EEG-Based Local–Global Dimensional Emotion Recognition Using Electrode Clusters, EEG Deformer, and Temporal Convolutional Network
Abstract
1. Introduction
2. Related Works
3. Proposed Method
3.1. Localized Feature Extraction at the Electrode-Cluster Level Using EEG Deformer
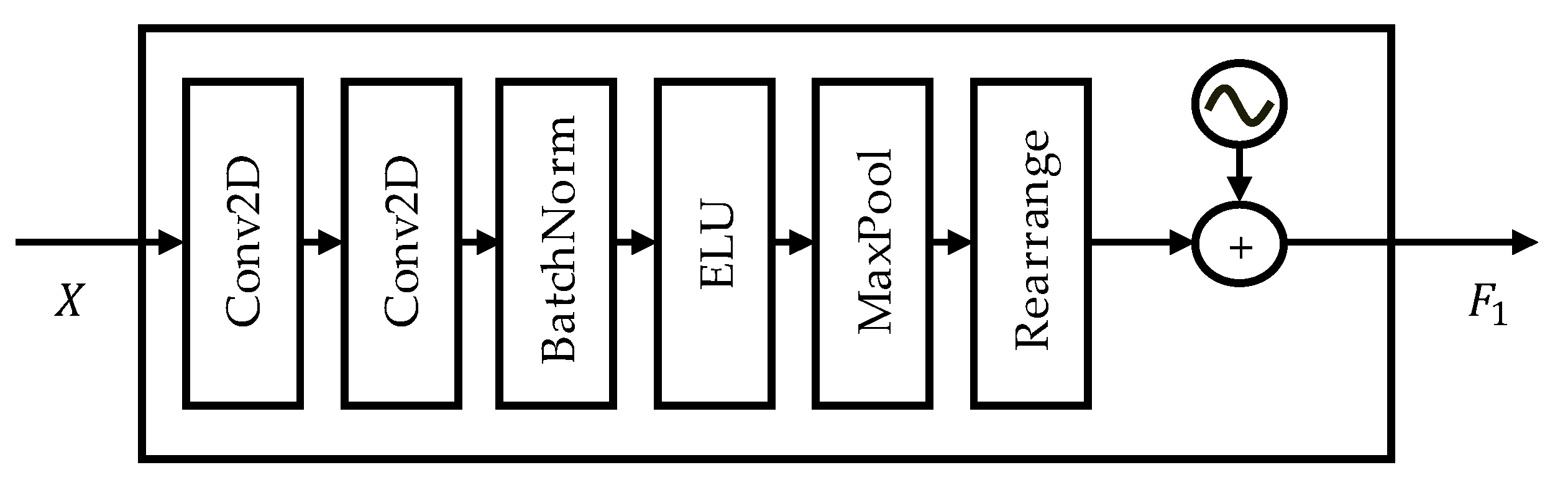
3.1.1. Shallow Feature Encoder (SFE)
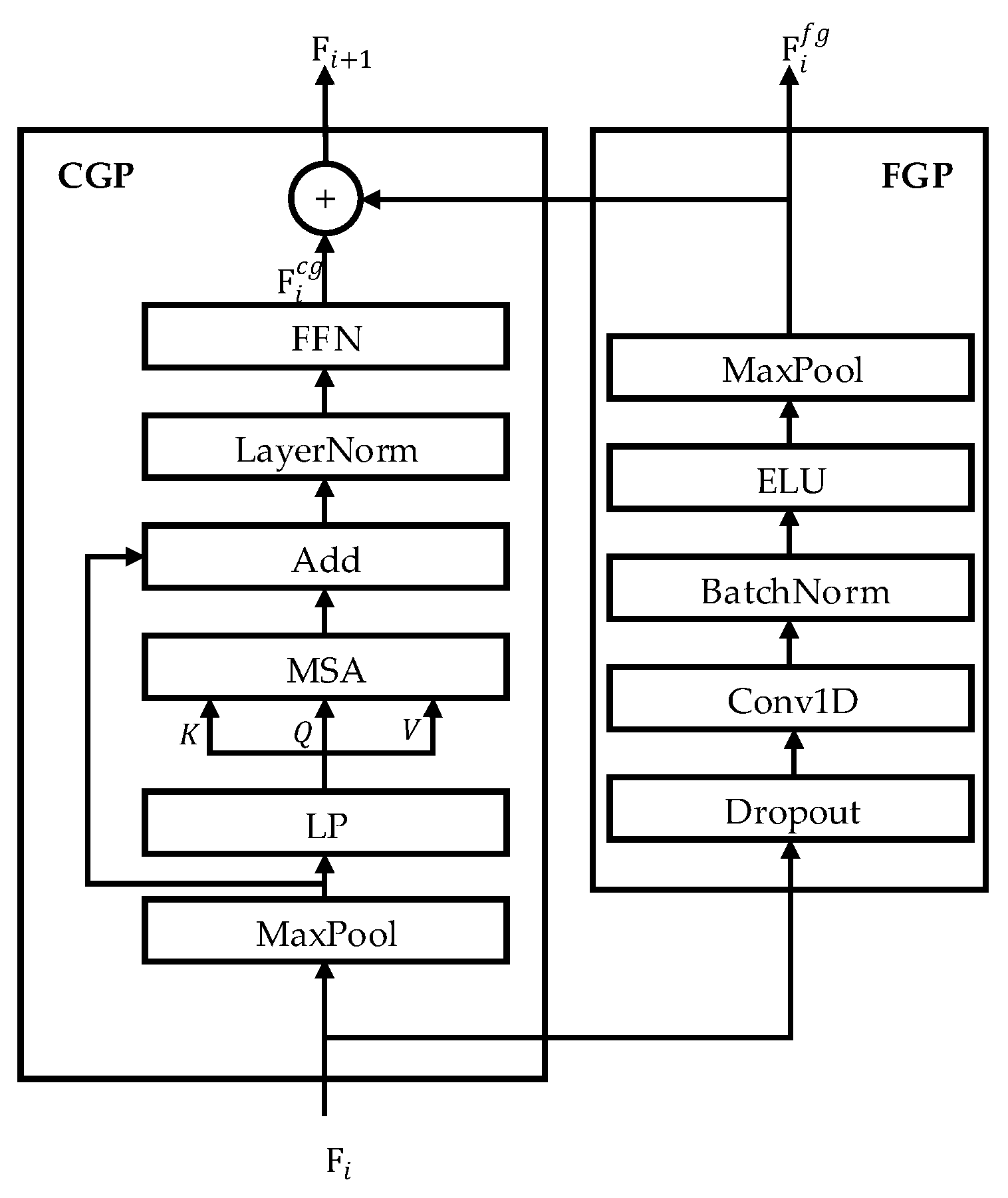
3.1.2. Hierarchical Coarse-to-Fine Transformer (HCT)
3.1.3. Dense Information Purification (DIP)
3.2. Global EEG Feature Integration via Bidirectional Cross Attention and Temporal Convolutional Network
| Algorithm 1. EEG-Based Local–Global Dimensional Emotion Recognition |
| Input: EEG data X ∈ ℝ^(C×T); Cluster groups {C1, C2, …, C9}; Emotion label y |
| Output: Predicted emotion ŷ |
| Step 0: EEG Preprocessing |
| Perform standard EEG preprocessing. |
| Output: Preprocessed EEG data X. |
Step 1: EEG Electrode-Clustering
|
Step 2: Local Cluster Feature Extraction (EEG-Deformer)
|
Step 3: Inter-Cluster Interaction Modeling in the Global Feature Integration
|
Step 4: Temporal Dynamics Modeling in the Global Feature Integration
|
Step 5: Dimensional Emotion Recognition
|
Step 6: Training Objective
|
Step 7: Inference
|
4. Experiment and Results
4.1. Evaluation Datasets
- DEAP [35]: The DEAP dataset was collected from 32 participants (16 males and 16 females) aged 19–37 for emotion recognition research. In addition to EEG, physiological signals such as heart rate and skin conductance were also recorded. Participants watched 40 music video clips while 32-channel EEG signals were recorded at a sampling rate of 512 Hz. Each trial lasted 63 s, including a 3-s baseline period. After each clip, participants rated arousal, valence, dominance, and liking on a 1–9 scale using the self-assessment manikin. Data from 24 participants (12 males and 12 females) with recordings of high quality were retained for analysis.
- MAHNOB-HCI [36]: The MAHNOB-HCI dataset is a multimodal resource that includes EEG and various physiological and visual modalities, such as heart rate, skin conductance, respiration, skin temperature, eye tracking, facial videos, and audio. EEG data were obtained from 27 participants (11 males and 16 females) using 32 channels at a sampling frequency of 256 Hz. The participants viewed 20 video clips (each lasting between 34 and 117 s) designed to elicit diverse emotional responses. After viewing each clip, each participant rated the arousal, valence, dominance, and predictability using a 9-point SAM scale. Data from 22 participants (11 males and 11 females) were used for the final analysis.
- SEED [37]: The SEED dataset comprises multimodal physiological signals, including EEG and eye-movement data, collected from 15 participants (7 males and 8 females). Each participant watched 15 Chinese film clips of about 4 min designed to induce positive, neutral, or negative emotions. EEG was recorded from 62 channels at 1 kHz and downsampled to 200 Hz for analysis. After each clip, participants rated their emotional state on a three-point scale (−1 = negative, 0 = neutral, 1 = positive). Data from 12 participants (6 males and 6 females) with complete sessions and high-quality recordings were included in the analysis.
4.2. Models Used for Performance Comparison
- CNN: This method comprises two convolutional layers with 3 × 3 filters, two 2 × 2 max-pooling layers, a dropout layer, two fully connected layers, and a softmax output layer.
- LSTM: Instead of a CNN, an LSTM network was employed. The first LSTM layer has 128 hidden units, followed by a second LSTM layer with 64 hidden units. This is followed by a dropout layer, a fully connected layer with 128 hidden units, and a softmax output layer.
- CRNN: A model combining a CNN and LSTM was applied to effectively extract spatiotemporal features from multichannel EEG signals. The model consists of two convolutional layers, pooling layers, an LSTM layer, and a final output layer.
- EEGNet [38]: This model is a lightweight CNN architecture that efficiently learns the spatiotemporal features of EEG signals using depthwise separable convolutions. It comprises convolutional blocks, batch normalization, pooling, dropout, fully connected layers, and a softmax output layer.
- EEG Conformer (EEG CF) [39]: This model extracts temporal and spatial features of EEG signals through one-dimensional convolution, learns global temporal dependencies via self-attention, and classifies EEG signals using a fully connected classifier.
- EEG Transformer (EEG TF) [40]: This model is a purely transformer-based architecture that effectively learns long-term dependencies in time-series EEG signals. It comprises embedding layers, positional encoding, self-attention blocks, fully connected layers, and a softmax output layer.
- EEG Deformer (EEG DF): This model extracts temporal features at multiple resolutions directly from multichannel EEG data without dividing the electrodes into predefined spatial clusters, enabling a detailed representation of complex spatiotemporal patterns.
- C2G DF-BCA-TCN: This model is the core method proposed in this study. The model combines an EEG Deformer, BCA, and TCN to extract spatiotemporal features, which are then fed into an MLP for emotion classification.
- C2G DF-TCN: After applying the EEG Deformer to each electrode cluster, the TCN is used without the BCA.
- C2G DF-LSTM: In the C2G EEG DF-TCN structure, LSTM is applied instead of TCN.
- C2G DF-BGRU: In the C2G structure, BGRU [41] is applied instead of TCN to enhance the global spatiotemporal features.
4.3. Experimental Results
4.4. Discussion and Study Limitations
5. Conclusions and Future Work
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Triberti, S.; Chirico, A.; La Rocca, G.; Riva, G. Developing emotional design: Emotions as cognitive processes and their role in the design of interactive technologies. Front. Psychol. 2017, 8, 1773. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhang, Y.; Tiwari, P.; Song, D.; Hu, B.; Yang, M.; Zhao, Z.; Kumar, N.; Marttinen, P. EEG based emotion recognition: A tutorial and review. arXiv 2022, arXiv:2203.11279. [Google Scholar] [CrossRef]
- Ezzameli, K.; Mahersia, H. Emotion recognition from unimodal to multimodal analysis: A review. Inf. Fusion. 2023, 99, 101847. [Google Scholar] [CrossRef]
- Ahmed, N.; Al Aghbari, Z.; Girija, S. A systematic survey on multimodal emotion recognition using learning algorithms. Intell. Syst. Appl. 2023, 17, 200171. [Google Scholar] [CrossRef]
- Alarcão, S.M.; Fonseca, M.J. Emotions recognition using EEG signals: A survey. IEEE Trans. Affect. Comput. 2017, 10, 374–393. [Google Scholar] [CrossRef]
- Liu, C.; Zhou, X.; Wu, Y.; Ding, Y.; Zhai, L.; Wang, K.; Jia, Z.; Liu, Y. A comprehensive survey on EEG-based emotion recognition: A graph-based perspective. arXiv 2024, arXiv:2408.06027. [Google Scholar]
- Craik, A.; He, Y.; Contreras-Vidal, J.L. Deep learning for electroencephalogram (EEG) classification tasks: A review. J. Neural Eng. 2019, 16, 031001. [Google Scholar] [CrossRef] [PubMed]
- Su, J.; Zhu, J.; Song, T.; Chang, H. Subject-independent EEG emotion recognition based on genetically optimized projection dictionary pair learning. Brain Sci. 2023, 13, 977. [Google Scholar] [CrossRef]
- Bagherzadeh, S.; Maghooli, K.; Shalbaf, A.; Maghsoudi, A. A hybrid EEG-based emotion recognition approach using wavelet convolutional neural networks and support vector machine. Basic. Clin. Neurosci. 2023, 14, 87. [Google Scholar] [CrossRef] [PubMed]
- Suhaimi, N.S.; Mountstephens, J.; Teo, J. EEG-based emotion recognition: A state-of-the-art review of current trends and opportunities. Comput. Intell. Neurosci. 2020, 2020, 8875426. [Google Scholar] [CrossRef]
- Ding, Y.; Li, Y.; Sun, H.; Liu, R.; Tong, C.; Liu, C.; Zhou, X.; Guan, C. EEG-Deformer: A dense convolutional transformer for brain-computer interfaces. IEEE J. Biomed. Health Inform. 2025, 29, 1909–1918. [Google Scholar] [CrossRef]
- Wang, X.; Guo, P.; Zhang, Y. Domain adaptation via bidirectional cross-attention transformer. arXiv 2022, arXiv:2201.05887. [Google Scholar] [CrossRef]
- Yang, L.; Liu, J. EEG-based emotion recognition using temporal convolutional network. In Proceedings of the 2019 IEEE 8th Data Driven Control and Learning Systems Conference (DDCLS), Dali, China, 24–27 May 2019; pp. 437–442. [Google Scholar] [CrossRef]
- Bilucaglia, M.; Duma, G.M.; Mento, G.; Semenzato, L.; Tressoldi, P.E. Applying machine learning EEG signal classification to emotion-related brain anticipatory activity. F1000Research 2021, 9, 173. [Google Scholar] [CrossRef]
- Johannesen, J.K.; Bi, J.; Jiang, R.; Kenney, J.G.; Chen, C.-M.A. Machine learning identification of EEG features predicting working memory performance in schizophrenia and healthy adults. Neuropsychiatr. Electrophysiol. 2016, 2, 3. [Google Scholar] [CrossRef]
- Zhou, M.; Tian, C.; Cao, R.; Wang, B.; Niu, Y.; Hu, T.; Guo, H.; Xiang, J. Epileptic seizure detection based on EEG signals and CNN. Front. Neuroinform. 2018, 12, 95. [Google Scholar] [CrossRef]
- Wang, P.; Jiang, A.; Liu, X.; Shang, J.; Zhang, L. LSTM-based EEG classification in motor imagery tasks. IEEE Trans. Neural Syst. Rehabil. Eng. 2018, 26, 2086–2095. [Google Scholar] [CrossRef]
- Li, X.; Song, D.; Zhang, P.; Yu, G.; Hou, Y.; Hu, B. Emotion recognition from multi-channel EEG data through convolutional recurrent neural network. In Proceedings of the 2016 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Shenzhen, China, 15–18 December 2016; pp. 352–359. [Google Scholar] [CrossRef]
- Chakravarthi, B.; Ng, S.C.; Ezilarasan, M.R.; Leung, M.-F. EEG-based emotion recognition using hybrid CNN and LSTM classification. Front. Comput. Neurosci. 2022, 16, 1019776. [Google Scholar] [CrossRef]
- Tao, W.; Li, C.; Song, R.; Cheng, J.; Liu, Y.; Wan, F.; Chen, X. EEG-based emotion recognition via channel-wise attention and self attention. IEEE Trans. Affect. Comput. 2022, 14, 382–393. [Google Scholar] [CrossRef]
- Hu, Z.; Chen, L.; Luo, Y.; Zhou, J. EEG-based emotion recognition using convolutional recurrent neural network with multi-head self-attention. Appl. Sci. 2022, 12, 11255. [Google Scholar] [CrossRef]
- Kim, H.-G.; Jeong, D.-K.; Kim, J.-Y. Emotional stress recognition using electroencephalogram signals based on a three-dimensional convolutional gated self-attention deep neural network. Appl. Sci. 2022, 12, 11162. [Google Scholar] [CrossRef]
- Zhang, P.; Min, C.; Zhang, K.; Xue, W.; Chen, J. Hierarchical spatiotemporal electroencephalogram feature learning and emotion recognition with attention-based antagonism neural network. Front. Neurosci. 2021, 15, 738167. [Google Scholar] [CrossRef]
- Wang, Z.; Wang, Y.; Hu, C.; Yin, Z.; Song, Y. Transformers for EEG-based emotion recognition: A hierarchical spatial information learning model. IEEE Sens. J. 2022, 22, 4359–4368. [Google Scholar] [CrossRef]
- Jeong, D.-K.; Kim, H.-G.; Kim, J.-Y. Emotion recognition using hierarchical spatiotemporal electroencephalogram information from local to global brain regions. Bioengineering 2023, 10, 1040. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Wang, M.; Ju, H.; Ding, W.; Zhang, D. AGBN-Transformer: Anatomy-guided brain network transformer for schizophrenia diagnosis. Biomed. Signal Process. Control 2025, 102, 107226. [Google Scholar] [CrossRef]
- Ding, Y.; Tong, C.; Zhang, S.; Jiang, M.; Li, Y.; Lim, K.J.; Guan, C. EmT: A novel transformer for generalized cross-subject EEG emotion recognition. IEEE Trans. Neural Netw. Learn. Syst. 2025, 36, 10381–10393. [Google Scholar] [CrossRef]
- Kalateh, S.; Estrada-Jimenez, L.A.; Nikghadam-Hojjati, S.; Barata, J. A systematic review on multimodal emotion recognition: Building blocks, current state, applications, and challenges. IEEE Access 2024, 12, 103976–104019. [Google Scholar] [CrossRef]
- Liang, Z.; Zhou, R.; Zhang, L.; Li, L.; Huang, G.; Zhang, Z.; Ishii, S. EEGFuseNet: Hybrid unsupervised deep feature characterization and fusion for high-dimensional EEG with an application to emotion recognition. IEEE Trans. Neural Syst. Rehabil. Eng. 2021, 29, 1913–1925. [Google Scholar] [CrossRef]
- Lan, Y.-T.; Liu, W.; Lu, B.-L. Multimodal emotion recognition using deep generalized canonical correlation analysis with an attention mechanism. In Proceedings of the 2020 International Joint Conference on Neural Networks (IJCNN), Glasgow, UK, 19–24 July 2020; pp. 1–6. [Google Scholar] [CrossRef]
- Ribas, G.C. The cerebral sulci and gyri. Neurosurg. Focus. 2010, 28, E2. [Google Scholar] [CrossRef]
- Yang, B.; Duan, K.; Fan, C.; Hu, C.; Wang, J. Automatic ocular artifacts removal in EEG using deep learning. Biomed. Signal Process. Control 2018, 43, 148–158. [Google Scholar] [CrossRef]
- Shah, A.; Kadam, E.; Shah, H.; Shinde, S.; Shingade, S. Deep residual networks with exponential linear unit. In Proceedings of the Third International Symposium on Computer Vision and the Internet, New York, NY, USA, 21–24 September 2016; pp. 59–65. [Google Scholar] [CrossRef]
- Irani, H.; Metsis, V. Positional encoding in transformer-based time series models: A survey. arXiv 2025, arXiv:2502.12370. [Google Scholar] [CrossRef]
- Koelstra, S.; Muhl, C.; Soleymani, M.; Lee, J.-S.; Yazdani, A.; Ebrahimi, T.; Pun, T.; Nijholt, A.; Patras, I. DEAP: A database for emotion analysis; using physiological signals. IEEE Trans. Affect. Comput. 2011, 3, 18–31. [Google Scholar] [CrossRef]
- Soleymani, M.; Lichtenauer, J.; Pun, T.; Pantic, M. A multimodal database for affect recognition and implicit tagging. IEEE Trans. Affect. Comput. 2011, 3, 42–55. [Google Scholar] [CrossRef]
- Zheng, W.-L.; Lu, B.-L. Investigating critical frequency bands and channels for EEG-based emotion recognition with deep neural networks. IEEE Trans. Auton. Ment. Dev. 2015, 7, 162–175. [Google Scholar] [CrossRef]
- Lawhern, V.J.; Solon, A.J.; Waytowich, N.R.; Gordon, S.M.; Hung, C.P.; Lance, B.J. EEGNet: A compact convolutional neural network for EEG-based brain–computer interfaces. J. Neural Eng. 2018, 15, 056013. [Google Scholar] [CrossRef]
- Song, Y.; Zheng, Q.; Liu, B.; Gao, X. EEG Conformer: Convolutional transformer for EEG decoding and visualization. IEEE Trans. Neural Syst. Rehabil. Eng. 2022, 31, 710–719. [Google Scholar] [CrossRef]
- Sun, J.; Xie, J.; Zhou, H. EEG classification with transformer-based models. In Proceedings of the 2021 IEEE 3rd Global Conference on Life Sciences and Technologies (LifeTech), Nara, Japan, 9–11 March 2021; pp. 92–93. [Google Scholar] [CrossRef]
- Chen, J.X.; Jiang, D.M.; Zhang, Y.N. A hierarchical bidirectional GRU model with attention for EEG-based emotion classification. IEEE Access 2019, 7, 118530–118540. [Google Scholar] [CrossRef]
- Zheng, W.-L.; Lu, B.-L. Personalizing EEG-based affective models with transfer learning. In Proceedings of the Twenty-Fifth International Joint Conference on Artificial Intelligence, New York, NY, USA, 9–15 July 2016; pp. 2732–2738. [Google Scholar]
- Zhang, H.; Zhou, Q.Q.; Chen, H.; Hu, X.Q.; Li, W.G.; Bai, Y.; Li, X.L. The applied principles of EEG analysis methods in neuroscience and clinical neurology. Mil. Med. Res. 2023, 10, 67. [Google Scholar] [CrossRef]
- Lim, R.Y.; Lew, W.-C.L.; Ang, K.K. Review of EEG affective recognition with a neuroscience perspective. Brain Sci. 2024, 14, 364. [Google Scholar] [CrossRef]
- Zhang, G.; Etemad, A. Partial label learning for emotion recognition from EEG. IEEE Trans. Affect. Comput. 2025, 16, 2381–2395. [Google Scholar] [CrossRef]
- Zhang, Y.; Pan, Y.; Zhang, Y.; Zhang, M.; Li, L.; Zhang, L.; Huang, G.; Su, L.; Liu, H.; Liang, Z.; et al. Unsupervised time-aware sampling network with deep reinforcement learning for EEG-based emotion recognition. IEEE Trans. Affect. Comput. 2023, 15, 1090–1103. [Google Scholar] [CrossRef]




| Rating Values (RVs) | Valence | Arousal | Dominance |
|---|---|---|---|
| Low | Low | Low | |
| High | High | High |
| Rating Values (RVs) | Valence | Arousal | Dominance |
|---|---|---|---|
| Negative | Activated | Controlled | |
| Neutral | Moderate | Moderate | |
| Positive | Deactivated | Overpowered |
| Methods | Two-Level CL | Three-Level CL | ||||
|---|---|---|---|---|---|---|
| VAL | ARO | DOM | VAL | ARO | DOM | |
| CNN | 69.7 (11.82) | 66.7 (9.50) | 70.1 (11.84) | 65.3 (10.09) | 64.7 (10.43) | 65.9 (8.94) |
| LSTM | 75.2 (11.56) | 72.3 (10.30) | 75.3 (9.11) | 71.1 (8.89) | 69.7 (11.13) | 71.6 (9.46) |
| CRNN | 78.3 (9.26) | 77.9 (10.72) | 78.5 (11.25) | 74.5 (10.56) | 73.2 (10.24) | 74.8 (10.62) |
| EEG TF | 79.1 (9.57) | 78.8 (10.53) | 79.6 (9.62) | 75.9 (10.26) | 75.6 (10.31) | 76.3 (10.72) |
| EEGNet | 79.2 (10.53) | 77.2 (10.26) | 78.3 (9.52) | 78.2 (9.57) | 75.8 (10.45) | 76.6 (10.64) |
| EEG CF | 83.3 (9.86) | 80.1 (9.57) | 82.8 (10.76) | 80.3 (10.62) | 78.9 (10.41) | 79.3 (10.46) |
| EEG DF | 86.8 (10.45) | 85.8 (9.65) | 87.6 (9.72) | 83.7 (10.64) | 82.9 (10.89) | 83.5 (10.58) |
| C2G DF-BCA-TCN | 94.7 (9.24) | 93.2 (9.65) | 94.6 (10.01) | 89.5 (9.32) | 89.1 (8.65) | 89.4 (9.88) |
| Methods | Two-Level CL | Three-Level CL | ||||
|---|---|---|---|---|---|---|
| VAL | ARO | DOM | VAL | ARO | DOM | |
| EEG DF | 87.4 (10.42) | 86.5 (9.61) | 88.5 (9.85) | 81.8 (10.34) | 80.8 (10.62) | 81.3 (10.73) |
| C2G DF-LSTM | 90.8 (9.78) | 89.5 (10.15) | 89.7 (9.76) | 85.8 (9.68) | 85.4 (10.27) | 86.5 (9.53) |
| C2G DF-BGRU | 92.1 (9.35) | 90.7 (10.24) | 91.2 (9.75) | 86.9 (10.28) | 86.3 (10.38) | 87.7 (10.26) |
| C2G DF-TCN | 92.9 (10.45) | 91.2 (10.61) | 91.9 (9.38) | 87.7 (9.65) | 87.2 (9.35) | 88.4 (10.23) |
| C2G DF-BCA-TCN | 95.4 (9.27) | 94.1 (10.02) | 95.6 (9.57) | 90.1 (9.63) | 89.4 (10.25) | 90.3 (9.86) |
| Methods | Four-Level CL (HAHV vs. LAHV vs. HALV vs. LALV) | Three-Level CL (VAL) | |
|---|---|---|---|
| DEAP | MAHNOB-HCI | SEED | |
| EEG DF | 77.5 (10.83) | 78.1 (9.85) | 85.7 (10.68) |
| C2G DF-LSTM | 81.1 (10.56) | 81.6 (10.62) | 89.4 (11.23) |
| C2G DF-BGRU | 82.3 (9.25) | 82.8 (10.83) | 90.9 (9.89) |
| C2G DF-TCN | 83.1 (10.12) | 83.7 (10.23) | 91.7 (9.87) |
| C2G DF-BCA-TCN | 85.3 (9.25) | 86.2 (10.05) | 94.5 (9.67) |
| Methods | Four-Level CL (HAHV vs. LAHV vs. HALV vs. LALV) | Three-Level CL (VAL) | |
|---|---|---|---|
| DEAP | MAHNOB-HCI | SEED | |
| EEG DF | 85.2 ± 11.5 (0.028) | 86.0 ± 10.3 (0.022) | 89.2 ± 10.9 (0.018) |
| C2G DF-LSTM | 89.0 ± 10.9 (0.020) | 89.6 ± 11.2 (0.016) | 92.7 ± 11.7 (0.012) |
| C2G DF-BGRU | 90.2 ± 10.0 (0.016) | 91.0 ± 11.0 (0.012) | 94.1 ± 10.5 (0.010) |
| C2G DF-TCN | 91.0 ± 10.6 (0.012) | 91.7 ± 10.8 (0.009) | 94.8 ± 10.2 (0.007) |
| C2G DF-BCA-TCN | 93.5 ± 9.7 (-) | 94.3 ± 10.1 (-) | 97.5 ± 9.9 (-) |
| Methods | Four-Level CL (HAHV vs. LAHV vs. HALV vs. LALV) | Three-Level CL (VAL) | |
|---|---|---|---|
| DEAP | MAHNOB-HCI | SEED | |
| w/o HCT + DIP | 71.5 (12.0) | 73.1 (11.5) | 76.5 (11.0) |
| w/o DIP | 78.7 (11.8) | 80.1 (11.0) | 82.9 (10.7) |
| EEG DF | 85.2 (11.5) | 86.0 (10.3) | 89.2 (10.9) |
| C2G DF-BCA | 91.0 (10.6) | 91.7 (10.8) | 94.8 (10.2) |
| C2G DF-BCA-TCN | 93.5 (9.7) | 94.3 (10.1) | 97.5 (9.9) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, H.-G.; Kim, J.-Y. EEG-Based Local–Global Dimensional Emotion Recognition Using Electrode Clusters, EEG Deformer, and Temporal Convolutional Network. Bioengineering 2025, 12, 1220. https://doi.org/10.3390/bioengineering12111220
Kim H-G, Kim J-Y. EEG-Based Local–Global Dimensional Emotion Recognition Using Electrode Clusters, EEG Deformer, and Temporal Convolutional Network. Bioengineering. 2025; 12(11):1220. https://doi.org/10.3390/bioengineering12111220
Chicago/Turabian StyleKim, Hyoung-Gook, and Jin-Young Kim. 2025. "EEG-Based Local–Global Dimensional Emotion Recognition Using Electrode Clusters, EEG Deformer, and Temporal Convolutional Network" Bioengineering 12, no. 11: 1220. https://doi.org/10.3390/bioengineering12111220
APA StyleKim, H.-G., & Kim, J.-Y. (2025). EEG-Based Local–Global Dimensional Emotion Recognition Using Electrode Clusters, EEG Deformer, and Temporal Convolutional Network. Bioengineering, 12(11), 1220. https://doi.org/10.3390/bioengineering12111220





