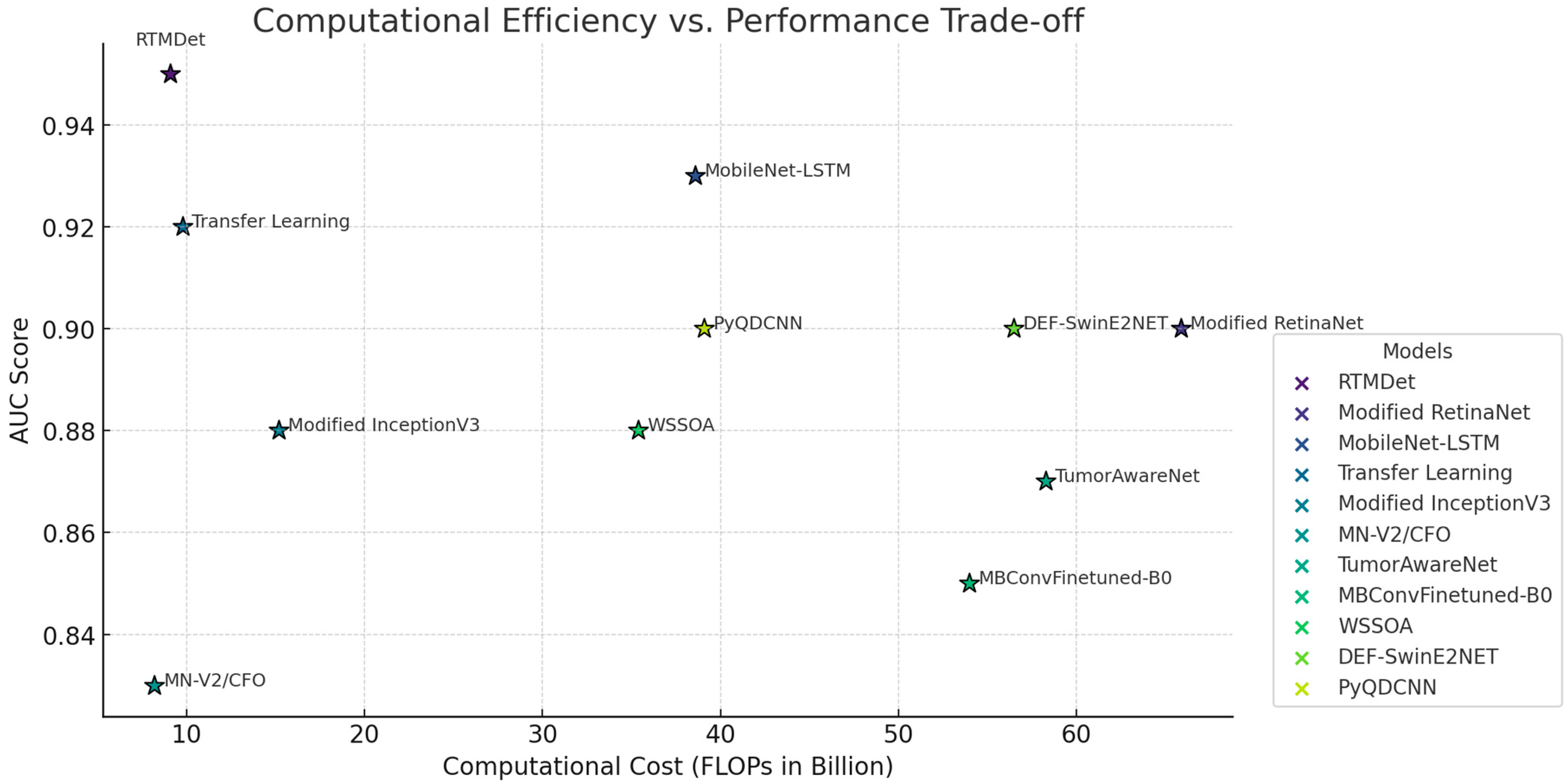
4.4. Comparison with State-of-the-Art Models
5. Discussion
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Leone, A.; Carbone, F.; Spetzger, U.; Vajkoczy, P.; Raffa, G.; Angileri, F.; Germanó, A.; Engelhardt, M.; Picht, T.; Colamaria, A.; et al. Preoperative mapping techniques for brain tumor surgery: A systematic review. Front. Oncol. 2025, 14, 1481430. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Li, S.; Hou, C.; Wang, Z.; He, W.; Zhang, W. Recent Advances in mRNA-Based Therapeutics for Neurodegenerative Diseases and Brain Tumors. Nanoscale 2025, 17, 3537–3548. [Google Scholar] [CrossRef] [PubMed]
- Avazov, K.; Mirzakhalilov, S.; Umirzakova, S.; Abdusalomov, A.; Cho, Y.I. Dynamic Focus on Tumor Boundaries: A Lightweight U-Net for MRI Brain Tumor Segmentation. Bioengineering 2024, 11, 1302. [Google Scholar] [CrossRef]
- Talukder, M.A.; Layek, M.A.; Hossain, M.A.; Islam, M.A.; Nur-e-Alam, M.; Kazi, M. ACU-Net: Attention-based convolutional U-Net model for segmenting brain tumors in fMRI images. Digital Health 2025, 11, 20552076251320288. [Google Scholar] [CrossRef] [PubMed]
- Abdusalomov, A.; Mirzakhalilov, S.; Umirzakova, S.; Shavkatovich Buriboev, A.; Meliboev, A.; Muminov, B.; Jeon, H.S. Accessible AI Diagnostics and Lightweight Brain Tumor Detection on Medical Edge Devices. Bioengineering 2025, 12, 62. [Google Scholar] [CrossRef]
- Khan, S.U.R.; Asif, S.; Zhao, M.; Zou, W.; Li, Y.; Li, X. Optimized deep learning model for comprehensive medical image analysis across multiple modalities. Neurocomputing 2025, 619, 129182. [Google Scholar] [CrossRef]
- Jayalakshmi, P.R.; Rajakumar, P.S. A Survey: Recent Advances and Clinical Applications. In Deep Learning in Medical Image Analysis; CRC Press: Boca Raton, FL, USA, 2025; pp. 59–78. [Google Scholar]
- Cheng, C.T.; Ooyang, C.H.; Liao, C.H.; Kang, S.C. Applications of deep learning in trauma radiology: A narrative review. Biomed. J. 2025, 48, 100743. [Google Scholar] [CrossRef]
- Zhao, Y.; Lv, W.; Xu, S.; Wei, J.; Wang, G.; Dang, Q.; Liu, Y.; Chen, J. Detrs beat yolos on real-time object detection. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Seattle, WA, USA, 16–22 June 2024; pp. 16965–16974. [Google Scholar]
- Wang, Y.; Chen, J.; Yang, B.; Chen, Y.; Su, Y.; Liu, R. RT-DETRmg: A lightweight real-time detection model for small traffic signs. J. Supercomput. 2025, 81, 307. [Google Scholar] [CrossRef]
- Hu, J.; Wei, Y.; Chen, W.; Zhi, X.; Zhang, W. CM-YOLO: Typical Object Detection Method in Remote Sensing Cloud and Mist Scene Images. Remote Sens. 2025, 17, 125. [Google Scholar] [CrossRef]
- Hekmat, A.; Zhang, Z.; Khan, S.U.R.; Shad, I.; Bilal, O. An attention-fused architecture for brain tumor diagnosis. Biomed. Signal Process. Control 2025, 101, 107221. [Google Scholar] [CrossRef]
- Chen, C.; Isa, N.A.M.; Liu, X. A review of convolutional neural network based methods for medical image classification. Comput. Biol. Med. 2025, 185, 109507. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Zhang, J.; Wei, S.; Li, Z. PFormer: An efficient CNN-Transformer hybrid network with content-driven P-attention for 3D medical image segmentation. Biomed. Signal Process. Control 2025, 101, 107154. [Google Scholar] [CrossRef]
- Schwehr, Z.; Achanta, S. Brain tumor segmentation based on deep learning, attention mechanisms, and energy-based uncertainty predictions. In Multimedia Tools and Applications; Springer: Berlin/Heidelberg, Germany, 2025; pp. 1–20. [Google Scholar]
- Mathivanan, S.K.; Sonaimuthu, S.; Murugesan, S.; Rajadurai, H.; Shivahare, B.D.; Shah, M.A. Employing deep learning and transfer learning for accurate brain tumor detection. Sci. Rep. 2024, 14, 7232. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, M.; Rani, G.; Kumar, A.; Kumar, P.; Manikandan, R.; Gandomi, A.H. Deep learning for enhanced brain Tumor Detection and classification. Results Eng. 2024, 22, 102117. [Google Scholar] [CrossRef]
- Khan, M.F.; Iftikhar, A.; Anwar, H.; Ramay, S.A. Brain tumor segmentation and classification using optimized deep learning. J. Comput. Biomed. Inform. 2024, 7, 632–640. [Google Scholar]
- Nhlapho, W.; Atemkeng, M.; Brima, Y.; Ndogmo, J.C. Bridging the Gap: Exploring Interpretability in Deep Learning Models for Brain Tumor Detection and Diagnosis from MRI Images. Information 2024, 15, 182. [Google Scholar] [CrossRef]
- Khaliki, M.Z.; Başarslan, M.S. Brain tumor detection from images and comparison with transfer learning methods and 3-layer CNN. Sci. Rep. 2024, 14, 2664. [Google Scholar] [CrossRef]
- Xu, L.; Mohammadi, M. Brain tumor diagnosis from MRI based on Mobilenetv2 optimized by contracted fox optimization algorithm. Heliyon 2024, 10, e23866. [Google Scholar] [CrossRef]
- Bodapati, J.D.; Balaji, B.B. TumorAwareNet: Deep representation learning with attention based sparse convolutional denoising autoencoder for brain tumor recognition. Multimed. Tools Appl. 2024, 83, 22099–22117. [Google Scholar] [CrossRef]
- Kavitha, P.; Dhinakaran, D.; Prabaharan, G.; Manigandan, M.D. Brain Tumor Detection for Efficient Adaptation and Superior Diagnostic Precision by Utilizing MBConv-Finetuned-B0 and Advanced Deep Learning. Int. J. Intell. Eng. Syst. 2024, 17, 632–644. [Google Scholar]
- Mandle, A.K.; Sahu, S.P.; Gupta, G.P. WSSOA: Whale social spider optimization algorithm for brain tumor classification using deep learning technique. Int. J. Inf. Technol. 2024, 1–17. [Google Scholar] [CrossRef]
- Malik, M.G.A.; Saeed, A.; Shehzad, K.; Iqbal, M. DEF-SwinE2NET: Dual enhanced features guided with multi-model fusion for brain tumor classification using preprocessing optimization. Biomed. Signal Process. Control 2025, 100, 107079. [Google Scholar]
- Jetlin, C.P. PyQDCNN: Pyramid QDCNNet for multi-level brain tumor classification using MRI image. Biomed. Signal Process. Control 2025, 100, 107042. [Google Scholar]



| Aspect | Description |
|---|---|
| Foundation of CNNs | Built on CNNs, the RTMDet utilizes multiple convolutional layers to extract and interpret spatial hierarchies in image data, enabling effective object recognition at various scales and orientations. |
| Integration of Depthwise Convolutional Blocks | Incorporates depthwise separable convolutions (DWConvBlocks), where depthwise convolutions handle spatial feature extraction and pointwise convolutions adjust dimensionality. This reduces computational complexity while maintaining high feature resolution. |
| Real-Time Processing Capabilities | Optimized for real-time performance with computational techniques such as batch normalization and skip connections. These enhancements improve training efficiency and gradient flow, preventing vanishing gradient issues in deep networks. |
| Adaptations for Medical Imaging | Fine-tuned for detecting brain tumors by adjusting filter sizes and strategically placing depthwise blocks. These refinements enhance sensitivity to medical-specific features, improving detection precision. |
| Deployment and Practical Applications | Designed for integration into medical imaging systems, the RTMDet enables real-time analysis and alerts for early disease detection. Beyond medical applications, its adaptable architecture supports various real-time object detection tasks. |
| Aspect | Details |
|---|---|
| Datasets Used | BraTS, ANDI (Brain Tumor Segmentation and Artificial and Natural Dataset for Intracranial Hemorrhage) |
| Preprocessing Steps | Skull stripping, image normalization, resampling, augmentation |
| Model Tested | RTMDet |
| Comparison Models | YOLOv5, YOLOv6, YOLOv7, YOLOv8, etc. |
| Key Metrics Evaluated | Average precision (AP), sensitivity, specificity, computational efficiency (FLOPs, parameters) |
| Main Outputs | RTMDet showed superior performance in speed and accuracy, with detailed results highlighted in the sensitivity and AP metrics. |
| Software and Tools | Description of software and analytical tools used for data processing and analysis. |
| Preprocessing Step | Description |
|---|---|
| Image Normalization | Ensures uniform intensity scales across different imaging modalities. MRI scans undergo Z-score normalization, adjusting pixel values to have a mean of zero and a standard deviation of one. CT scans use windowing to highlight intensity ranges relevant to hemorrhages. |
| Resampling | Standardizes resolution across all images to ensure consistent voxel sizes. This step enables the model to learn scale-invariant features and prevents variations in voxel dimensions from affecting training. |
| Skull Stripping | Removes non-brain tissues, particularly in MRI scans, to focus the model’s attention on brain structures where tumors are located. Automated algorithms leveraging anatomical atlases or deep learning methods are used for precision. |
| Augmentation | Enhances model robustness and prevents overfitting by applying rotation, flipping, and slight translations. These transformations help the model generalize better to variations present in clinical settings. |
| Model | Dataset | Input Shape | Params (M) | FLOPs (G) | AP | Epochs |
|---|---|---|---|---|---|---|
| Yolov5 | BraTS + ANDI | 240 × 240 | 7.2 | 9.6 | 0.816 | 300 |
| Yolov6 | BraTS + ANDI | 240 × 240 | 17.1 | 22.2 | 0.83 | 300 |
| Yolov7 | BraTS + ANDI | 240 × 240 | 34.9 | 45.7 | 0.82 | 300 |
| Yolov8 | BraTS + ANDI | 240 × 240 | 42.4 | 54.2 | 0.781 | 300 |
| Yolov9 | BraTS + ANDI | 240 × 240 | 44.2 | 57.8 | 0.842 | 300 |
| Yolov10 | BraTS + ANDI | 240 × 240 | 48.6 | 62.3 | 0.85 | 300 |
| Yolov11 | BraTS + ANDI | 240 × 240 | 52.3 | 67.1 | 0.858 | 300 |
| Baseline | BraTS + ANDI | 240 × 240 | 8.99 | 14.8 | 0.846 | 300 |
| RTMDet (Ours) | BraTS + ANDI | 240 × 240 | 6.76 | 9.65 | 0.969 | 300 |
| Model | Sensitivity | Specificity | AUC | Parameters (Millions) | FLOPs (Billions) |
|---|---|---|---|---|---|
| RTMDet (Ours) | 0.95 | 0.94 | 0.95 | 6.5 | 9.1 |
| Modified RetinaNet [5] | 0.90 | 0.88 | 0.90 | 61.5 | 65.9 |
| Attention-Fused MobileNet-LSTM [12] | 0.93 | 0.90 | 0.93 | 44.0 | 38.6 |
| Transfer learning-based [16] | 0.92 | 0.91 | 0.92 | 7.3 | 9.8 |
| Modified InceptionV3 [17] | 0.87 | 0.85 | 0.88 | 25.6 | 15.2 |
| MN-V2/CFO [21] | 0.83 | 0.82 | 0.83 | 7.0 | 8.2 |
| TumorAwareNet [22] | 0.89 | 0.89 | 0.87 | 50.4 | 58.3 |
| Modified MBConvFinetuned-B0 [23] | 0.84 | 0.85 | 0.85 | 32.8 | 54.0 |
| WSSOA [24] | 0.88 | 0.85 | 0.88 | 21.9 | 35.4 |
| DEF-SwinE2NET [25] | 0.90 | 0.90 | 0.88 | 48.0 | 56.5 |
| PyQDCNN [26] | 0.91 | 0.90 | 0.90 | 29.6 | 39.1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bakhtiyorov, S.; Umirzakova, S.; Musaev, M.; Abdusalomov, A.; Whangbo, T.K. Real-Time Object Detector for Medical Diagnostics (RTMDet): A High-Performance Deep Learning Model for Brain Tumor Diagnosis. Bioengineering 2025, 12, 274. https://doi.org/10.3390/bioengineering12030274
Bakhtiyorov S, Umirzakova S, Musaev M, Abdusalomov A, Whangbo TK. Real-Time Object Detector for Medical Diagnostics (RTMDet): A High-Performance Deep Learning Model for Brain Tumor Diagnosis. Bioengineering. 2025; 12(3):274. https://doi.org/10.3390/bioengineering12030274
Chicago/Turabian StyleBakhtiyorov, Sanjar, Sabina Umirzakova, Musabek Musaev, Akmalbek Abdusalomov, and Taeg Keun Whangbo. 2025. "Real-Time Object Detector for Medical Diagnostics (RTMDet): A High-Performance Deep Learning Model for Brain Tumor Diagnosis" Bioengineering 12, no. 3: 274. https://doi.org/10.3390/bioengineering12030274
APA StyleBakhtiyorov, S., Umirzakova, S., Musaev, M., Abdusalomov, A., & Whangbo, T. K. (2025). Real-Time Object Detector for Medical Diagnostics (RTMDet): A High-Performance Deep Learning Model for Brain Tumor Diagnosis. Bioengineering, 12(3), 274. https://doi.org/10.3390/bioengineering12030274






