Review of Artificial Intelligence Techniques for Breast Cancer Detection with Different Modalities: Mammography, Ultrasound, and Thermography Images
Abstract
1. Introduction
2. Medical Image Analysis
2.1. Mammography Images
- Digital Database for Screening Mammography (DDSM): This dataset includes 2620 mammograms that were originally on film and later scanned and divided into 43 volumes.
- Curated Breast Imaging Subset of the DDSM (CBIS-DDSM): This is an improved version of DDSM with bounding boxes, better mass segmentation, and decompressed images. It contains 10,239 mammograms, each with corresponding mask images.
- INBreast: This contains 410 images from 115 patients. In 90 cases, both breasts had cancer. It includes breast mass, calcifications, asymmetries, and distortions.
- Mini-MIAS: A dataset with 322 mammogram images and ground truth markers indicating possible abnormalities.
- BCDR (Breast Cancer Digital Repository):
- ○
- BCDR-FM: Film-based mammography database.
- ○
- BCDR-DM: Full-field digital mammography database.
- ○
- Both include normal and abnormal cases, along with clinical details.
- ○
- The BCDR-FM has 1010 cases (998 women, 12 men), 104 lesions, and 3703 mammograms taken from 1125 studies in Mediolateral Oblique (MLO) and Craniocaudal (CC) views.
2.2. Ultrasound Images
- Breast ultrasound images (BUSI) dataset contains 780 grayscale ultrasound images of 600 female patients: normal–133 images, benign–437 images, and malignant–210 images.
- Open access series of breast ultrasonic data (OASBUD) contains 200 ultrasound scans of 78 women aged between 25 and 75 years: 100 breast lesions (52 malignant, 48 benign).
- UDIAT breast ultrasound dataset (UDIAT) contains 163 ultrasound images: benign–109 images and malignant–54 images.
- BrEaST ultrasound dataset contains 256 breast ultrasound scans from 256 patients, including 266 benign and malignant segmented lesions.
- QAMEBI ultrasound database contains 232 breast ultrasound images, including 123 benign and 109 malignant breast lesions.
- BUS-BRA breast ultrasound dataset contains 1875 anonymized images from 1064 female patients, divided into 722 benign and 342 malignant cases.
2.3. Thermography Images
2.4. Publicly Available Datasets
- Breast Cancer Data Repository (BCDR)
- Digital Database for Screening Mammography (DDSM)
- INBreast
- Mammographic Image Analysis Society (MIAS)/Mini-MIAS
- Wisconsin Breast Cancer Dataset (WBCD)
- Wisconsin Diagnosis Breast Cancer (WDBC)
- Image Retrieval in Medical Applications (IRMA)
- Breast Cancer Histopathological Image (BreakHis)
- Breast Ultrasound Images Dataset (BUSI)
- Open Access Series of Breast Ultrasonic Data (OASBUD)
- UDIAT Breast Ultrasound Dataset (UDIAT)
- BrEaST Ultrasound Dataset
- The Mastology Research with Infrared Image (DMR-IR)
- NIRAMAI Health Analytix
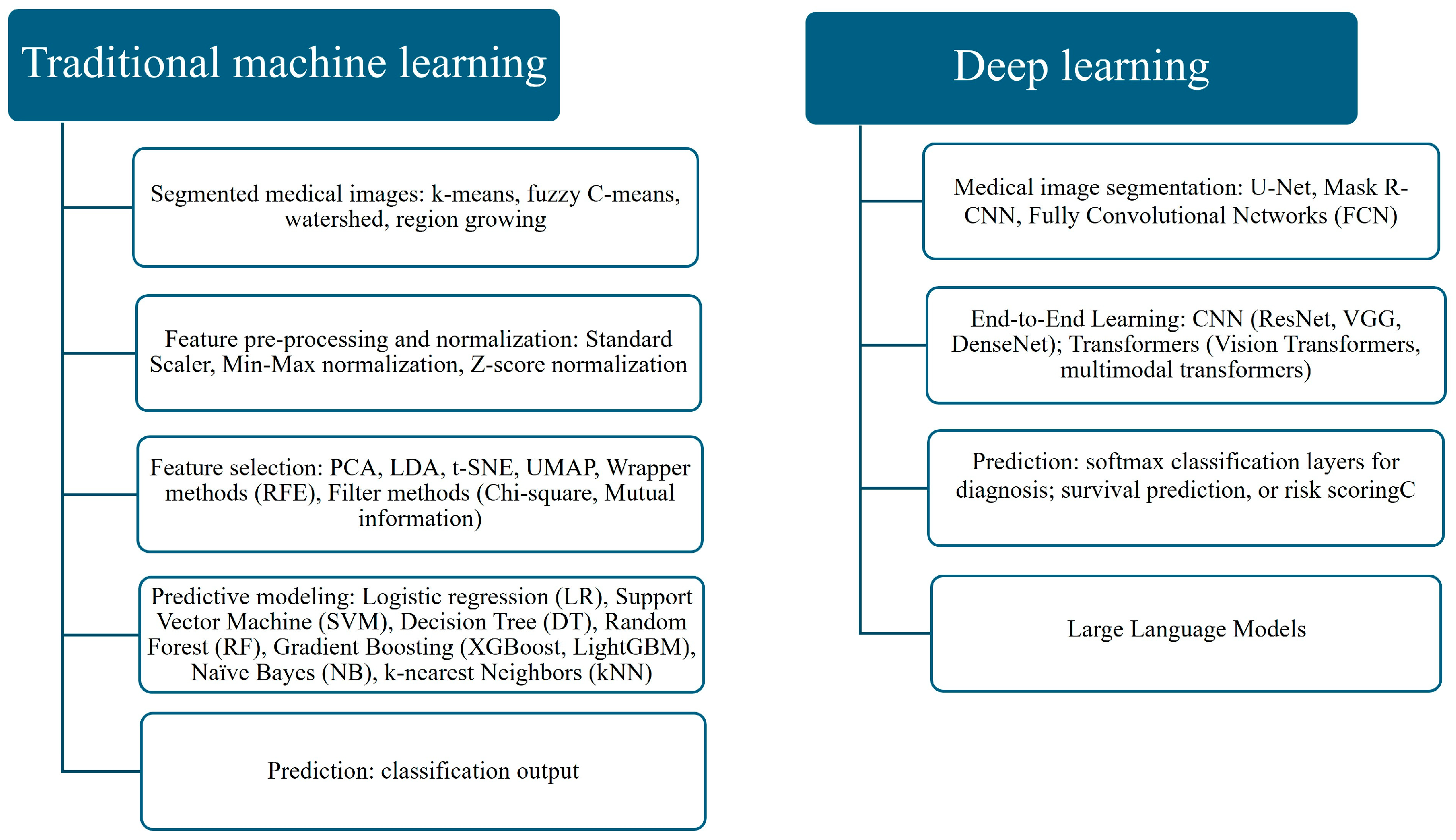
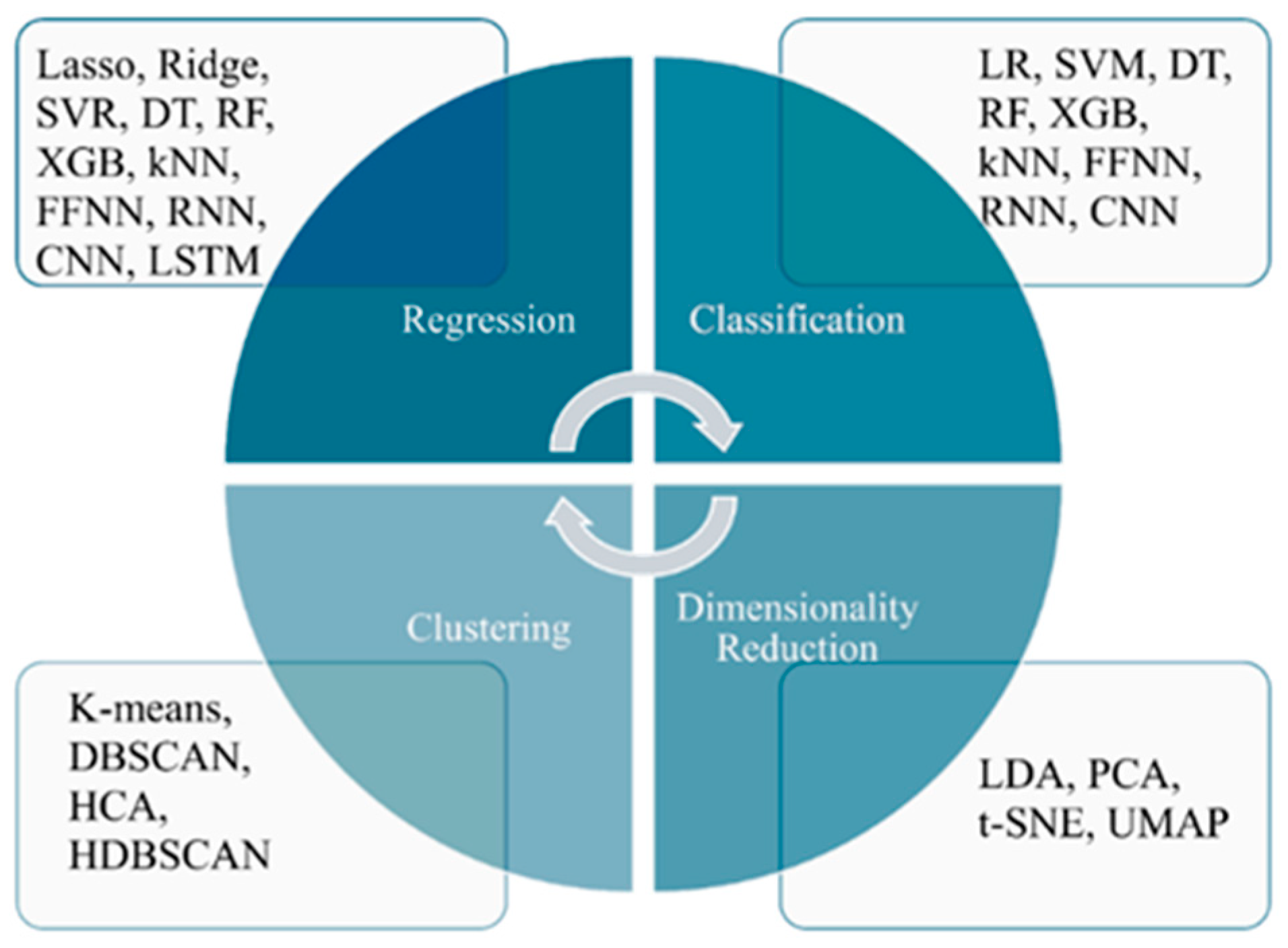
3. Machine Learning and Deep Learning Techniques
3.1. Traditional Machine Learning and Deep Learning
- The DL approach, which involves deep feature extraction or end-to-end learning directly from images.
3.2. Large Language Models
3.3. Multimodal Large Language Models
3.4. Explainable AI for Medical Diagnosis and Medical Image Recognition
- (1)
- LIME (local interpretable model) approximates the complex model locally around a prediction using a simple interpretable model (like a linear model). It perturbs the input data and observes changes in prediction to estimate feature importance [68].
- (2)
- SHAP (SHapley Additive exPlanations) is based on cooperative game theory (Shapley values). It measures each feature’s contribution by calculating its marginal impact across all possible feature combinations [69].
- (3)
- (4)
- (5)
- Counterfactual explanations find the smallest change to the input that would result in a different prediction [74].
- (6)
- Anchors identify a set of rules (“anchors”) that “lock in” a prediction. If these rules are met, the model’s prediction is likely to stay the same [75].
- (7)
- Surrogate models train a simpler, interpretable model (like a decision tree) to mimic the predictions of a more complex model [76].
- (8)
- Models using knowledge representation schemes. It is possible to use one of the previous methods to identify the critical parts (for diagnosis) of a medical image and subsequently to calculate various characteristic quantities of these subsets of the image. Then, a Bayesian Network (BN) can be constructed with informational nodes representing those quantities. The causal structure of the BN reveals immediately the most influential factors for the classification and how these factors interact and influence each other [77,78,79,80].
3.5. What Clinicians Should Know
4. CAD Systems Performance Evaluation
5. Literature Search Methodology
- “breast cancer” AND (“mammography” OR “ultrasound” OR “thermography”);
- “artificial intelligence” OR “machine learning” OR “deep learning”;
- “explainable AI” OR “XAI”;
- “large language models” OR “LLMs” OR “multimodal AI”;
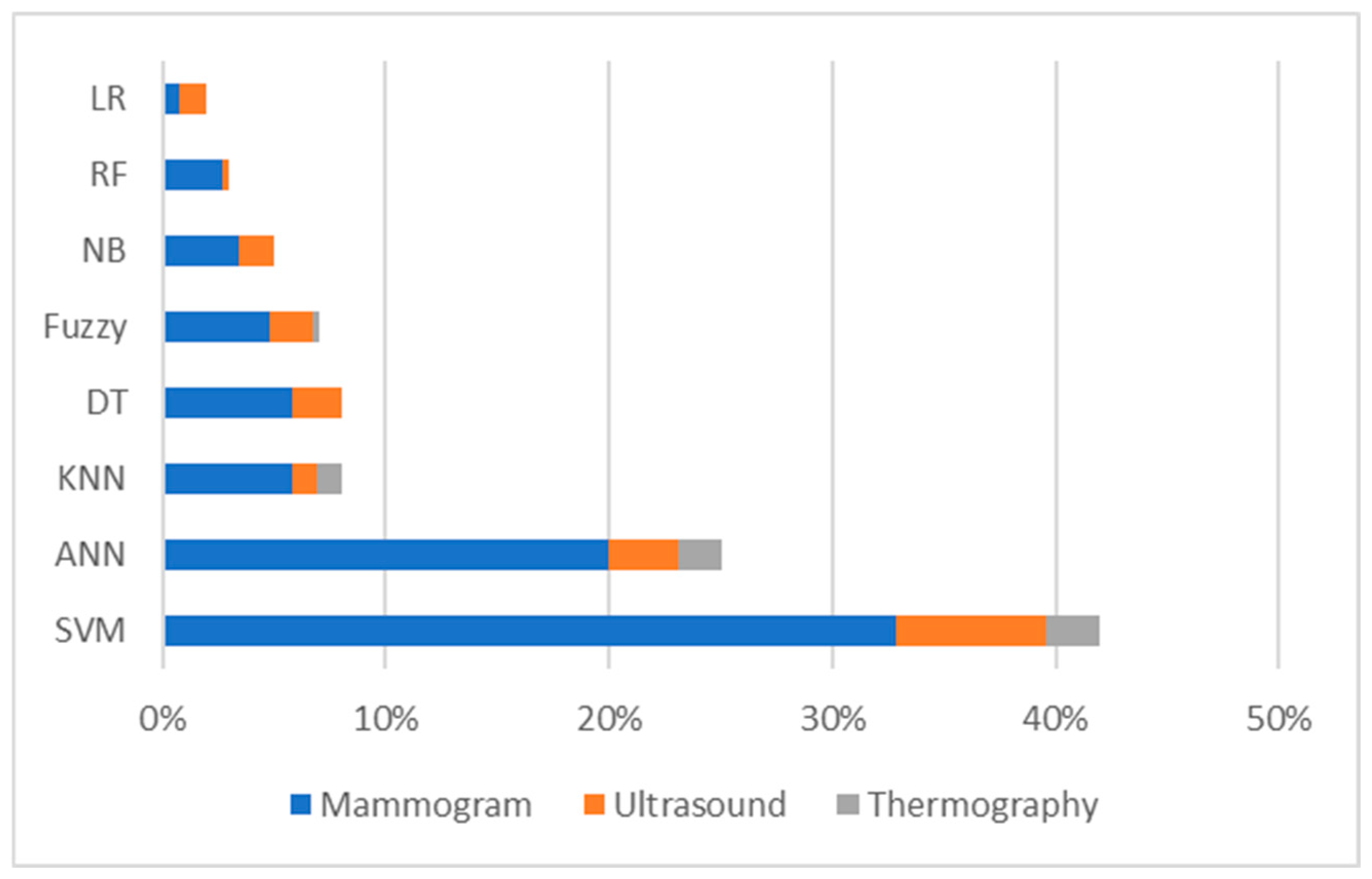
6. Breast Cancer Detection and Classification Using Machine Learning and Deep Learning
7. Artificial Intelligence for Breast Cancer Detection: Machine Learning, Deep Learning, and Hybrid Algorithms
7.1. Machine Learning, Deep Learning, and Hybrid Algorithms for Breast Cancer Detection Using Mammograms
| Reference | ML, DL, Hybrid Algorithms | Dataset | Performance Evaluation |
|---|---|---|---|
| Sha, Z.; Hu, L.; Rouyendegh, B.D. (2020) [87] | Hybrid: CNN, SVM | MIAS, DDSM | Sn is 96%, Sp is 93%, PPV is 85%, NPV is 97%, ACC is 92%. |
| Sapate, S.; Talbar, S.; Mahajan, A.; Sable, N.; Desai, S.; Thakur, M. (2020) [88] | ML: fuzzy c-means algorithm; SVM; | Tata Memorial Centre (TMC), Mumbai, India; BIRADS | Sn is 75.91% at 0.69 FPs/I and Sn is 73.65% at 0.72 FPs/I. |
| Mansour, S.; Kamal, R.; Hussein, S.A.; Emara, M.; Kassab, Y.; Taha, S.N.; Gomaa, M.M. (2025) [89] | AI | Internal dataset | Sn is 73.4%, Sp is 89%, ACC is 78.4%. |
| Malherbe, K. (2025) [90] | AI Breast | Internal dataset: Daspoort PoliClinic in Gauteng, South Africa | 97.04% were negative, 2.46% were positive, and a single patient was classified with a BIRADS2 score. |
| Hernström, V.; Josefsson, V.; Sartor, H.; Schmidt, D.; Larsson, A.-M.; Hofvind, S.; Andersson, I.; Rosso, A.; Hagberg, O.; Lång, K. (2025) [91] | AI system | Swedish national screening program, women recruited at four screening sites in southwest Sweden (Malmö, Lund, Landskrona, and Trelleborg) | AI-supported screening led to a 29% increase in cancer detection. |
| Nour, A.; Boufama, B. (2025) [92] | CNN: U-Net deep learning model; ACM | Chinese Mammography Database (CMMD) | ACC is 0.9734; validation Loss is 0.037; average Dice Coefficient is 0.813; average intersection over Union is 0.891. |
| Umamaheswari, T.; Babu, Y.M.M. (2024) [93] | Hybrid DL model combining CNN (EfficientNetB7) and Transformer (ViT): ViT-MAENB7 model | The Complete Mini-DDSM Dataset; | ACC is 96.6%; recall is 96.6%; Sp is 96.6%; Pr is 93.4%; F1-score is 94.9%. |
| Mannarsamy, V.; Mahalingam, P.; Kalivarathan, T.; Amutha, K.; Paulraj, R.K.; Ramasamy, S. (2025) [94] | SIFT-CNN; Fuzzy-based decision tree | CBIS-DDSM dataset | For normal cases: ACC is 98.98%; Sp is 96.28%; Sn is 94.78%. For benign cases: ACC is 99.74%; Sp is 95.76%; Sn is 93.64%. For malignant cases: ACC is 98.89%; Sp is 93.59%; Sn is 95.82%. |
| Puttegowda, K.; Veeraprathap V; Kumar, H.S.R.; Sudheesh, K.V.; Prabhavathi, K.; Vinayakumar, R.; Tabianan, K. (2025) [95] | ML: Faster R-CNN; YOLOv3; RetinaNet | DDSM, INbreast, AIIMS | For normal cases: recall is 99%, Sp is 98.79%, Pr is 98.59%, F1-score is 98.95%, AUC is 99.58%. For benign cases: recall is 93.56%, Sp is 98.57%, Pr is 96.38%, F1-score is 94.67%, AUC is 97.59%. For malignant cases: recall is 92.78%, Sp is 97.43%, Pr is 94.71%, F1-score is 92.43%, AUC is 99.87%. |
| Ahmad, J.; Akram. S.; Jaffar. A.; Ali, Z.; Bhatti, S.M.; Ahmad, A. et al. (2024) [96] | CAD system: ML (SVM) & computer vision techniques | CBIS-DDSM | ACC is 99.16%, Sn is 97.13%, Sp is 99.30%. |
| Gudur, R.; Patil, N.; Thorat, S. (2024) [97] | Integration of CNN, ResNet50, RNN | RSNA | ACC is 97%; AUC is 0.68; Pr is 60%; recall is 80%; F1-score is 0.18. |
| Mahmood, T.; Saba, T.; Rehman, A.; Alamri, F.S. (2024) [98] | DCNN-based models: CNN + LSTM and CNN + SVM | MIAS, INbreast | ACC is 98%; Pr is 97%; recall is 97%; Sn is 97%; F1-score is 0.97. |
| Muduli, D.; Dash, R.; Majhi, B. (2020) [99] | LWT; MFO-ELM | MIAS, DDSM | ACC is 98.80%, AUC is 0.99. |
7.2. Machine Learning, Deep Learning, and Hybrid Algorithms for Breast Cancer Detection Using Ultrasound Images
| Reference | ML, DL, Hybrid Algorithms | No. of Dataset Images | Performance Evaluation |
|---|---|---|---|
| Liu, Y.; Ren, L.; Cao, X.; Tong, Y. (2020) [100] | SVM with edge-based features (SMC, SMCP, and SMCSD) | 192 | ACC is 82.69%, Sn is 66.67%, Sp is 93.55%, PPV is 87.5%, NPV is 80.56% |
| Ametefe, D.S.; John, D.; Aliu, A.A.; Ametefe, G.D.; Hamid, A.; Darboe, T. (2025) [101] | Deep transfer learning (3 pre-trained CNNs: VGG16, VGG19, and EfficientNet) and U-Net | 780 | ACC: VGG16—95%, VGG19—95.5%, EfficientNetB3—85.8%; Sn: VGG16—94.1%, VGG19—94.06%, EfficientNetB3—86.8%; Sp: VGG16—96.6%, VGG19—97%, EfficientNetB3—85.3%; Pr: VGG16—96.4%, VGG19—96.9%, EfficientNetB3—85.8%; F1-score: VGG16—95.3%, VGG19—95.5%, EfficientNetB3—85.9% |
| Wang, C.; Guo, Y.; Chen, H.; Guo, Q.; He, H.; Chen, L.; Zhang, Q. (2025) [102] | GCN-based network, ABUS-Net | 547 | AUC is 82.6%; ACC is 86.4%; Pr is 92.6%; Sn is 71.4%; Sp is 96.2%; F1-score is 80.6% |
| Kiran, A.; Ramesh, J.V.N.; Rahat, I.S.; Khan, M.A.U.; Hossain, A.; Uddin, R. (2024) [103] | Hybrid approach: EfficientNetB3 and k-Nearest Neighbors | 780 | ACC, Pr, recall, and F1-score all at 100% |
| Tian, R.; Lu, G.; Tang, S.; Sang, L.; Ma, H.; Qian, W.; Yang, W. (2024) [104] | Transfer learning | 1050 | ACC is 0.964; AUC is 0.981 |
7.3. Machine Learning, Deep Learning, and Hybrid Algorithms for Breast Cancer Detection Using Thermograms
| Reference | ML, DL, Hybrid Algorithms | Dataset | Performance Evaluation |
|---|---|---|---|
| Ekici, S.; Jawzal, H. (2020) [105] | CNN | Mastology research dataset | ACC is 98.95% |
| de Freitas Barbosa, V.A.; de Santana, M.A.; Andrade, M.K.S.; de Lima, R.d. C.F.; dos Santos, W.P. (2020) [106] | DWAN | Mastology research dataset | Sn is 0.95 |
| Cabıoğlu, Ç.; and Oğul, H. (2020) [107] | CNN | DMR | ACC is 94.3% |
| Sánchez-Ruiz, D.; Olmos-Pineda, I.; Olvera-López, J.A. (2020) [108] | ROI ANN | Mastology research dataset | ACC is 90.2% Sn is 89.34% Sp is 91% |
| Resmini, R.; da Silva, L.F.; Medeiros, P.R.T.; Araujo, A.S.; Muchaluat-Saade, D.C.; Conci, A. (2021) [109] | K-star; SVM | DMR | ACC is 94.61%; AUC is 94.87% |
| Allugunti, V.R. (2022) [110] | CNN, SVM, RF | 1000 images (from Kaggle) | ACC: CNN is 99.67%; SVM is 89.84%; RF is 90.55% |
| Mohamed, E.A.; Rashed, E.A.; Gaber, T.; Karam, O. (2022) [111] | CNN U_NET | DMR_IR | ACC is 99.3% Sn is 100% Sp is 98.67% |
| Civilibal, S.; Cevik, K.K.; Bozkurt, A. (2023) [112] | R-CNN with transfer learning | 76 images of women | ACC is 97%; Pr is 96.1%; recall is 1.0; F1-score is 0.98 |
| Ramacharan, S.; Margala, M.; Shaik, A.; Chakrabarti, P.; Chakrabarti, T. (2024) [113] | HERA-Net, integrating VGG19, U-Net, GRU, ResNet-50 | DMR | ACC is 99.86; Sn is 100%; Sp is 99.81% |
7.4. Multimodal Large Language Models, Large Language Models for Breast Cancer Diagnosis
8. Discussion
9. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Latha, H.P.; Ravi, S.; Saranya, A. Breast Cancer Detection Using Machine Learning in Medical Imaging–A Survey. Procedia Comput. Sci. 2024, 239, 2235–2242. [Google Scholar] [CrossRef]
- National Comprehensive Cancer Network. NCCN Clinical Practice Guidelines in Oncology: Breast Cancer Screening and Diagnosis, Version 2. 2025. Available online: https://www.nccn.org/professionals/physician_gls/pdf/breast-screening.pdf (accessed on 3 January 2025).
- World Health Organization. Breast Cancer: Screening and Early Detection; WHO: Geneva, Switzerland, 2023; Available online: https://www.who.int/news-room/fact-sheets/detail/breast-cancer (accessed on 3 January 2025).
- Gradishar, W.J.; Anderson, B.O.; Abraham, J.; Aft, R.; Agnese, D.; Allison, K.H.; Blair, S.L.; Burstein, H.J.; Dang, C.; Elias, A.D.; et al. Breast cancer, version 3.2020, NCCN clinical practice guidelines in oncology. J. Compr. Cancer Netw. 2020, 18, 452–478. [Google Scholar] [CrossRef]
- Bevers, T.B.; Niell, B.L.; Baker, J.L.; Bennett, D.L.; Bonaccio, E.; Camp, M.S.; Chikarmane, S.; Conant, E.F.; Eghtedari, M.; Flanagan, M.R.; et al. NCCN Guidelines® Insights: Breast Cancer Screening and Diagnosis, Version 1.2023. J. Natl. Compr. Canc. Netw. 2023, 21, 900–909. [Google Scholar] [CrossRef] [PubMed]
- Oeffinger, K.C.; Fontham, E.T.H.; Etzioni, R.; Herzig, A.; Michaelson, J.S.; Shih, Y.-C.T.; Walter, L.C.; Church, T.R.; Flowers, C.R.; LaMonte, S.J.; et al. Breast cancer screening for women at average risk: 2015 guideline update from the american cancer society. JAMA 2015, 314, 1599–1614. [Google Scholar] [CrossRef]
- Cardoso, F.; Kyriakides, S.; Ohno, S.; Penault-Llorca, F.; Poortmans, P.; Rubio, I.T.; Zackrisson, S.; Senkus, E.; ESMO Guidelines Committee. Early breast cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up†. Ann. Oncol. 2019, 30, 1194–1220. [Google Scholar] [CrossRef]
- Lee, C.H.; Dershaw, D.D.; Kopans, D.; Evans, P.; Monsees, B.; Monticciolo, D.; Brenner, R.J.; Bassett, L.; Berg, W.; Feig, S.; et al. Breast cancer screening with imaging: Recommendations from the Society of Breast Imaging and the ACR on the use of mammography, breast MRI, breast ultrasound, and other technologies for the detection of clinically occult breast cancer. J. Am. Coll. Radiol. 2010, 7, 18–27. [Google Scholar] [CrossRef]
- Siu, A.L.; U.S. Preventive Services Task Force. Screening for breast cancer: U.S. Preventive services task force recommendation statement. Ann. Intern. Med. 2016, 164, 279–296. [Google Scholar] [CrossRef]
- Tagliafico, A.S.; Piana, M.; Schenone, D.; Lai, R.; Massone, A.M.; Houssami, N. Overview of radiomics in breast cancer diagnosis and prognostication. Breast 2020, 49, 74–80. [Google Scholar] [CrossRef]
- Baltzer, P.A.T.; Kapetas, P.; Marino, M.A.; Clauser, P. New diagnostic tools for breast cancer. Memo 2017, 10, 175–180. [Google Scholar] [CrossRef] [PubMed]
- Borgstede, J.P.; Bagrosky, B.M. Screening of High-risk Patients. In Early Diagnosis and Treatment of Cancer Series: Breast Cancer; Elsevier: Amsterdam, The Netherlands, 2011; pp. 141–149. [Google Scholar] [CrossRef]
- Lowry, K.P.; Trentham-Dietz, A.; Schechter, C.B.; Alagoz, O.; Barlow, W.E.; Burnside, E.S.; Conant, E.F.; Hampton, J.M.; Huang, H.; Kerlikowske, K.; et al. Long-term outcomes and cost-effectiveness of breast Cancer Screening with digital breast tomosynthesis in the United States. J. Natl. Cancer Inst. 2020, 112, 582–589. [Google Scholar] [CrossRef] [PubMed]
- Løberg, M.; Lousdal, M.L.; Bretthauer, M.; Kalager, M. Benefits and harms of mammography screening. Breast Cancer Res. 2015, 17, 63. [Google Scholar] [CrossRef] [PubMed]
- Obeagu, E.I.; Obeagu, G.U. Breast cancer: A review of risk factors and diagnosis. Medicine 2024, 103, e36905. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, D.A.; Lee, T.; Seely, D. A comparative review of thermography as a breast cancer screening technique. Integr. Cancer Ther. 2009, 8, 9–16. [Google Scholar] [CrossRef]
- USF Digital Mammography Home Page. Available online: http://www.eng.usf.edu/cvprg/Mammography/Database.html (accessed on 3 June 2025).
- DDSM Mammography. Available online: https://www.kaggle.com/datasets/skooch/ddsm-mammography (accessed on 3 June 2025).
- Moreira, I.C.; Amaral, I.; Domingues, I.; Cardoso, A.; Cardoso, M.J.; Cardoso, J.S. INbreast: Toward a full-field digital mammographic database. Acad. Radiol. 2012, 19, 236–248. [Google Scholar] [CrossRef]
- Suckling, J. The Mini-MIAS Database of Mammograms. Available online: http://peipa.essex.ac.uk/info/mias.html (accessed on 3 June 2025).
- Breast Cancer Digital Repository. Available online: https://bcdr.ceta-ciemat.es/information/about (accessed on 6 June 2025).
- BREAST-CANCER-SCREENING-DBT-The Cancer Imaging Archive (TCIA). Available online: https://www.cancerimagingarchive.net/collection/breast-cancer-screening-dbt/ (accessed on 16 January 2025).
- CMMD-The Cancer Imaging Archive (TCIA). Available online: https://www.cancerimagingarchive.net/collection/cmmd/ (accessed on 12 January 2025).
- Loizidou, K.; Skouroumouni, G.; Pitris, C.; Pitris, C. Breast Micro-calcifications Dataset with Precisely Annotated Sequential Mammograms. Eur. Radiol. Exp. 2021, 5, 40. [Google Scholar] [CrossRef]
- Halling-Brown, M.D.; Warren, L.M.; Ward, D.; Lewis, E.; Mackenzie, A.; Wallis, M.G.; Wilkinson, L.S.; Given-Wilson, R.M.; McAvinchey, R.; Young, K.C. OPTIMAM mammography image database: A large-scale resource of mammography images and clinical data. Radiol. Artif. Intell. 2021, 3, e200103. [Google Scholar] [CrossRef]
- CDD-CESM-The Cancer Imaging Archive (TCIA). Available online: https://www.cancerimagingarchive.net/collection/cdd-cesm/ (accessed on 16 January 2025).
- Shin, S.Y.; Lee, S.; Yun, I.D.; Jung, H.Y.; Heo, Y.S.; Kim, S.M.; Lee, K.M. A novel cascade classifier for automatic microcalcification detection. PLoS ONE 2015, 10, e0143725. [Google Scholar] [CrossRef]
- Digital Mammography Dataset for Breast Cancer Diagnosis Research (DMID) DMID.rar. Available online: https://figshare.com/articles/dataset/_b_Digital_mammography_Dataset_for_Breast_Cancer_Diagnosis_Research_DMID_b_DMID_rar/24522883/1 (accessed on 3 January 2025).
- VinDr-Mammo: A Large-Scale Benchmark Dataset for Computer-Aided Detection and Diagnosis in Full-Field Digital Mammography v1.0.0. Available online: https://physionet.org/content/vindr-mammo/1.0.0/images/#files-panel (accessed on 3 January 2025).
- Mammographic Mass-UCI Machine Learning Repository. Available online: https://archive.ics.uci.edu/dataset/161/mammographic+mass (accessed on 3 January 2025).
- VICTRE-The Cancer Imaging Archive (TCIA). Available online: https://www.cancerimagingarchive.net/collection/victre/ (accessed on 3 January 2025).
- Chen, X.; Zhang, K.; Abdoli, N.; Gilley, P.W.; Wang, X.; Liu, H.; Zheng, B.; Qiu, Y. Transformers improve breast cancer diagnosis from unregistered multi-view mammograms. Diagnostics 2022, 12, 1549. [Google Scholar] [CrossRef]
- Ambra|Home. Available online: https://mamografiaunifesp.ambrahealth.com/ (accessed on 3 January 2025).
- Matsoukas, C.; Hernandez, A.B.I.; Liu, Y.; Dembrower, K.; Miranda, G.; Konuk, E.; Fredin Haslum, J.; Zouzos, A.; Lindholm, P.; Strand, F.; et al. CSAW-S. In Proceedings of the International Conference on Machine Learning (ICML), Online, 13–18 July 2020; Zenodo: Geneva, Switzerland, 2020. [Google Scholar] [CrossRef]
- Safe Haven (DaSH)|Research|The University of Aberdeen. Available online: https://www.abdn.ac.uk/research/digital-research/dash.php (accessed on 3 January 2025).
- Catalano, O.; Fusco, R.; Carriero, S.; Tamburrini, S.; Granata, V. Ultrasound findings after breast cancer radiation therapy: Cutaneous, pleural, pulmonary, and cardiac changes. Korean J. Radiol. 2024, 25, 982–991. [Google Scholar] [CrossRef]
- Kim, Y.E.; Cha, J.H.; Kim, H.H.; Shin, H.J.; Chae, E.Y.; Choi, W.J. The accuracy of mammography, ultrasound, and magnetic resonance imaging for the measurement of invasive breast cancer with extensive intraductal components. Clin. Breast Cancer 2023, 23, 45–53. [Google Scholar] [CrossRef] [PubMed]
- Shao, Y.; Hashemi, H.S.; Gordon, P.; Warren, L.; Wang, J.; Rohling, R.; Salcudean, S. Breast cancer detection using multimodal time series features from ultrasound shear wave absolute vibroelastography. IEEE J. Biomed. Health Inform. 2022, 26, 704–714. [Google Scholar] [CrossRef]
- Geisel, J.; Raghu, M.; Hooley, R. The role of ultrasound in breast cancer screening: The case for and against ultrasound. Semin. Ultrasound CT MR 2018, 39, 25–34. [Google Scholar] [CrossRef]
- Zanello, P.A.; Robim, A.F.C.; de Oliveira, T.M.G.; Junior, J.E.; de Andrade, J.M.; Monteiro, C.R.; Filho, J.M.S.; Carrara, H.H.A.; Muglia, V.F. Breast ultrasound diagnostic performance and outcomes for mass lesions using Breast Imaging Reporting and Data System category 0 mammogram. Clinics 2011, 66, 443–448. [Google Scholar] [CrossRef]
- Al-Dhabyani, W.; Gomaa, M.; Khaled, H.; Fahmy, A. Dataset of breast ultrasound images. Data Brief 2020, 28, 104863. [Google Scholar] [CrossRef]
- Piotrzkowska-Wróblewska, H.; Dobruch-Sobczak, K.; Byra, M.; Nowicki, A. Open access database of raw ultrasonic signals acquired from malignant and benign breast lesions. Med. Phys. 2017, 44, 6105–6109. [Google Scholar] [CrossRef]
- UDIAT Diagnostic Centre, Parc Taulí Corporation, Sabadell, Spain. Available online: https://www.kaggle.com/datasets?search=udiat (accessed on 3 January 2025).
- OpenAI; Achiam, J.; Adler, S.; Agarwal, S.; Ahmad, L.; Akkaya, I.; Aleman, F.L.; Almeida, D.; Altenschmidt, J.; Altman, S.; et al. GPT 4 Technical Report. arXiv 2024, arXiv:2303.08774. [Google Scholar]
- Ardakani, A.A.; Mohammadi, A.; Mirza-Aghazadeh-Attari, M.; Acharya, U.R. Acharya, An open-access breast lesion ultrasound image database: Applicable in artificial intelligence studies. Comput. Biol. Med. 2023, 152, 106438. [Google Scholar] [CrossRef]
- Hamyoon, H.; Chan, W.Y.; Mohammadi, A.; Kuzan, T.Y.; Mirza-Aghazadeh-Attari, M.; Leong, W.L.; Altintoprak, K.M.; Vijayananthan, A.; Rahmat, K.; Ab Mumin, N.; et al. Artificial intelligence, BI-RADS evaluation and morphometry: A novel combination to diagnose breast cancer using ultrasonography, results from multi-center cohorts. Eur. J. Radiol. 2022, 157, 110591. [Google Scholar] [CrossRef] [PubMed]
- Homayoun, H.; Chan, W.Y.; Kuzan, T.Y.; Leong, W.L.; Altintoprak, K.M.; Mohammadi, A.; Vijayananthan, A.; Rahmat, K.; Leong, S.S.; Mirza-Aghazadeh-Attari, M.; et al. Applications of machine-learning algorithms for prediction of benign and malignant breast lesions using ultrasound radiomics signatures: A multi-center study. Biocybern. Biomed. Eng. 2022, 42, 921–933. [Google Scholar] [CrossRef]
- Gómez-Flores, W.; Gregorio-Calas, M.J.; Coelho de Albuquerque Pereira, W. BUS-BRA: A breast ultrasound dataset for assessing computer-aided diagnosis systems. Med. Phys. 2024, 51, 3110–3123. [Google Scholar] [CrossRef] [PubMed]
- Vreugdenburg, T.D.; Willis, C.D.; Mundy, L.; Hiller, J.E. A systematic review of elastography, electrical impedance scanning, and digital infrared thermography for breast cancer screening and diagnosis. Breast Cancer Res. Treat. 2013, 137, 665–676. [Google Scholar] [CrossRef]
- Lashkari, A.; Pak, F.; Firouzmand, M. Full intelligent cancer classification of thermal breast images to assist physician in clinical diagnostic applications. J. Med. Signals Sens. 2016, 6, 12–24. [Google Scholar] [CrossRef]
- FDA. FDA Warns Thermography Should not Be Used in Place of Mammography to Detect, Diagnose, or Screen for Breast Cancer: FDA Safety Communication. US Food & Drug Administration. 2019. Available online: https://www.fda.gov/news-events/press-announcements/fda-issues-warning-letter-clinic-illegally-marketing-unapproved-thermography-device-warns-consumers (accessed on 16 June 2025).
- Mashekova, A.; Zhao, Y.; Ng, E.Y.; Zarikas, V.; Fok, S.C.; Mukhmetov, O. Mukhmetov, Early detection of the breast cancer using infrared technology—A comprehensive review. Therm. Sci. Eng. Prog. 2022, 27, 101142. [Google Scholar] [CrossRef]
- Shen, Y.; Heacock, L.; Elias, J.; Hentel, K.D.; Reig, B.; Shih, G.; Moy, L. ChatGPT and other large language models are double-edged swords for medical diagnosis. Nat. Med. 2023, 29, 1124–1126. [Google Scholar] [CrossRef]
- Nath, M.K.; Sundararajan, K.; Mathivanan, S.; Thandapani, B. Analysis of breast cancer classification and segmentation techniques: A comprehensive review. Inform. Med. Unlocked 2025, 56, 101642. [Google Scholar] [CrossRef]
- Aerts, H.J.W.L.; Velazquez, E.R.; Leijenaar, R.T.H.; Parmar, C.; Grossmann, P.; Carvalho, S.; Bussink, J.; Monshouwer, R.; Haibe-Kains, B.; Rietveld, D.; et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat. Commun. 2014, 5, 4006. [Google Scholar] [CrossRef] [PubMed]
- Castiglioni, I.; Gallivanone, F.; Soda, P.; Avanzo, M.; Stancanello, J.; Aiello, M.; Interlenghi, M.; Salvatore, M. AI based applications in hybrid imaging: How to build smart and truly multiparametric decision models for radiomics. Eur. J. Nucl. Med. Mol. Imaging 2019, 46, 2673–2699. [Google Scholar] [CrossRef]
- Sala, E.; Mema, E.; Himoto, Y.; Veeraraghavan, H.; Brenton, J.; Snyder, A.; Weigelt, B.; Vargas, H. Unravelling tumour heterogeneity using next-generation imaging: Radiomics, radiogenomics, and habitat imaging. Clin. Radiol. 2017, 72, 3–10. [Google Scholar] [CrossRef]
- Lambin, P.; Leijenaar, R.T.H.; Deist, T.M.; Peerlings, J.; de Jong, E.E.C.; van Timmeren, J.; Sanduleanu, S.; Larue, R.T.H.M.; Even, A.J.G.; Jochems, A.; et al. Radiomics: The bridge between medical imaging and personalized medicine. Nat. Rev. Clin. Oncol. 2017, 14, 749–762. [Google Scholar] [CrossRef]
- Li, Y.; Wu, F.-X.; Ngom, A. A review on machine learning principles for multi-view biological data integration. Brief. Bioinform. 2016, 19, 325–340. [Google Scholar] [CrossRef]
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; van der Laak, J.A.W.M.; van Ginneken, B.; Sánchez, C.I. A survey on deep learning in medical image analysis. Med. Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef]
- Willemink, M.J.; Koszek, W.A.; Hardell, C.; Wu, J.; Fleischmann, D.; Harvey, H.; Folio, L.R.; Summers, R.M.; Rubin, D.L.; Lungren, M.P. Preparing medical imaging data for machine learning. Radiology 2020, 295, 4–15. [Google Scholar] [CrossRef]
- Sahiner, B.; Pezeshk, A.; Hadjiiski, L.M.; Wang, X.; Drukker, K.; Cha, K.H.; Summers, R.M.; Giger, M.L. Deep learning in medical imaging and radiation therapy. Med. Phys. 2019, 46, e1–e36. [Google Scholar] [CrossRef] [PubMed]
- Castiglioni, I.; Rundo, L.; Codari, M.; Di Leo, G.; Salvatore, C.; Interlenghi, M.; Gallivanone, F.; Cozzi, A.; D’Amico, N.C.; Sardanelli, F. AI applications to medical images: From machine learning to deep learning. Phys. Medica Eur. J. Med. Phys. 2021, 83, 9–24. [Google Scholar] [CrossRef] [PubMed]
- Ghorbian, M.; Ghobaei-Arani, M.; Ghorbian, S. Transforming breast cancer diagnosis and treatment with large language Models: A comprehensive survey. Methods 2025, 239, 85–110. [Google Scholar] [CrossRef]
- Singhal, K.; Azizi, S.; Tu, T.; Mahdavi, S.S.; Wei, J.; Chung, H.W.; Scales, N.; Tanwani, A.; Cole-Lewis, H.; Pfohl, S.; et al. Large language models encode clinical knowledge. Nature 2023, 620, 172–180. [Google Scholar] [CrossRef]
- Sallam, M. ChatGPT utility in healthcare education, research, and practice: Systematic review on the promising perspectives and valid concerns. Healthcare 2023, 11, 887. [Google Scholar] [CrossRef] [PubMed]
- Biswas, S. Role of ChatGPT in public health. Ann. Biomed. Eng. 2023, 51, 868–872. [Google Scholar] [CrossRef]
- Ribeiro, M.T.; Singh, S.; Guestrin, C. “Why should I trust you?”: Explaining the predictions of any classifier. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; pp. 1135–1144. [Google Scholar] [CrossRef]
- Lundberg, S.M.; Lee, S.-I. A unified approach to interpreting model predictions. Adv. Neural Inf. Process. Syst. 2017, 30, 4765–4774. [Google Scholar]
- Simonyan, K.; Vedaldi, A.; Zisserman, A. Deep inside convolutional networks: Visualising image classification models and saliency maps. arXiv 2013, arXiv:1312.6034. [Google Scholar]
- Sundararajan, M.; Taly, A.; Yan, Q. Axiomatic attribution for deep networks. In Proceedings of the 34th International Conference on Machine Learning, Sydney, Australia, 6–11 August 2017; pp. 3319–3328. [Google Scholar]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Fisher, A.; Rudin, C.; Dominici, F. All models are wrong, but many are useful: Learning a variable’s importance by studying an entire class of prediction models simultaneously. J. Mach. Learn. Res. 2019, 20, 1–81. [Google Scholar]
- Wachter, S.; Mittelstadt, B.; Russell, C. Counterfactual explanations without opening the black box: Automated decisions and the GDPR. Harv. J. Law Technol. 2017, 31, 841–887. [Google Scholar] [CrossRef]
- Ribeiro, M.T.; Singh, S.; Guestrin, C. Anchors: High-precision model-agnostic explanations. In Proceedings of the 32nd AAAI Conference on Artificial Intelligence, New Orleans, LA, USA, 2–7 February 2018; pp. 1527–1535. [Google Scholar]
- Craven, M.W.; Shavlik, J.W. Extracting tree-structured representations of trained networks. Adv. Neural Inf. Process. Syst. 1996, 8, 24–30. Available online: https://proceedings.neurips.cc/paper/1995/file/45f31d16b1058d586fc3be7207b58053-Paper.pdf (accessed on 3 January 2025).
- Zarikas, V.; Georgakopoulos, S.V. Interpretable Medical Image Diagnosis Methodology using Convolutional Neural Networks and Bayesian Networks. In Proceedings of the 2024 9th International Conference on Mathematics and Computers in Sciences and Industry (MCSI), Rhodes Island, Greece, 22–24 August 2024; pp. 128–133. [Google Scholar] [CrossRef]
- Aidossov, N.; Zarikas, V.; Zhao, Y.; Mashekova, A.; Ng, E.Y.K.; Mukhmetov, O.; Mirasbekov, Y.; Omirbayev, A. An Integrated Intelligent System for Breast Cancer Detection at Early Stages Using IR Images and Machine Learning Methods with Explainability. SN Comput. Sci. 2023, 4, 184. [Google Scholar] [CrossRef]
- Aidossov, N.; Zarikas, V.; Mashekova, A.; Zhao, Y.; Ng, E.Y.K.; Midlenko, A.; Mukhmetov, O. Evaluation of Integrated CNN, Transfer Learning, and BN with Thermography for Breast Cancer Detection. Appl. Sci. 2023, 13, 600. [Google Scholar] [CrossRef]
- Omirbayev, A.; Aidossov, N.; Zarikas, V.; Mashekova, A.; Zhao, Y.; Ng, Y.K. Breast cancer diagnosis using thermograms and Bayesian and Convolutional Neural Networks. In Proceedings of the Iupesm World Congress on Medical Physics and Biomedical Engineering (IUPESM WC2022), Singapore, 12–17 June 2022; Available online: https://wc2022.org/wc202 (accessed on 3 January 2025).
- Abdel-Nasser, M.; Rashwan, H.A.; Puig, D.; Moreno, A. Analysis of tissue abnormality and breast density in mammographic images using a uniform local directional pattern. Expert Syst. Appl. 2015, 42, 9499–9511. [Google Scholar] [CrossRef]
- Touvron, H.; Lavril, T.; Izacard, G.; Martinet, X.; Lachaux, M.A.; Lacroix, T.; Rozière, B.; Goyal, N.; Hambro, E.; Azhar, F.; et al. LLaMA: Open and Efficient Foundation Language Models. arXiv 2023, arXiv:2302.13971. [Google Scholar] [CrossRef]
- Alshuhri, M.S.; Al Musawi, S.G.; Al Alwany, A.A.; Uinarni, H.; Rasulova, I.; Rodrigues, P.; Alkhafaji, A.T.; Alshanberi, A.M.; Alawadi, A.H.; Abbas, A.H. Artificial intelligence in cancer diagnosis: Opportunities and challenges. Pathol. Res. Pract. 2024, 253, 154996. [Google Scholar] [CrossRef] [PubMed]
- Khalighi, S.; Reddy, K.; Midya, A.; Pandav, K.B.; Madabhushi, A.; Abedalthagafi, M. Artificial intelligence in neuro oncology: Advances and challenges in brain tumor diagnosis, prognosis, and precision treatment. NPJ Precis. Oncol. 2024, 8, 80. [Google Scholar] [CrossRef]
- Bhayana, R. Chatbots and Large Language Models in Radiology: A Practical Primer for Clinical and Research Applications. Radiology 2024, 310, e232756. [Google Scholar] [CrossRef]
- Habchi, Y.; Kheddar, H.; Himeur, Y.; Belouchrani, A.; Serpedin, E.; Khelifi, F.; Chowdhury, M.E.H. Advanced deep learning and large language models: Comprehensive insights for cancer detection. Image Vis. Comput. 2025, 157, 105495. [Google Scholar] [CrossRef]
- Sha, Z.; Hu, L.; Rouyendegh, B.D. Deep learning and optimization algorithms for automatic breast cancer detection. Int. J. Imaging Syst. Technol. 2020, 30, 495–506. [Google Scholar] [CrossRef]
- Sapate, S.; Talbar, S.; Mahajan, A.; Sable, N.; Desai, S.; Thakur, M. Breast cancer diagnosis using abnormalities on ipsilateral views of digital mammograms. Biocybern. Biomed. Eng. 2020, 40, 290–305. [Google Scholar] [CrossRef]
- Mansour, S.; Kamal, R.; Hussein, S.A.; Emara, M.; Kassab, Y.; Taha, S.N.; Gomaa, M.M. Enhancing detection of previously missed non-palpable breast carcinomas through artificial intelligence. Eur. J. Radiol. Open 2025, 14, 100629. [Google Scholar] [CrossRef] [PubMed]
- Malherbe, K. Revolutionizing Breast Cancer Screening: Integrating Artificial Intelligence with Clinical Examination for Targeted Care in South Africa. J. Radiol. Nurs. 2025, 44, 195–202. [Google Scholar] [CrossRef]
- Hernström, V.; Josefsson, V.; Sartor, H.; Schmidt, D.; Larsson, A.-M.; Hofvind, S.; Andersson, I.; Rosso, A.; Hagberg, O.; Lång, K. Screening performance and characteristics of breast cancer detected in the Mammography Screening with Artificial Intelligence trial (MASAI): A randomised, controlled, parallel-group, non-inferiority, single-blinded, screening accuracy study. Lancet Digit. Health 2025, 7, 175–183. [Google Scholar] [CrossRef]
- Nour, A.; Boufama, B. Hybrid deep learning and active contour approach for enhanced breast lesion segmentation and classification in mammograms. Intell.-Based Med. 2025, 11, 100224. [Google Scholar] [CrossRef]
- Umamaheswari, T.; Babu, Y.M.M. ViT-MAENB7: An innovative breast cancer diagnosis model from 3D mammograms using advanced segmentation and classification process. Comput. Methods Programs Biomed. 2024, 257, 108373. [Google Scholar] [CrossRef] [PubMed]
- Mannarsamy, V.; Mahalingam, P.; Kalivarathan, T.; Amutha, K.; Paulraj, R.K.; Ramasamy, S. Sift-BCD: SIFT-CNN integrated machine learning-based breast cancer detection. Biomed. Signal Process. Control 2025, 106, 107686. [Google Scholar] [CrossRef]
- Puttegowda, K.; Veeraprathap, V.; Kumar, H.S.R.; Sudheesh, K.V.; Prabhavathi, K.; Vinayakumar, R.; Tabianan, K. Enhanced Machine Learning Models for Accurate Breast Cancer Mammogram Classification. Glob. Transit. 2025, 7, 276–295. [Google Scholar] [CrossRef]
- Ahmad, J.; Akram, S.; Jaffar, A.; Ali, Z.; Bhatti, S.M.; Ahmad, A.; Rehman, S.U. Deep learning empowered breast cancer diagnosis: Advancements in detection and classification. PLoS ONE 2024, 19, e0304757. [Google Scholar] [CrossRef] [PubMed]
- Gudur, R.; Patil, N.; Thorat, S.T. Early Detection of Breast Cancer using Deep Learning in Mammograms. J. Pioneer. Med. Sci. 2024, 13, 18–27. [Google Scholar] [CrossRef]
- Mahmood, T.; Saba, T.; Rehman, A.; Alamri, F.S. Harnessing the power of radiomics and deep learning for improved breast cancer diagnosis with multiparametric breast mammography. Expert Syst. Appl. 2024, 249, 123747. [Google Scholar] [CrossRef]
- Muduli, D.; Dash, R.; Majhi, B. Automated breast cancer detection in digital mammograms: A moth flame optimization based ELM approach. Biomed. Signal Process. Control 2020, 59, 101912. [Google Scholar] [CrossRef]
- Liu, Y.; Ren, L.; Cao, X.; Tong, Y. Breast tumors recognition based on edge feature extraction using support vector machine. Biomed. Signal Process. Control 2020, 58, 101825. [Google Scholar] [CrossRef]
- Ametefe, D.S.; John, D.; Aliu, A.A.; Ametefe, G.D.; Hamid, A.; Darboe, T. Advancing breast cancer diagnosis: Integrating deep transfer learning and U-Net segmentation for precise classification and delineation of ultrasound images. Results Eng. 2025, 26, 105047. [Google Scholar] [CrossRef]
- Wang, C.; Guo, Y.; Chen, H.; Guo, Q.; He, H.; Chen, L.; Zhang, Q. ABUS-Net: Graph convolutional network with multi-scale features for breast cancer diagnosis using automated breast ultrasound. Expert Syst. Appl. 2025, 273, 126978. [Google Scholar] [CrossRef]
- Kiran, A.; Ramesh, J.V.N.; Rahat, I.S.; Khan, M.A.U.; Hossain, A.; Uddin, R. Advancing breast ultrasound diagnostics through hybrid deep learning models. Comput. Biol. Med. 2024, 180, 108962. [Google Scholar] [CrossRef]
- Tian, R.; Lu, G.; Tang, S.; Sang, L.; Ma, H.; Qian, W.; Yang, W. Benign and malignant classification of breast tumor ultrasound images using conventional radiomics and transfer learning features: A multicenter retrospective study. Med. Eng. Phys. 2024, 125, 104117. [Google Scholar] [CrossRef]
- Ekici, S.; Jawzal, H. Breast cancer diagnosis using thermography and convolutional neural networks. Med. Hypotheses 2020, 137, 109542. [Google Scholar] [CrossRef] [PubMed]
- De Freitas Barbosa, V.A.; de Santana, M.A.; Andrade, M.K.S.; de Lima, R.d.C.F.; dos Santos, W.P. Deep-wavelet neural networks for breast cancer early diagnosis using mammary thermographies. In Deep Learning for Data Analytics; Elsevier: Hoboken, NJ, USA, 2020; pp. 99–124. [Google Scholar]
- Cabıoğlu, Ç.; Oğul, H. Computer-aided breast cancer diagnosis from thermal images using transfer learning. In Proceedings of the International Work-Conference on Bioinformatics and Biomedical Engineering, Granada, Spain, 30 September–2 October 2020; Springer: Berlin/Heidelberg, Germany, 2020; pp. 716–726. [Google Scholar]
- Sánchez-Ruiz, D.; Olmos-Pineda, I.; Olvera-López, J.A. Automatic region of interest segmentation for breast thermogram image classification. Pattern Recognit. Lett. 2020, 135, 72–81. [Google Scholar] [CrossRef]
- Resmini, R.; da Silva, L.F.; Medeiros, P.R.; Araujo, A.S.; Muchaluat-Saade, D.C.; Conci, A. A hybrid methodology for breast screening and cancer diagnosis using thermography. Comput. Biol. Med. 2021, 135, 104553. [Google Scholar] [CrossRef]
- Allugunti, V.R. Breast cancer detection based on thermographic images using machine learning and deep learning algorithms. Int. J. Eng. Comput. Sci. 2022, 4, 49–56. [Google Scholar] [CrossRef]
- Mohamed, E.A.; Rashed, E.A.; Gaber, T.; Karam, O. Deep learning model for fully automated breast cancer detection system from thermograms. PLoS ONE 2022, 17, e0262349. [Google Scholar] [CrossRef]
- Civilibal, S.; Cevik, K.K.; Bozkurt, A. A deep learning approach for automatic detection, segmentation and classification of breast lesions from thermal images. Expert Syst. Appl. 2023, 212, 118774. [Google Scholar] [CrossRef]
- Ramacharan, S.; Margala, M.; Shaik, A.; Chakrabarti, P.; Chakrabarti, T. Advancing Breast Cancer Diagnosis: The Development and Validation of the HERA-Net Model for Thermographic Analysis. Comput. Mater. Contin. 2024, 81, 3731–3760. [Google Scholar] [CrossRef]
- Guo, D.; Lu, C.; Chen, D.; Yuan, J.; Duan, Q.; Xue, Z.; Liu, S.; Huang, Y. A multimodal breast cancer diagnosis method based on Knowledge-Augmented Deep Learning. Biomed. Signal Process. Control 2024, 90, 105843. [Google Scholar] [CrossRef]
- Rao, A.; Kim, J.; Kamineni, M.; Pang, M.; Lie, W.; Dreyer, K.J.; Succi, M.D. Evaluating GPT as an Adjunct for Radiologic Decision Making: GPT-4 Versus GPT-3.5 in a Breast Imaging Pilot. J. Am. Coll. Radiol. 2023, 20, 990–997. [Google Scholar] [CrossRef]
- Miao, S.; Jia, H.; Cheng, K.; Hu, X.; Li, J.; Huang, W.; Wang, R. Deep learning radiomics under multimodality explore association between muscle/fat and metastasis and survival in breast cancer patients. Brief. Bioinform. 2022, 23, bbac432. [Google Scholar] [CrossRef]
- Choi, H.S.; Song, J.Y.; Shin, K.H.; Chang, J.H.; Jang, B.-S. Developing prompts from large language model for extracting clinical information from pathology and ultrasound reports in breast cancer. Radiat. Oncol. J. 2023, 41, 209–216. [Google Scholar] [CrossRef]
- Sorin, V.; Klang, E.; Sklair-Levy, M.; Cohen, I.; Zippel, D.B.; Lahat, N.B.; Konen, E.; Barash, Y. Large language model (ChatGPT) as a support tool for breast tumor board. npj Breast Cancer 2023, 9, 44. [Google Scholar] [CrossRef] [PubMed]
- Nakach, F.-Z.; Idri, A.; Goceri, E. A comprehensive investigation of multimodal deep learning fusion strategies for breast cancer classification. Artif. Intell. Rev. 2024, 57, 327. [Google Scholar] [CrossRef]
- Haver, H.L.; Ambinder, E.B.; Bahl, M.; Oluyemi, E.T.; Jeudy, J.; Yi, P.H. Appropriateness of Breast Cancer Prevention and Screening Recommendations Provided by ChatGPT. Radiology 2023, 307, 1–4. [Google Scholar] [CrossRef]
- Grigore, I.; Popa, C.-A. MambaDepth: Enhancing Long-range Dependency for Self-Supervised Fine-Structured Monocular Depth Estimation. arXiv 2024, arXiv:2406.04532. [Google Scholar] [CrossRef]
- Griewing, S.; Lechner, F.; Gremke, N.; Lukac, S.; Janni, W.; Wallwiener, M.; Wagner, U.; Hirsch, M.; Kuhn, S. Proof-of-concept study of a small language model chatbot for breast cancer decision support–a transparent, source-controlled, explainable and data-secure approach. J. Cancer Res. Clin. Oncol. 2024, 150, 451. [Google Scholar] [CrossRef]
- AlSaad, R.; Abd-alrazaq, A.; Boughorbel, S.; Ahmed, A.; Renault, M.A.; Damseh, R.; Sheikh, J. Multimodal Large Language Models in Health Care: Applications, Challenges, and Future Outlook. J. Med. Internet Res. 2024, 26, e59505. [Google Scholar] [CrossRef] [PubMed]
- Sorin, V.; Glicksberg, B.S.; Artsi, Y.; Barash, Y.; Konen, E.; Nadkarni, G.N.; Klang, E. Utilizing large language models in breast cancer management: Systematic review. J. Cancer Res. Clin. Oncol. 2024, 150, 140. [Google Scholar] [CrossRef]
| Dataset Name | Image Modality | URL |
|---|---|---|
| Mammography Dataset | ||
| MIAS | Mammogram | https://www.repository.cam.ac.uk/handle/1810/250394 (accessed on 3 January 2025) |
| mini-MIAS | Mammogram | http://peipa.essex.ac.uk/info/mias.html (accessed on 3 January 2025) |
| BCDR | Mammogram | https://bcdr.ceta-ciemat.es/information/about (accessed on 3 January 2025) |
| DDSM | Mammogram | https://www.kaggle.com/datasets/skooch/ddsm-mammography (accessed on 11 October 2025) |
| INBreast | Mammogram | https://biokeanos.com/source/INBreast (accessed on 11 October 2025) |
| WBCD | Multimodality | https://archive.ics.uci.edu/ml/datasets/Breast+Cancer+Wisconsin+(Diagnostic) (accessed on 3 January 2025) |
| WDBC | Multimodality | http://networkrepository.com/breast-cancer-wisconsin-wdbc.php (accessed on 3 January 2025) |
| IRMA | Mammogram | https://data.world/datasets/irma (accessed on 3 January 2025) |
| Breast-Cancer-Screening-DBT | Mammogram | https://www.cancerimagingarchive.net/collection/breast-cancer-screening-dbt/ (accessed on 11 October 2025) |
| CMMD | Mammogram | https://www.cancerimagingarchive.net/collection/cmmd/ (accessed on 11 October 2025) |
| Breast Micro-Calcifications Dataset | Mammogram | https://data.europa.eu/data/datasets/oai-zenodo-org-5036062?locale=en (accessed on 3 January 2025) |
| OPTIMAM Mammography Image Database | Mammogram | https://medphys.royalsurrey.nhs.uk/omidb/ (accessed on 3 January 2025) |
| Digital Mammography Dataset for Breast Cancer Diagnosis Research (DMID) | Mammogram | https://figshare.com/articles/dataset/_b_Digital_mammography_Dataset_for_Breast_Cancer_Diagnosis_Research_DMID_b_DMID_rar/24522883/1 (accessed on 3 January 2025) |
| Mammographic Mass—UCI Machine Learning Repository | Mammogram | https://archive.ics.uci.edu/dataset/161/mammographic+mass (accessed on 3 January 2025) |
| ICTRE—The Cancer Imaging Archive (TCIA) | Mammogram | https://www.cancerimagingarchive.net/collection/victre/ (accessed on 3 January 2025) |
| Ambra UNIFESP Mammography | Mammogram | https://mamografiaunifesp.ambrahealth.com/ (accessed on 3 January 2025) |
| CSAW-S | Mammogram | https://zenodo.org/records/4030660 (accessed on 11 October 2025) |
| Safe Haven (DaSH) | Mammogram | https://www.abdn.ac.uk/research/digital/platforms/safe-haven-dash/accessing-data/available-datasets/ (accessed on 3 January 2025) |
| Ultrasound Dataset | ||
| BUSI | Ultrasound | https://www.kaggle.com/datasets/aryashah2k/breast-ultrasound-images-dataset (accessed on 3 January 2025) |
| OASBUD | Ultrasound | https://zenodo.org/records/545928#.X0xKf8hKg2z (accessed on 3 January 2025) |
| UDIAT | Ultrasound | upon request (accessed on 3 January 2025) |
| BrEaST | Ultrasound | https://www.cancerimagingarchive.net/collection/breast-lesions-usg/?utm_source (accessed on 3 January 2025) |
| QAMEBI | Ultrasound | https://qamebi.com/breast-ultrasound-images-database/ (accessed on 3 January 2025) |
| BUS-BRA | Ultrasound | https://www.kaggle.com/datasets/orvile/bus-bra-a-breast-ultrasound-dataset (accessed on 3 January 2025) |
| Thermography Dataset | ||
| The Mastology Research with Infrared Image (DMR-IR) | thermograms | https://www.kaggle.com/datasets/asdeepak/thermal-images-for-breast-cancer-diagnosis-dmrir (accessed on 11 October 2025) |
| NIRAMAI Health Analytix | thermograms | https://niramai.com/about/thermalytix/ (accessed on 3 January 2025) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mashekova, A.; Zhao, M.Y.; Zarikas, V.; Mukhmetov, O.; Aidossov, N.; Ng, E.Y.K.; Wei, D.; Shapatova, M. Review of Artificial Intelligence Techniques for Breast Cancer Detection with Different Modalities: Mammography, Ultrasound, and Thermography Images. Bioengineering 2025, 12, 1110. https://doi.org/10.3390/bioengineering12101110
Mashekova A, Zhao MY, Zarikas V, Mukhmetov O, Aidossov N, Ng EYK, Wei D, Shapatova M. Review of Artificial Intelligence Techniques for Breast Cancer Detection with Different Modalities: Mammography, Ultrasound, and Thermography Images. Bioengineering. 2025; 12(10):1110. https://doi.org/10.3390/bioengineering12101110
Chicago/Turabian StyleMashekova, Aigerim, Michael Yong Zhao, Vasilios Zarikas, Olzhas Mukhmetov, Nurduman Aidossov, Eddie Yin Kwee Ng, Dongming Wei, and Madina Shapatova. 2025. "Review of Artificial Intelligence Techniques for Breast Cancer Detection with Different Modalities: Mammography, Ultrasound, and Thermography Images" Bioengineering 12, no. 10: 1110. https://doi.org/10.3390/bioengineering12101110
APA StyleMashekova, A., Zhao, M. Y., Zarikas, V., Mukhmetov, O., Aidossov, N., Ng, E. Y. K., Wei, D., & Shapatova, M. (2025). Review of Artificial Intelligence Techniques for Breast Cancer Detection with Different Modalities: Mammography, Ultrasound, and Thermography Images. Bioengineering, 12(10), 1110. https://doi.org/10.3390/bioengineering12101110











