Computational Investigation of Smooth Muscle Cell Plasticity in Atherosclerosis and Vascular Calcification: Insights from Differential Gene Expression Analysis of Microarray Data
Abstract
1. Introduction
2. Methods
2.1. Retrieval and Processing of GEO Microarray Datasets
2.2. Differentially Expressed Genes (DEGs) and PPI Analyses
2.3. Reactome Pathway Enrichment Analysis
2.4. Random Forest Analysis
3. Results
3.1. Strategy
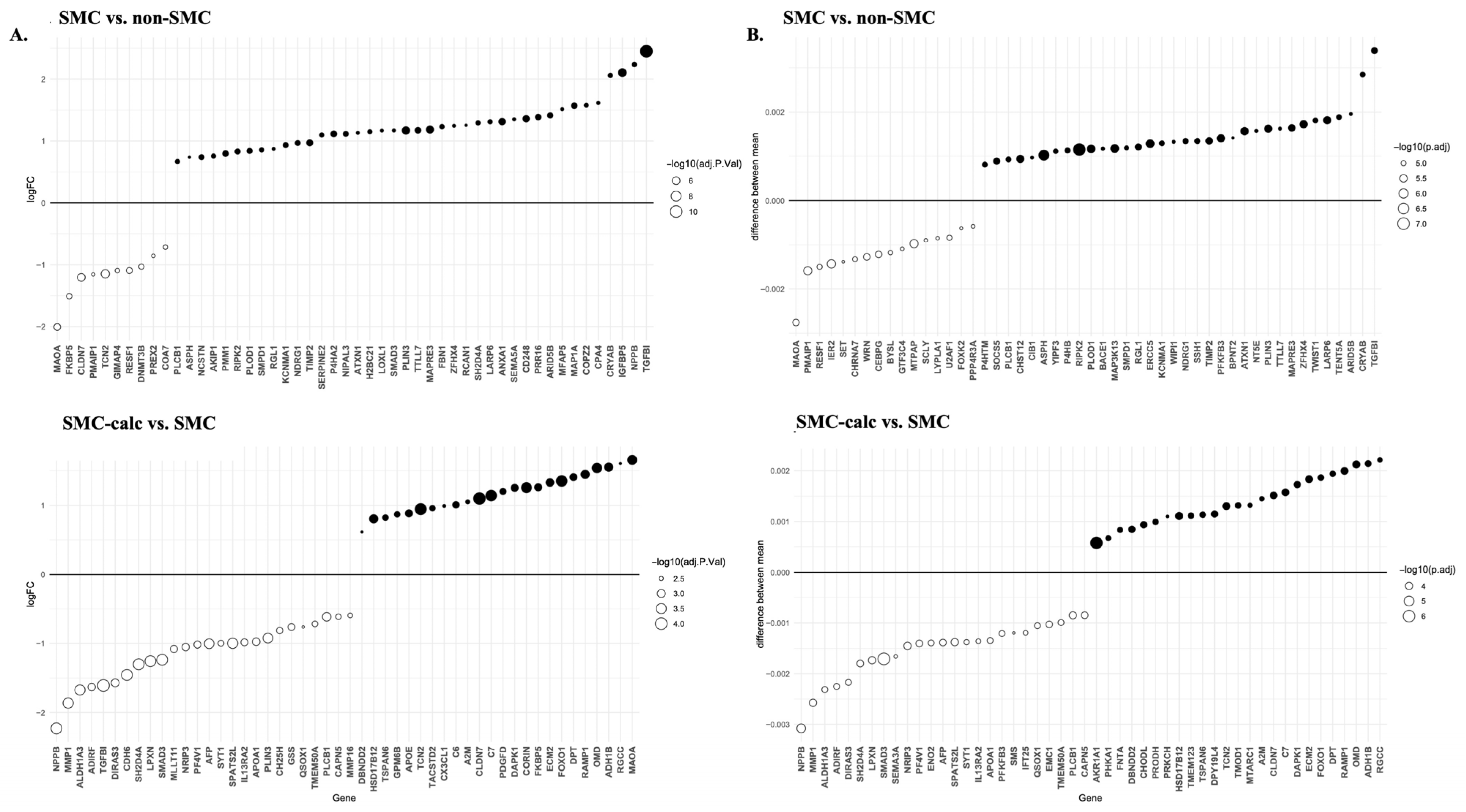
3.2. Identification of DEGs and PPI Network Analysis
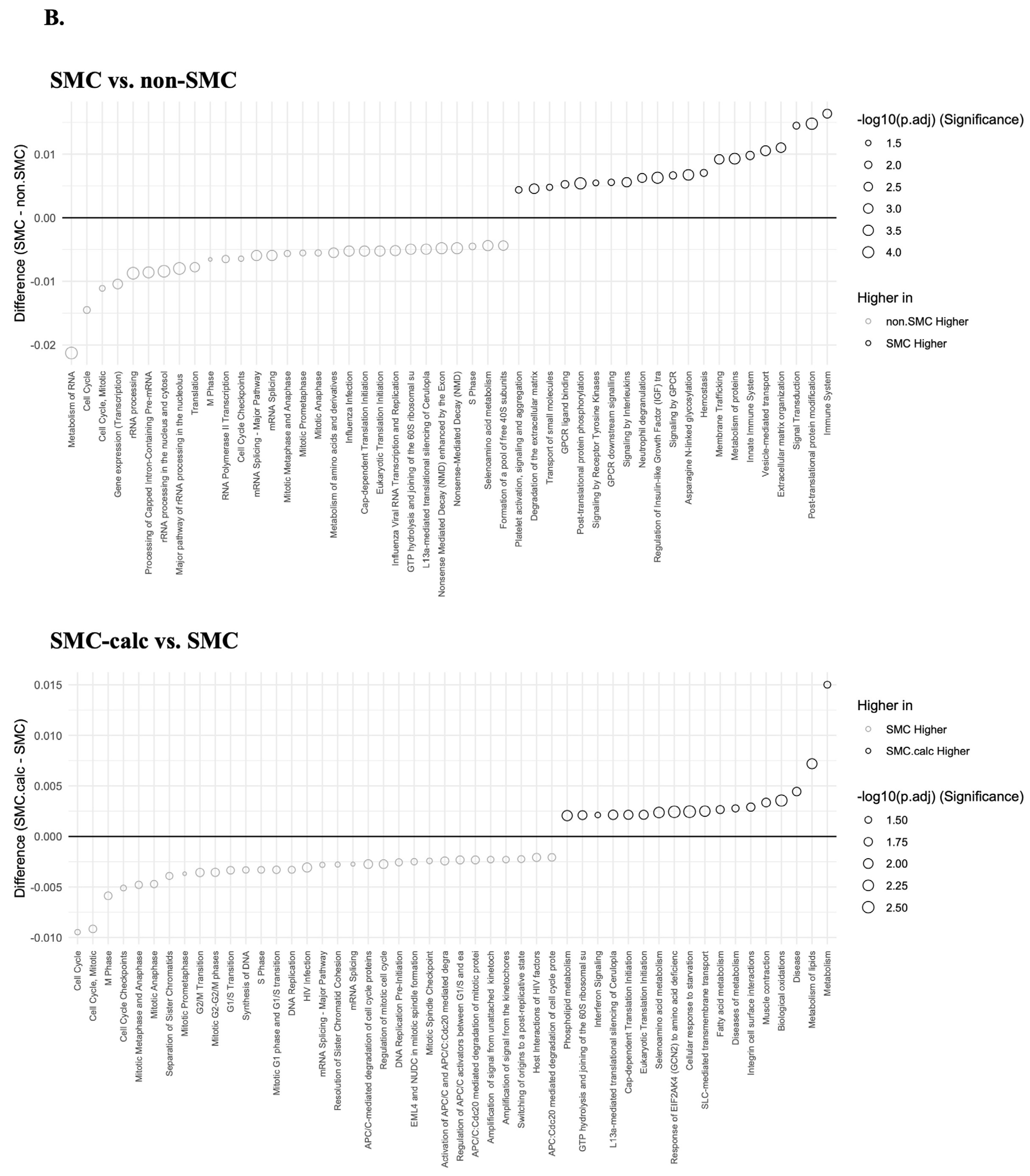
3.3. Reactome Pathway
3.4. Random Forest Algorithm
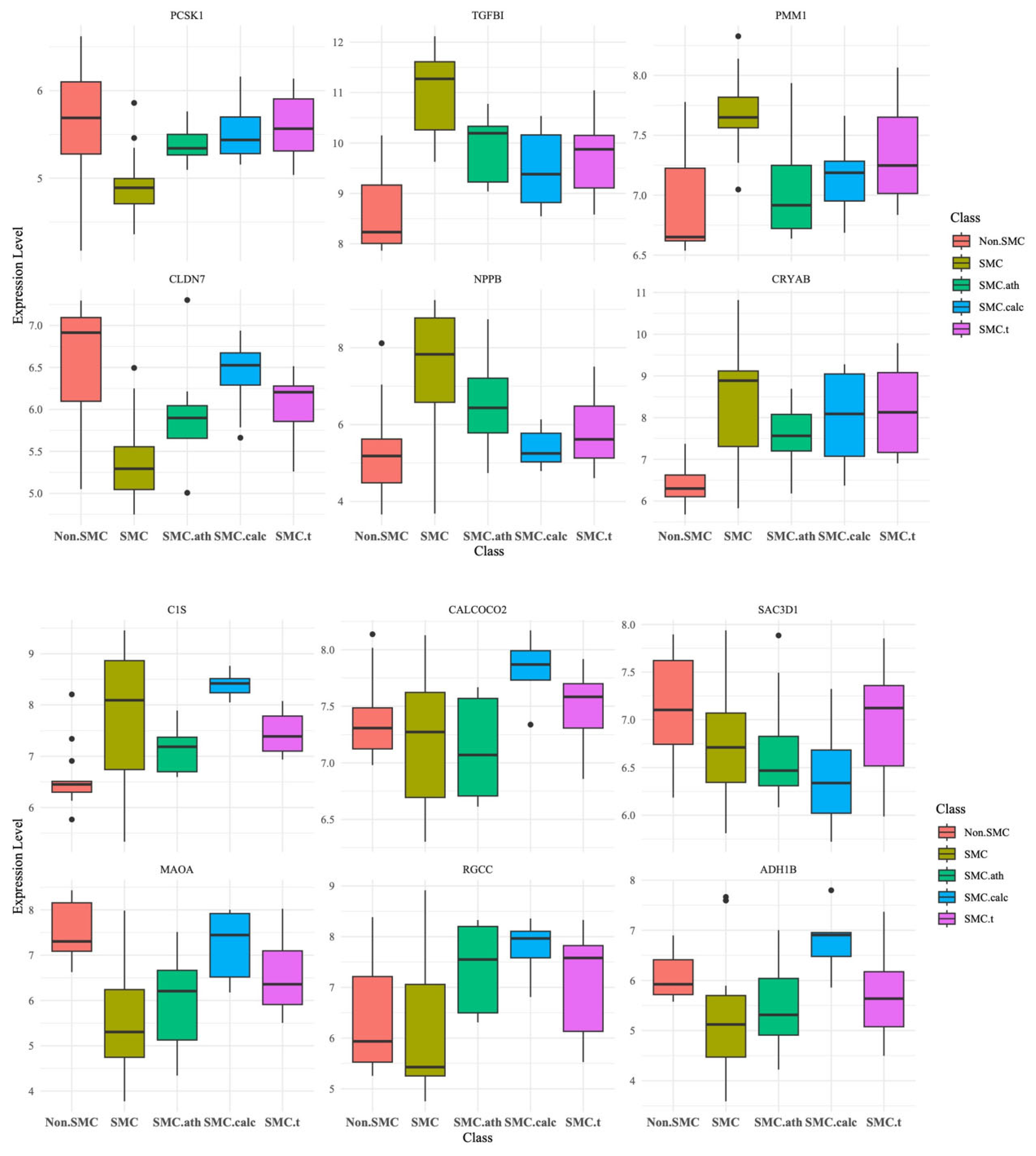
3.5. Candidate Genes
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chamley-Campbell, J.; Campbell, G.R.; Ross, R. The smooth muscle cell in culture. Physiol. Rev. 1979, 59, 1–61. [Google Scholar] [CrossRef]
- Slomp, J.; Gittenberger-de Groot, A.C.; Glukhova, M.A.; Conny van Munsteren, J.; Kockx, M.M.; Schwartz, S.M.; Koteliansky, V.E. Differentiation, dedifferentiation, and apoptosis of smooth muscle cells during the development of the human ductus arteriosus. Arterioscler. Thromb. Vasc. Biol. 1997, 17, 1003–1009. [Google Scholar] [CrossRef]
- Cao, G.; Lu, Z.; Gu, R.; Xuan, X.; Zhang, R.; Hu, J.; Dong, H. Deciphering the Intercellular Communication Between Immune Cells and Altered Vascular Smooth Muscle Cell Phenotypes in Aortic Aneurysm From Single-Cell Transcriptome Data. Front. Cardiovasc. Med. 2022, 9, 936287. [Google Scholar] [CrossRef] [PubMed]
- Rensen, S.S.; Doevendans, P.A.; van Eys, G.J. Regulation and characteristics of vascular smooth muscle cell phenotypic diversity. Neth. Heart J. 2007, 15, 100–108. [Google Scholar] [CrossRef]
- Sorokin, V.; Vickneson, K.; Kofidis, T.; Woo, C.C.; Lin, X.Y.; Foo, R.; Shanahan, C.M. Role of Vascular Smooth Muscle Cell Plasticity and Interactions in Vessel Wall Inflammation. Front. Immunol. 2020, 11, 599415. [Google Scholar] [CrossRef]
- Badimon, J.J.; Fuster, V.; Chesebro, J.H.; Badimon, L. Coronary atherosclerosis. A multifactorial disease. Circulation 1993, 87, II3–II16. [Google Scholar]
- Libby, P.; Schwartz, D.; Brogi, E.; Tanaka, H.; Clinton, S.K. A cascade model for restenosis. A special case of atherosclerosis progression. Circulation 1992, 86, III47–III52. [Google Scholar]
- Leopold, J.A. Vascular calcification: Mechanisms of vascular smooth muscle cell calcification. Trends Cardiovasc. Med. 2015, 25, 267–274. [Google Scholar] [CrossRef]
- Durham, A.L.; Speer, M.Y.; Scatena, M.; Giachelli, C.M.; Shanahan, C.M. Role of smooth muscle cells in vascular calcification: Implications in atherosclerosis and arterial stiffness. Cardiovasc. Res. 2018, 114, 590–600. [Google Scholar] [CrossRef]
- Chi, J.T.; Rodriguez, E.H.; Wang, Z.; Nuyten, D.S.; Mukherjee, S.; van de Rijn, M.; van de Vijver, M.J.; Hastie, T.; Brown, P.O. Gene expression programs of human smooth muscle cells: Tissue-specific differentiation and prognostic significance in breast cancers. PLoS Genet. 2007, 3, 1770–1784. [Google Scholar] [CrossRef]
- Shu, B.; Zhuo, M.; Liu, Z.; Lu, Z.; Qian, M. Cholesterol induces dedifferentiation of vascular smooth muscle cells by regulating monocyte chemotactic protein-1-induced protein 1. Int. J. Clin. Exp. Pathol. 2019, 12, 3258–3267. [Google Scholar]
- Oka, M.; Shimo, S.; Ohno, N.; Imai, H.; Abekura, Y.; Koseki, H.; Miyata, H.; Shimizu, K.; Kushamae, M.; Ono, I.; et al. Dedifferentiation of smooth muscle cells in intracranial aneurysms and its potential contribution to the pathogenesis. Sci. Rep. 2020, 10, 8330. [Google Scholar] [CrossRef]
- Pan, H.; Xue, C.; Auerbach, B.J.; Fan, J.; Bashore, A.C.; Cui, J.; Yang, D.Y.; Trignano, S.B.; Liu, W.; Shi, J.; et al. Single-Cell Genomics Reveals a Novel Cell State During Smooth Muscle Cell Phenotypic Switching and Potential Therapeutic Targets for Atherosclerosis in Mouse and Human. Circulation 2020, 142, 2060–2075. [Google Scholar] [CrossRef]
- Zhang, Z.; Huang, J.; Wang, Y.; Shen, W. Transcriptome analysis revealed a two-step transformation of vascular smooth muscle cells to macrophage-like cells. Atherosclerosis 2022, 346, 26–35. [Google Scholar] [CrossRef]
- Edgar, R.; Domrachev, M.; Lash, A.E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002, 30, 207–210. [Google Scholar] [CrossRef]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 28. [Google Scholar] [CrossRef]
- Davis, S.; Meltzer, P.S. GEOquery: A bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics 2007, 23, 1846–1847. [Google Scholar] [CrossRef]
- Foong, L.C. How-to-Analyze-GEO-Microarray-Data. Available online: https://github.com/Lindseynicer/How-to-analyze-GEO-microarray-data (accessed on 1 December 2024).
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2025; Available online: https://www.R-project.org/ (accessed on 1 December 2024).
- Patsch, C.; Challet-Meylan, L.; Thoma, E.C.; Urich, E.; Heckel, T.; O’Sullivan, J.F.; Grainger, S.J.; Kapp, F.G.; Sun, L.; Christensen, K.; et al. Generation of vascular endothelial and smooth muscle cells from human pluripotent stem cells. Nat. Cell Biol. 2015, 17, 994–1003. [Google Scholar] [CrossRef]
- Janjanam, J.; Zhang, B.; Mani, A.M.; Singh, N.K.; Traylor, J.G., Jr.; Orr, A.W.; Rao, G.N. LIM and cysteine-rich domains 1 is required for thrombin-induced smooth muscle cell proliferation and promotes atherogenesis. J. Biol. Chem. 2018, 293, 3088–3103. [Google Scholar] [CrossRef]
- Alves, R.D.; Eijken, M.; van de Peppel, J.; van Leeuwen, J.P. Calcifying vascular smooth muscle cells and osteoblasts: Independent cell types exhibiting extracellular matrix and biomineralization-related mimicries. BMC Genom. 2014, 15, 965. [Google Scholar] [CrossRef]
- Kayisli, U.A.; Basar, M.; Guzeloglu-Kayisli, O.; Semerci, N.; Atkinson, H.C.; Shapiro, J.; Summerfield, T.; Huang, S.J.; Prelle, K.; Schatz, F.; et al. Long-acting progestin-only contraceptives impair endometrial vasculature by inhibiting uterine vascular smooth muscle cell survival. Proc. Natl. Acad. Sci. USA 2015, 112, 5153–5158. [Google Scholar] [CrossRef] [PubMed]
- Rigassi, L.; Barchiesi Bozzolo, F.; Lucchinetti, E.; Zaugg, M.; Fingerle, J.; Rosselli, M.; Imthurn, B.; Jackson, E.K.; Dubey, R.K. 2-Methoxyestradiol blocks the RhoA/ROCK1 pathway in human aortic smooth muscle cells. Am. J. Physiol. Endocrinol. Metab. 2015, 309, E995–E1007. [Google Scholar] [CrossRef]
- Minta, J.; Jungwon Yun, J.; St. Bernard, R. Microarray analysis of ox-LDL (oxidized low-density lipoprotein)-regulated genes in human coronary artery smooth muscle cells. Cell Biol. Int. Rep. 2010, 17, e00007. [Google Scholar] [CrossRef]
- Ji, H.; Atchison, L.; Chen, Z.; Chakraborty, S.; Jung, Y.; Truskey, G.A.; Christoforou, N.; Leong, K.W. Transdifferentiation of human endothelial progenitors into smooth muscle cells. Biomaterials 2016, 85, 180–194. [Google Scholar] [CrossRef] [PubMed]
- Bhasin, M.; Yuan, L.; Keskin, D.B.; Otu, H.H.; Libermann, T.A.; Oettgen, P. Bioinformatic identification and characterization of human endothelial cell-restricted genes. BMC Genom. 2010, 11, 342. [Google Scholar] [CrossRef]
- Shalhoub, V.; Shatzen, E.M.; Ward, S.C.; Young, J.I.; Boedigheimer, M.; Twehues, L.; McNinch, J.; Scully, S.; Twomey, B.; Baker, D.; et al. Chondro/osteoblastic and cardiovascular gene modulation in human artery smooth muscle cells that calcify in the presence of phosphate and calcitriol or paricalcitol. J. Cell. Biochem. 2010, 111, 911–921. [Google Scholar] [CrossRef]
- Puig, O.; Yuan, J.; Stepaniants, S.; Zieba, R.; Zycband, E.; Morris, M.; Coulter, S.; Yu, X.; Menke, J.; Woods, J.; et al. A gene expression signature that classifies human atherosclerotic plaque by relative inflammation status. Circ. Cardiovasc. Genet. 2011, 4, 595–604. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B 1995, 57, 12. [Google Scholar] [CrossRef]
- Parks, D.H.; Tyson, G.W.; Hugenholtz, P.; Beiko, R.G. STAMP: Statistical analysis of taxonomic and functional profiles. Bioinformatics 2014, 30, 3123–3124. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING database in 2023: Protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023, 51, D638–D646. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Enright, A.J.; Van Dongen, S.; Ouzounis, C.A. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 2002, 30, 1575–1584. [Google Scholar] [CrossRef]
- Xu, S.; Hu, E.; Cai, Y.; Xie, Z.; Luo, X.; Zhan, L.; Tang, W.; Wang, Q.; Liu, B.; Wang, R.; et al. Using clusterProfiler to characterize multiomics data. Nat. Protoc. 2024, 19, 3292–3320. [Google Scholar] [CrossRef]
- Yu, G.; He, Q.Y. ReactomePA: An R/Bioconductor package for reactome pathway analysis and visualization. Mol. Biosyst. 2016, 12, 477–479. [Google Scholar] [CrossRef]
- Milacic, M.; Beavers, D.; Conley, P.; Gong, C.; Gillespie, M.; Griss, J.; Haw, R.; Jassal, B.; Matthews, L.; May, B.; et al. The Reactome Pathway Knowledgebase 2024. Nucleic Acids Res. 2024, 52, D672–D678. [Google Scholar] [CrossRef] [PubMed]
- Liaw, A.; Wiener, M. Classification and Regression by randomForest. R News 2002, 2, 5. [Google Scholar]
- Kuhn, M. Building Predictive Models in R Using the caret Package. J. Stat. Softw. 2008, 28, 26. [Google Scholar] [CrossRef]
- Owens, G.K.; Kumar, M.S.; Wamhoff, B.R. Molecular regulation of vascular smooth muscle cell differentiation in development and disease. Physiol. Rev. 2004, 84, 767–801. [Google Scholar] [CrossRef]
- Bennett, M.R.; Sinha, S.; Owens, G.K. Vascular Smooth Muscle Cells in Atherosclerosis. Circ. Res. 2016, 118, 692–702. [Google Scholar] [CrossRef]
- Suur, B.E.; Chemaly, M.; Lindquist Liljeqvist, M.; Djordjevic, D.; Stenemo, M.; Bergman, O.; Karlof, E.; Lengquist, M.; Odeberg, J.; Hurt-Camejo, E.; et al. Therapeutic potential of the Proprotein Convertase Subtilisin/Kexin family in vascular disease. Front. Pharmacol. 2022, 13, 988561. [Google Scholar] [CrossRef]
- Chen, G.; Khalil, N. TGF-beta1 increases proliferation of airway smooth muscle cells by phosphorylation of map kinases. Respir. Res. 2006, 7, 2. [Google Scholar] [CrossRef]
- Kapustin, A.; Tsakali, S.S.; Whitehead, M.; Chennell, G.; Wu, M.Y.; Molenaar, C.; Kutikhin, A.; Bogdanov, L.; Sinitsky, M.; Rubina, K.; et al. Extracellular vesicles stimulate smooth muscle cell migration by presenting collagen VI. bioRxiv 2023. [Google Scholar] [CrossRef]
- Mardinoglu, A.; Stancakova, A.; Lotta, L.A.; Kuusisto, J.; Boren, J.; Bluher, M.; Wareham, N.J.; Ferrannini, E.; Groop, P.H.; Laakso, M.; et al. Plasma Mannose Levels Are Associated with Incident Type 2 Diabetes and Cardiovascular Disease. Cell Metab. 2017, 26, 281–283. [Google Scholar] [CrossRef]
- West, J.J.; Golloshi, R.; Cho, C.Y.; Wang, Y.; Stevenson, P.; Stein-O’Brien, G.; Fertig, E.J.; Ewald, A.J. Claudin 7 suppresses invasion and metastasis through repression of a smooth muscle actin program. J. Cell Biol. 2024, 223, e202311002. [Google Scholar] [CrossRef]
- Jordan, J.; Birkenfeld, A.L.; Melander, O.; Moro, C. Natriuretic Peptides in Cardiovascular and Metabolic Crosstalk: Implications for Hypertension Management. Hypertension 2018, 72, 270–276. [Google Scholar] [CrossRef]
- Worssam, M.D.; Lambert, J.; Oc, S.; Taylor, J.C.K.; Taylor, A.L.; Dobnikar, L.; Chappell, J.; Harman, J.L.; Figg, N.L.; Finigan, A.; et al. Cellular mechanisms of oligoclonal vascular smooth muscle cell expansion in cardiovascular disease. Cardiovasc. Res. 2023, 119, 1279–1294. [Google Scholar] [CrossRef] [PubMed]
- Speidl, W.S.; Kastl, S.P.; Huber, K.; Wojta, J. Complement in atherosclerosis: Friend or foe? J. Thromb. Haemost. 2011, 9, 428–440. [Google Scholar] [CrossRef]
- Xie, X.; Li, F.; Wang, Y.; Wang, Y.; Lin, Z.; Cheng, X.; Liu, J.; Chen, C.; Pan, L. Molecular basis of ubiquitin recognition by the autophagy receptor CALCOCO2. Autophagy 2015, 11, 1775–1789. [Google Scholar] [CrossRef]
- Khuda, S.E.; Yoshida, M.; Xing, Y.; Shimasaki, T.; Takeya, M.; Kuwahara, K.; Sakaguchi, N. The Sac3 homologue shd1 is involved in mitotic progression in mammalian cells. J. Biol. Chem. 2004, 279, 46182–46190. [Google Scholar] [CrossRef]
- Han, M.E.; Kim, J.Y.; Kim, G.H.; Park, S.Y.; Kim, Y.H.; Oh, S.O. SAC3D1: A novel prognostic marker in hepatocellular carcinoma. Sci. Rep. 2018, 8, 15608. [Google Scholar] [CrossRef]
- Xenodochidis, C.; Staneva, D.; Vasileva, B.; Draganova, M.; Miloshev, G.; Georgieva, M.; Zagorchev, P. The Photobiomodulation of MAO-A Affects the Contractile Activity of Smooth Muscle Gastric Tissues. Biomolecules 2022, 13, 32. [Google Scholar] [CrossRef]
- Cui, X.B.; Luan, J.N.; Dong, K.; Chen, S.; Wang, Y.; Watford, W.T.; Chen, S.Y. RGC-32 (Response Gene to Complement 32) Deficiency Protects Endothelial Cells From Inflammation and Attenuates Atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 2018, 38, e36–e47. [Google Scholar] [CrossRef]
- Misior, A.M.; Deshpande, D.A.; Loza, M.J.; Pascual, R.M.; Hipp, J.D.; Penn, R.B. Glucocorticoid- and protein kinase A-dependent transcriptome regulation in airway smooth muscle. Am. J. Respir. Cell Mol. Biol. 2009, 41, 24–39. [Google Scholar] [CrossRef]
- Llorian, M.; Gooding, C.; Bellora, N.; Hallegger, M.; Buckroyd, A.; Wang, X.; Rajgor, D.; Kayikci, M.; Feltham, J.; Ule, J.; et al. The alternative splicing program of differentiated smooth muscle cells involves concerted non-productive splicing of post-transcriptional regulators. Nucleic Acids Res. 2016, 44, 8933–8950. [Google Scholar] [CrossRef] [PubMed]
- Green, I.D.; Liu, R.; Wong, J.J.L. The Expanding Role of Alternative Splicing in Vascular Smooth Muscle Cell Plasticity. Int. J. Mol. Sci. 2021, 22, 10213. [Google Scholar] [CrossRef]
- Tian, L.; Chen, F.; Macosko, E.Z. The expanding vistas of spatial transcriptomics. Nat. Biotechnol. 2023, 41, 773–782. [Google Scholar] [CrossRef]




| GSM | Groups * | GEO | Platform | Genes Nos | References |
|---|---|---|---|---|---|
| GSM1435047, GSM1435048, GSM1435062, GSM1435063, GSM1435064 | SMC | GSE59326 | GPL16025 | 45033 | [26] |
| GSM1435049, GSM1435050, GSM1435051 | non-SMC | ||||
| GSM1435052, GSM1435053 | |||||
| GSM2802493, GSM2802494, GSM2802495 | SMC | GSE104498 | GPL6884 | 48803 | [22] |
| GSM2802499, GSM2802500, GSM2802501 | SMC-ath | ||||
| GSM2802508, GSM2802509, GSM2802510 | SMC-t | ||||
| GSM921590, GSM921591, GSM921592 GSM921593 | SMC | GSE37558 | GPL6947 | 48803 | [20] |
| GSM921600, GSM921601, GSM921602 GSM921603, GSM921604, GSM921605 | SMC-calc | ||||
| GSM1342582, GSM1342583, GSM1342584 | SMC | GSE55736 | GPL10558 | 47231 | [24] |
| GSM1342585, GSM1342586, GSM1342587 GSM1342588, GSM1342589, GSM1342590 | SMC-t | ||||
| GSM307999, GSM308000, GSM308001 | non-SMC | GSE12261 | GPL570 | 54675 | [28] |
| GSM308008, GSM308009, GSM308010 | SMC-t | ||||
| GSM894977 | SMC | GSE36487 | GPL96 | 22283 | [25] |
| GSM894978, GSM894979 | SMC-ath | ||||
| GSM1895345, GSM1895346, GSM1895347 | non-SMC | GSE73469 | GPL571 | 22277 | [23] |
| GSM1895351, GSM1895352, GSM1895353 GSM1895354, GSM1895355, GSM1895356 | SMC | ||||
| GSM530367, GSM530369 | non-SMC | GSE21212 | GPL3921 | 22277 | [21] |
| GSM530379, GSM530381 | SMC | ||||
| GSM301134, GSM301167, GSM301204 | SMC | GSE11917 | GPL570 | 54675 | [29] |
| GSM301149, GSM301157, GSM301182 GSM301190, GSM301217, GSM301227 | SMC-calc | ||||
| GSM571578, GSM571580, GSM571583 | SMC-ath | GSE23303 | GPL4372 | 39096 | [27] |
| Non-SMC | SMC | SMC-ath | SMC-calc | SMC-t | |
|---|---|---|---|---|---|
| PCSK1 | 62.733 | 100.000 | 46.721 | 78.199 | 53.004 |
| TGFBI | 38.601 | 43.406 | 23.182 | 26.441 | 31.568 |
| ALDH1A3 | 31.619 | 41.766 | 28.116 | 33.278 | 27.762 |
| SH2D4A | 29.233 | 39.417 | 27.021 | 23.853 | 35.339 |
| HSD17B12 | 29.627 | 37.103 | 20.031 | 34.060 | 21.032 |
| CLDN7 | 26.459 | 35.539 | 19.963 | 31.110 | 0.000 |
| PLIN3 | 35.008 | 34.847 | 24.163 | 29.595 | 24.152 |
| CFH | 35.554 | 34.247 | 10.693 | 25.307 | 28.402 |
| RIPK2 | 31.629 | 31.768 | 19.310 | 25.073 | 24.740 |
| SMAD3 | 25.245 | 31.564 | 24.974 | 30.729 | 13.388 |
| Non-SMC | SMC | SMC-ath | SMC-calc | SMC-t | |
|---|---|---|---|---|---|
| PCSK1 | 62.733 | 100.000 | 46.721 | 78.199 | 53.004 |
| C1S | 42.738 | 21.013 | 34.337 | 51.975 | 29.658 |
| LUM | 34.521 | 12.726 | 22.710 | 46.343 | 26.576 |
| THY1 | 25.047 | 16.462 | 13.380 | 40.420 | 30.583 |
| PPIC | 34.105 | 20.113 | 21.731 | 37.905 | 23.531 |
| DNAAF5 | 52.480 | 23.263 | 28.099 | 37.518 | 28.962 |
| NNMT | 32.963 | 25.275 | 24.937 | 35.604 | 21.126 |
| HSD17B12 | 29.627 | 37.103 | 20.031 | 34.060 | 21.032 |
| ALDH1A3 | 31.619 | 41.766 | 28.116 | 33.278 | 27.762 |
| CLDN7 | 26.459 | 35.539 | 19.963 | 31.110 | 0.000 |
| Non-SMC | SMC | SMC-ath | SMC-calc | SMC-t | MeanDecrease Accuracy | MeanDecrease Gini | |
|---|---|---|---|---|---|---|---|
| PMM1 | 1.726 | 2.175 | 1.001 | 1.001 | −0.640 | 2.605 | 0.046 |
| CLDN7 | 1.989 | 2.164 | 0.000 | 0.000 | −1.388 | 2.104 | 0.040 |
| CAMK2N1 | 1.729 | 1.989 | −1.001 | 0.000 | 0.000 | 2.044 | 0.023 |
| GAMT | 0.426 | 1.986 | 0.714 | 1.343 | 1.001 | 2.039 | 0.035 |
| CDH6 | 0.000 | 1.967 | 1.001 | 1.343 | 1.001 | 1.721 | 0.019 |
| ZNF148 | 1.388 | 1.966 | 0.000 | 0.000 | 0.000 | 1.898 | 0.023 |
| INHBB | 1.314 | 1.828 | −0.905 | 1.001 | 1.301 | 1.801 | 0.032 |
| PCSK1 | 1.001 | 1.801 | 0.000 | 1.001 | 0.000 | 1.731 | 0.030 |
| HIP1 | 0.000 | 1.734 | 1.416 | −1.001 | −1.001 | 1.796 | 0.026 |
| HYOU1 | 1.001 | 1.731 | 0.000 | 0.000 | −1.001 | 1.307 | 0.017 |
| Non-SMC | SMC | SMC-ath | SMC-calc | SMC-t | MeanDecrease Accuracy | MeanDecrease Gini | |
|---|---|---|---|---|---|---|---|
| CALCOCO2 | −1.001 | −0.137 | 0.000 | 2.022 | 0.000 | 1.590 | 0.042 |
| SAC3D1 | 1.001 | 0.000 | 1.001 | 1.688 | 0.000 | 1.408 | 0.016 |
| AARS1 | 1.001 | −1.001 | 0.000 | 1.674 | −1.001 | 1.378 | 0.020 |
| KRT7 | 1.001 | 1.685 | 0.000 | 1.669 | −1.001 | 1.949 | 0.018 |
| CYP27A1 | −1.001 | 1.257 | 0.000 | 1.667 | 0.000 | 1.664 | 0.017 |
| MRPL34 | 1.001 | 0.294 | −1.001 | 1.635 | 1.001 | 1.449 | 0.031 |
| RECQL4 | 1.214 | −0.175 | 0.000 | 1.597 | 0.000 | 1.408 | 0.014 |
| RTCB | 1.407 | 0.000 | 0.000 | 1.416 | 0.000 | 1.614 | 0.015 |
| YEATS4 | 1.388 | 1.001 | 0.000 | 1.416 | 0.000 | 1.658 | 0.016 |
| NCAPD2 | 1.343 | 1.411 | 1.001 | 1.416 | 0.000 | 1.414 | 0.016 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, D.; Kuo, J.; Lin, C.-H. Computational Investigation of Smooth Muscle Cell Plasticity in Atherosclerosis and Vascular Calcification: Insights from Differential Gene Expression Analysis of Microarray Data. Bioengineering 2025, 12, 1223. https://doi.org/10.3390/bioengineering12111223
Liu D, Kuo J, Lin C-H. Computational Investigation of Smooth Muscle Cell Plasticity in Atherosclerosis and Vascular Calcification: Insights from Differential Gene Expression Analysis of Microarray Data. Bioengineering. 2025; 12(11):1223. https://doi.org/10.3390/bioengineering12111223
Chicago/Turabian StyleLiu, Daniel, Jimmy Kuo, and Chorng-Horng Lin. 2025. "Computational Investigation of Smooth Muscle Cell Plasticity in Atherosclerosis and Vascular Calcification: Insights from Differential Gene Expression Analysis of Microarray Data" Bioengineering 12, no. 11: 1223. https://doi.org/10.3390/bioengineering12111223
APA StyleLiu, D., Kuo, J., & Lin, C.-H. (2025). Computational Investigation of Smooth Muscle Cell Plasticity in Atherosclerosis and Vascular Calcification: Insights from Differential Gene Expression Analysis of Microarray Data. Bioengineering, 12(11), 1223. https://doi.org/10.3390/bioengineering12111223






