1. Introduction
Atrial fibrillation (AF) is the most common cardiac arrhythmia [
1], with its global incidence and prevalence on the rise [
2]. The presence of AF promotes thrombus formation, which may lead to stroke [
3] and systemic embolism [
4], and increases the risk of other cardiovascular diseases such as heart failure [
3], myocardial infarction [
5], and sudden cardiac death [
6]. Currently, ECG signals serve as the gold standard for clinical AF diagnosis [
7]. On an ECG, AF is characterized by irregular ventricular rhythms, absence of P waves, and presence of F waves [
8]. According to guidelines, AF episodes lasting at least 30 s on ECG can be diagnosed as clinical AF [
9]. AF is inherently progressive, meaning that it tends to worsen over time without intervention [
10]. Therefore, early diagnosis of AF is crucial for timely intervention [
11].
However, due to the often asymptomatic [
12] and paroxysmal nature of AF, clinical screening for AF remains challenging. While palpitations, dyspnea, fatigue, and chest discomfort are common symptoms, a substantial proportion of AF patients remain asymptomatic, posing major challenges for timely detection and management. Recent evidence indicates that approximately 27% of AF patients are asymptomatic, with estimates ranging from 20% to 50% across populations [
13]. Importantly, asymptomatic AF carries a risk of thromboembolic and cardiovascular events comparable to, or even higher than, symptomatic AF, largely due to delayed diagnosis and treatment. Moreover, asymptomatic status has been associated with specific clinical profiles—including male sex, diabetes mellitus, chronic kidney disease, and prior stroke or transient ischemic attack—that may predispose individuals to unrecognized AF episodes. Relying solely on infrequent and short-term rapid ECG assessments (30 s) for detecting AF may lead to underdiagnosis. It is further emphasized that patients at higher risk of AF prediction require long-term ECG monitoring [
14] to increase the chances of early AF detection. In addition, there is still a need to rely on medical experts for interpreting ECG data. Manual visual inspection of ECG recordings is also a highly time-consuming and error-prone task. This significantly reduces the chances of timely detection of AF. Therefore, reliable automated methods must be developed for analyzing and interpreting long-term ECG recordings, particularly for asymptomatic individuals. Automated beat-level AF localization algorithms could facilitate earlier detection and personalized intervention, helping to prevent progression and reduce the burden of AF-related complications.
In recent years, machine learning (ML) and deep learning (DL) have been extensively developed in the field of AF detection [
15]. Machine learning methods focus on classifying ECG segments using manually extracted features obtained through feature extraction and feature selection, with generally low computational costs in training and testing [
16]. It is worth noting that the process of developing effective handcrafted features and finding the best-performing feature combinations is time-consuming and labor-intensive, heavily relying on manual operations. In the majority of ML-based AF detection approaches, QRS wave detection is a mandatory initial step. The performance limitations of QRS detectors and the presence of noise can affect the performance of the final ML model. Furthermore, there are significant differences in electrocardiographic signals among different patients, making it challenging to find effective ECG morphological features. Therefore, machine learning methods based on handcrafted features alone are insufficient for accurate AF detection. More and more studies are employing DL methods to achieve more accurate prediction results, albeit potentially requiring higher computational costs. Most DL-based methods utilize end-to-end frameworks with raw data as input to automatically extract features, allowing the model to learn feature embeddings most suitable for specific classification tasks. Deep learning demonstrates tremendous potential in automatically detecting and classifying arrhythmias from electrocardiographic signals and exhibits proficiency at a level comparable to that of medical experts in classifying various arrhythmias.
However, there are still some overlooked issues in existing methods that require further research. Firstly, the majority of existing AF detection methods generate a single prediction label for an ECG segment without providing beat-level prediction labels. An ECG segment may contain both AF and non-AF parts, leading to ambiguity in the feature space and misleading the model to embed them into overlapping feature spaces. Secondly, AF beat detection in long-term ECG recordings remains a challenge. The research on beat-level AF detection is significantly less explored and its performance needs improvement. Existing AF detection models expect isolated heartbeat signals as input for beat-level detection. Although shorter input segment lengths can achieve more fine-grained detection results, shorter ECG segments mean fewer modeled heartbeat dependencies, which often lead to poorer detection performance. Additionally, AF detection methods that use a single heartbeat signal or RRI sequence as input still require explicit heartbeat localization. Traditional threshold-based QRS detection algorithms still have limitations in various arrhythmia scenarios, and erroneous localization results may further restrict the detection capabilities.
To address these issues, this paper models the AF detection task as an object detection problem to achieve beat-level prediction. An AF object detection model takes fixed-length ECG segments as input and outputs classification results for AF beats along with position predictions. By capturing heartbeat dependencies, the model can enhance the classification performance of individual beats. Considering all beat classification results collectively also allows for segment-level classification predictions. The DETR architecture [
17] has achieved significant success in the field of image object detection. Inspired by this, we introduce the DETR architecture for object detection in 1D ECG signals.
To be specific, this paper presents a novel AF object detection model named AF- DETR, which consists of a CNN feature extraction backbone, a transformer encoder, a dual-query transformer decoder, and several prediction heads. The AF-DETR model utilizes the CNN backbone to extract rhythm and morphology information from ECG signals and feeds a compact ECG feature representation into the transformer encoder to capture dependencies between heartbeats. The refined encoder ECG features are further decoded by the decoder to predict heartbeat positions and categories. In the decoder, decoder queries are divided into content and positional parts, and position queries are designed using 2D bounding boxes which represent heartbeat positions. Through iterative updates, the decoder progressively corrects the bounding boxes and captures crucial classification information, ultimately achieving accurate heartbeat localization and classification predictions. Additionally, a denoising training method is introduced during the training process to stabilize bipartite matching and accelerate model convergence. Positive and negative noise samples are constructed from the same ground truth to help the model avoid duplicate outputs of the same target. Cross-dataset testing is conducted using five datasets to evaluate beat-level localization and classification performance as well as segment-level classification performance, demonstrating the strong AF detection capabilities of the model. The contributions of this paper are as follows:
- (1)
A novel AF-DETR model with a CNN-Transformer architecture is proposed to model the AF detection as an object detection problem, thereby achieving localization and classification of AF heartbeats.
- (2)
Deriving decoder positional queries from 2D bounding boxes representing heartbeat positions, enabling the decoder to iteratively update bounding boxes layer by layer, thus accelerating model convergence and effectively localizing AF heartbeats.
- (3)
Denoising training is introduced to stabilize bipartite matching. Concretely, a contrastive denoising mechanism is conducted to help the model avoid duplicate detections of the same heartbeat by adding positive and negative noise samples.
- (4)
The proposed AF-DETR model achieves accurate detection of AF heartbeats in cross-dataset testing on four external datasets and demonstrates excellent generalization performance. Additionally, it performs comparably to existing methods in segment-level classification performance.
The remaining parts of this study are arranged as follows.
Section 2 introduces the related work.
Section 3 outlines the methods used in this study.
Section 4 presents the datasets, experiments and results.
Section 5 discusses the proposed method.
Section 6 summarizes this study.
2. Related Work
The AF detection approaches can be roughly categorized into three types: methods based on ventricular rhythm information, methods based on atrial morphology information, and methods based on overall ECG information.
Methods based on ventricular rhythm typically involve extracting RR interval sequences and computing various handcrafted features, including multiple time-domain and frequency-domain features [
18], various entropy features [
19,
20,
21], Poincare plots [
22,
23], and Lorenz plots [
24], to construct machine learning models such as support vector machine (SVM) [
25,
26,
27], decision tree [
28], k-nearest neighbor (KNN) [
29], and random forest (RF) [
30,
31,
32]. A few studies have constructed DL-based models with RR interval sequences as input [
33,
34]. Due to the good anti-interference performance of R-peak detection, methods based on irregular ventricular rhythm information generally exhibit good generalization performance. However, they are difficult to implement in short time series, and the irregular behavior of AF overlaps with other arrhythmias [
35], leading to false positive detection in cases with frequent premature beats. Additionally, in some cases, the RR intervals of AF can also be regular, further limiting the effectiveness of rhythm-based methods.
Methods based on atrial morphology typically involve extracting P-wave/F-wave morphological features to determine the disappearance of P-waves and the appearance of F-waves [
36,
37]. To extract features from atrial activity, some studies isolate F- waves by eliminating the QRS complex from the ECG signal [
38]. However, due to the small amplitude of P-waves and their susceptibility to various types of noise [
39], the performance of AF detection algorithms based on atrial morphology information is not very robust. Therefore, the morphology-based methods are often used in conjunction with rhythm-based methods [
40,
41] to further improve the accuracy of AF detection.
Methods based on overall ECG information do not explicitly extract ventricular rhythm information or atrial morphology information but instead employ deep neural networks to achieve end-to-end AF detection. CNN backbones are commonly used for automatic feature extraction from ECG signals [
42,
43]. Some studies further refine the features extracted by CNN backbones using LSTM [
44,
45,
46,
47], GRU [
48], and transformer encoders [
49].
Traditional AF detection methods typically generate segment-level AF detection results. When finer-grained prediction labels are desired, reducing the length of input segments is a common approach, but this reduces the available heartbeat dependencies, leading to a sharp decrease in classification performance. This paper aims to achieve beat-level AF detection without reducing the length of input segments, using an object detection approach.
3. Method
3.1. Problem Definition
Beat-level atrial fibrillation detection can be expressed as an object detection task to predict the type and location of the target heartbeat. The AF-DETR architecture models object detection as a set prediction problem that directly infers predictions. The fixed size is set to be significantly larger than the number of objects in the sample, and an additional class label Non-obj is set to indicate cases where there is no object in the predicted bounding box.
A normalized ECG segment with a sampling frequency of and a duration of seconds is used as a sample, which can be defined as , where the sample length is . The ground truth set of objects of the ECG sample is expressed as and each element can be represented as , where is the class label, is the ground truth box, is the central coordinate, and is the width relative to the sample length.
The AF-DETR model mainly consists of a backbone, a transformer encoder, and a dual-query transformer decoder. Samples are fed into the CNN-based backbone to extract a compact ECG feature representation. Then, the CNN features are refined by combining positional encoding and the encoder. The encoded ECG features are decoded to objects in parallel by transformer encoder. The output of the final layer of the decoder generates the prediction label and the 2D prediction box, respectively, through the classification prediction head and the bounding box prediction head.
Since the predicted objects are not directly matched with the ground truth objects, the loss is calculated according to the optimal bipartite matching between the ground truth objects and the predicted objects during training. The AF-DETR model directly makes
predictions based on the input sample, and each prediction corresponds to only one GT object or Non-obj. To match the prediction set, the ground truth set
is considered as a set of size
filled with
(Non-obj). The Hungarian algorithm can be used to efficiently compute an optimal bipartite matching between these two sets from the cost matrix of objects, which means to find a permutation of
elements
with the lowest cost:
where
is a pair-wise matching cost between the ground truth
and the prediction
. The matching cost considers the location cost and the classification cost, respectively. For the prediction
, the predicted box is defined as
, and the prediction probability of
class is defined as
. The negative prediction probability is used as the classification cost, and the location cost is considered using the negative 1D generalized intersection over union (GIoU) and the
L1 norm of the bounding boxes. The 1D GIoU and
L1 norms for ground truth and prediction boxes can be calculated as:
The symbol
refers to the length of the box, and
means the length of the smallest interval that covers both bounding boxes. Then, the matching cost adjusted by the hyperparameters
and
can be expressed as:
For the loss function, it is also necessary to consider both classification loss and positioning loss, and the total loss can be expressed as:
where
is the classification loss coefficient. The classification loss function adopts weighted multi-classification cross entropy loss to alleviate class imbalance, which can be expressed as:
where a typical value
is used when
, and
is used in other cases. In addition, a linear combination of
L1 loss and 1D GIoU loss is used as the positioning loss to improve the positioning accuracy. It is calculated as:
where
and
are the hyperparameters for adjusting the loss weight.
3.2. CNN Backbone
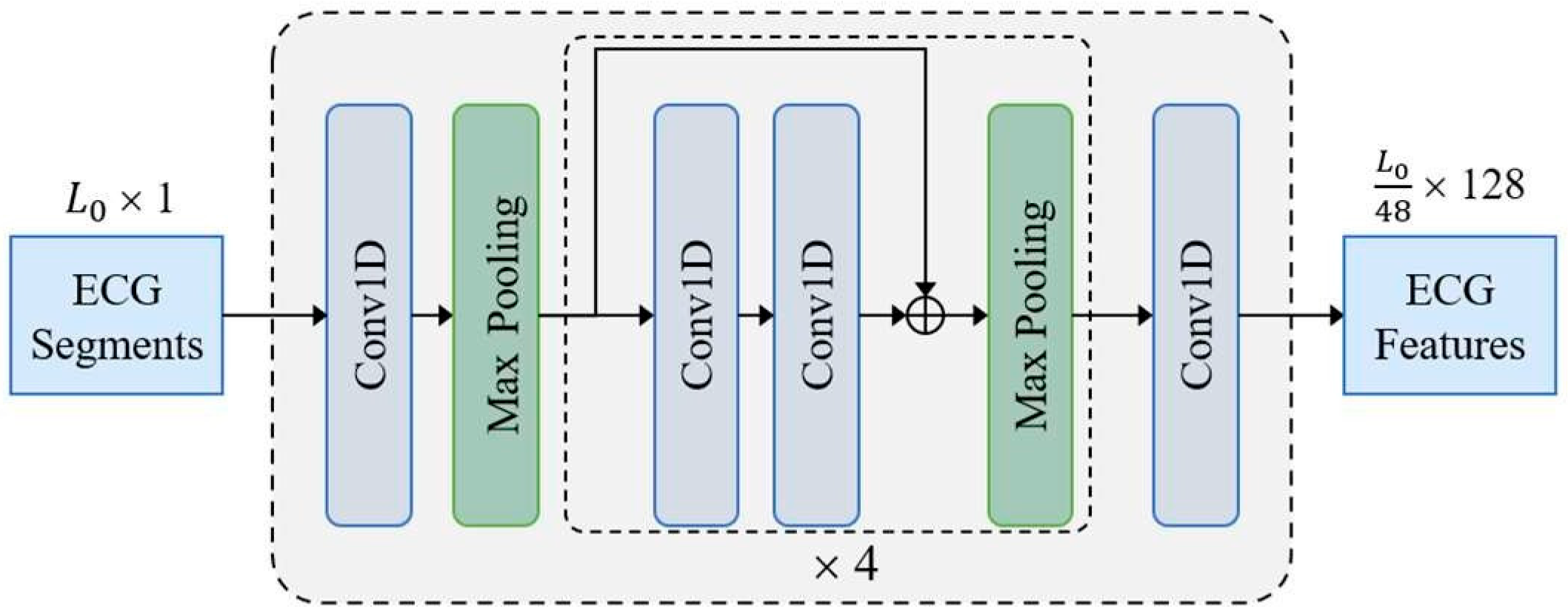
The CNN backbone constructed with 1D residual blocks is shown in
Figure 1. The input sample
is fed to a 1D convolution layer and four residual blocks to acquire a compact ECG feature representation. Finally, the number of feature channels is adjusted to
D = 128 through a 1D convolution layer with kernel size of 1. Except the last convolutional layer, all convolutional layers use a kernel size of 3 followed by a batch normalization layer and a ReLU activation layer. Each residual block contains two convolution layers and doubles the number of channels in the first convolution layer. In addition to setting the first max pooling layer with stride size of 3 and kernel size of 3, all max pooling layers have a stride size of 2 and a kernel size of 2. The ECG features extracted by the CNN backbone are fed to transformer for final predictions.
3.3. Transformer Encoder
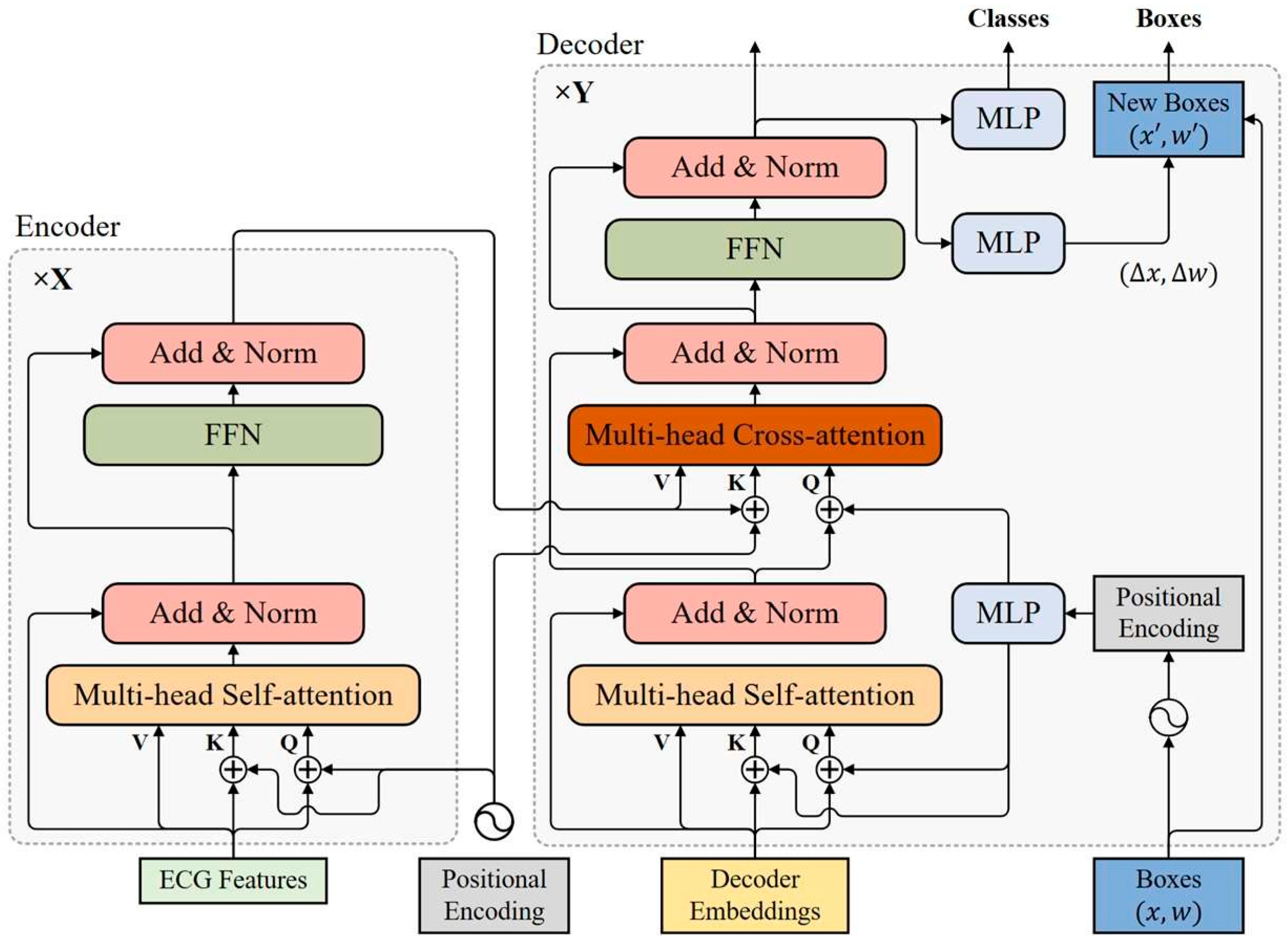
The transformer encoder consists of multiple encoder layers with the same structure, all of which are capable of encoding ECG features at the same resolution as the input. As shown in
Figure 2, each encoder layer has a standard architecture consisting of an attention module and a feed forward network (FFN) with a residual connection around them, and followed by a layer normalization module. The feed forward network consists of two fully connected layers, which are connected using the ReLU activation layer.
3.3.1. Positional Encoding
Because of the permutation invariance of transformer architecture, fixed positional encoding is added to the input of each attention module to capture positional dependencies, which helps complement the location representation capabilities of model. The sinusoidal function is used to construct a positional encoding vector on the same scale. The positional encoding function PE:
, which maps position
to a D-dimensional sinusoidal embedding, is defined as:
where symbol
in the encoder refers to the position of the encoding vector in the time dimension. The subscripts
and
represent the index in the encoding vector, The parameter
is a hand-designed temperature, and the typical value used in the encoder is
.
3.3.2. Multi-Head Self-Attention Module
In each encoding layer, ECG features are combined with positional encoding to generate a query
, a key
, and a value
, respectively, as inputs to the self-attention module. In the self-attention module, the query
, key
, and value
are linearly projected
times to obtain
groups
,
, and
, respectively, preparing for multi-head self-attention. A typical setting is
and the weight matrices
are learnable parameters of linear projection layers. The scaled dot-product attention function for each head can be expressed as:
The output of all heads is concatenated in the feature dimension and fed to a linear layer with a weight parameter
to produce the final output, which can be expressed as:
where
is the output of the
i-th head. The symbol
represents the concatenation function.
3.4. Transformer Decoder
The transformer decoder uses multiple decoder layers to decode N targets in parallel. Each decoding layer uses multi-head self-attention modules to update queries and cross-attention modules to probe the objects based on the similarity of queries and keys. The decoder updates the query layer by layer and continuously approximates the target ground-truth objects. Considering that the keys in the cross-attention module contain both the content part (encoded ECG features) and the location part (positional embedding), a dual-query mode consisting of content queries and positional queries is introduced, where the initial content query uses decoder embeddings.
3.4.1. Positional Query
In order to make the positional similarity of queries and keys in the cross-attention module more consistent, the bounding boxes are directly learned in each decoder layer and positional queries are derived from these bounding boxes with sinusoidal positional encoding functions. The
q-th box of N bounding boxes is defined as
, where
and
. The corresponding content query and positional query are represented as
and
, respectively, where
is the dimension of decoder queries. The positional query
derived from the given bounding box
is:
where the symbol
represents the positional encoding function
that maps a float number to a
D-dimensional sinusoidal embedding. A hyperparameter setting T = 20 is used in decoder. The symbol
represents a multi-layer perceptron
, which projects a
2D vector into
D-dimensional vector. It includes two fully connected layers and its parameters are shared between all decoder layers.
For the initial queries of the first decoding layer, the decoder embeddings and initial bounding boxes need to be prepared, respectively. Here, the decoder embeddings are designed to be learnable embeddings, while the initial bounding boxes adopt a fixed box width and a uniformly distributed center point.
3.4.2. Self-Attention Module of Decoder
The self-attention module in the decoder is used to query updating the cross-attention module. All queries, keys, and values use the same content item
, while queries and keys contain an additional positional item
. The input of the self-attention module in the
d-th decoding layer can be expressed as:
3.4.3. Cross-Attention Module of Decoder
The cross-attention module in the decoder also uses a multi-head attention mechanism. In the cross-attention module, positional and content information are combined as object queries to extract object information from the encoded ECG features. The input of the cross-attention module in the d-th decoder layer can be expressed as:
where
is the feature vector at position
in the encoded ECG feature,
S is the output of the self-attention module in the decoding layer
d, and
LN represents the layer normalization function.
3.4.4. Iterative Bounding Box Updating
The pattern of deriving positional queries from bounding boxes makes it possible to update bounding boxes layer by layer. With the exception of the first decoding layer, each decoder layer updates positional queries based on the updated bounding boxes of the previous layer. Each layer predicts the relative offset of the bounding box
for the next layer through the unshared bounding box prediction head
, which helps to further reduce optimization difficulty. All bounding box prediction heads contains three fully connected layers. Given a bounding box
provided by the (
d − 1)-th decoder layer, the d-th decoder layer updates the bounding box as:
where the symbols
and
represent the sigmoid function and the inverse sigmoid function, respectively.
3.4.5. Predicted Output
In addition to using the updated bounding boxes of each decoder layer as its box prediction, the normalized output of each decoder layer is fed to an unshared classification prediction head to make classification predictions. All classification prediction heads consist of two fully connected layers. Hungarian matching is applied to predicted results of each decoder layer against the ground truth objects, and auxiliary loss of each decoder layer is built to update parameters, which helps speed up model convergence. For stable training, the bounding box prediction head and classification prediction head of each decoder layer use only its auxiliary loss for parameter updating. The predictions of the last decoder layer are the predictions of the whole model. When inference is performed, predictions with a Non-obj label can be omitted, and other valid predictions need to be retained.
3.5. Denoising Training
In order to mitigate the instability caused by Hungarian matching, denoising (DN) training is introduced, which is able to reconstruct the object from the noised ground truth. Therefore, random noise is added to the bounding box and class label of each ground truth object to construct additional noisy query. Similar to the ordinary query, the noise query is also composed of content query and positional query. The noised labels generate the initial content queries of the decoder, and the noised bounding boxes generates the initial positional queries. Noised bounding boxes and noised labels are constructed as follows:
- (1)
Noised bounding boxes: The method of center shifting is adopted for adding noise to bounding boxes. Random noise satisfying is added in the center point of the box, so that the center of the shifted box is still in the original box. The random noise is controlled by the noise scale parameter . Contrastive denoising (CDN) training is further considered to help the model avoid duplicate predictions of the same object. Contrast denoising training generates positive and negative queries by setting two different parameters and . A positive query which satisfies is expected to reconstruct its corresponding ground truth box. Negative queries satisfy , and they are expected to be predicted as Non-obj.
- (2)
Noised labels: Noised labels are acquired by randomly flipping some ground truth labels to other labels, and the proportion of label flipping is controlled the hyperparameter . For the noise query, label embedding generated by the noised label is used as the content part.
In fact, multiple groups of noisy queries can be constructed. It is important to note that despite the addition of noise, the noise query contains information about the true object. In order to prevent information leakage, when the self-attention module of the decoder calculates the attention weight matrix, it is necessary to use attention mask isolation between ordinary queries and multi-group noise queries. The predictions corresponding to noise queries can bypass bipartite matching and directly calculate its reconstruction loss with the corresponding ground truth objects. The reconstruction loss of noise queries is similar to that of ordinary queries. Noise queries are only added to the decoder during training and removed during inference.
5. Discussion
5.1. Overview of the Proposed Method
In this work, an AF object detection model called AF-DETR is proposed to accurately localize AF heartbeats in single-lead ECG records. In the transformer decoder, decoder positional queries are constructed using 2D bounding boxes which represent the heartbeat positions, enabling iterative updates of the bounding boxes for precise heartbeat localization and classification prediction. Additionally, a denoising training method is introduced to stabilize bipartite matching and accelerate model convergence, while contrasting denoising mechanisms to avoid redundant predictions of the same heartbeat. Cross-dataset testing results demonstrate the excellent detection accuracy and generalization performance of the proposed method, making it applicable to various datasets without the need for parameter adjustments in practical applications.
5.2. Method Evaluation
The DETR architecture is introduced for object detection in 1D ECG signals. The research results suggest that the DETR architecture has the potential to improve the accuracy of physiological signal analysis, which could have a broader impact on the field of physiological signal diagnosis.
For AF heartbeat detection, both classification performance and localization accuracy are equally important. In the AF-DETR model, 2D bounding boxes composed of center points and box widths are introduced to derive positional queries. The positional queries are encoded using sine positional encoding, similar to the positional encoding used for encoder features, ensuring the similarity between the position queries and the positional information in the encoder features. Deriving queries using coordinates enables the iterative updating of bounding boxes, and the progressively updated bounding boxes allow the construction of auxiliary losses using outputs from each decoding layer. These strategies accelerate model convergence and enhance both localization and classification accuracy.
Due to the stochastic nature of the training process, slight changes in the cost matrix can lead to significant variations in matching results, resulting in unstable bipartite matching and unstable model training. Denoise training can reduce the instability of bipartite matching to accelerate model convergence. In fact, denoise training bypasses bipartite matching because the correspondence between the noise query prediction and the truth value is known. By training the model to reconstruct boxes from noised boxes close to ground truth, denoise training allows the model to focus more on the nearby region of each query, preventing potential prediction conflicts between queries. Additionally, we construct positive and negative noise queries for the same ground truth object, which helps the model differentiate subtle differences between bounding boxes and avoids redundant predictions.
Considering factors that affect AF detection performance during model training is crucial, as it determines whether the trained model can generalize to unseen individuals. Employing appropriate strategies can enhance model performance. Firstly, the diversity of samples in the training set significantly impacts model performance. Typically, the number of participants in the dataset is directly related to sample diversity, as ECG samples from the same participant over a short period tend to be highly similar. Data augmentation methods are often used to enhance sample diversity. Secondly, while considering sample diversity, the balance of samples should not be overlooked. Imbalanced samples can reduce the model ability to recognize minority classes. However, its effects can be mitigated by sampling strategies or special loss functions. Additionally, differences between training and testing data in terms of lead configurations can lead to prediction failures. Therefore, in this work, we applied a polarity inversion (vertical flipping) augmentation to emulate physiological polarity differences between leads. As verified by the ablation in
Section 4.3.3, the augmentation yields negligible impact on F-wave visibility while slightly improving cross-lead generalization. Future work will further incorporate lead-aware augmentations such as baseline wander, narrowband interference, and electrode noise to better reflect real-world ambulatory artifacts
Although the AF-DETR framework uses a fixed 400 ms heartbeat-box width, the additional ablation demonstrated that the performance is largely insensitive to moderate variations in box size. The adaptive-RR configuration provided only minimal gains, indicating that the empirical width adequately captures the P-QRS-T morphology under both bradycardic and tachycardic conditions. Therefore, the fixed-width design offers an effective trade-off between physiological coverage and computational simplicity.
Cross-database evaluation further confirmed that AF-DETR generalizes robustly across datasets with differing sampling rates and labeling styles, maintaining consistent performance within ±0.7% AF F1.
The additional analysis on MITDB demonstrated that premature atrial and ventricular ectopy are the dominant causes of false-positive AF detections. The use of focal loss or class-weighted cross-entropy significantly improved both precision and probability calibration, suggesting that ectopy-aware training objectives could further enhance model reliability in future studies.
The sensitivity and time-consecutive analyses verified that the 50% segment-level criterion approximates the clinical 30 s AF definition with high fidelity, confirming that the beat-level predictions of AF-DETR possess adequate temporal continuity for clinical interpretation.
5.3. Limitation and Future Work
Although there are many benefits, this study still has certain limitations. Firstly, the AF-DETR model did not consider the impact of different levels of noise on its performance. Another limitation is that there is still room for improvement in the positioning performance of the model, because the positioning performance affects the classification performance of the heartbeat. Lastly, the model still needs validation in more realistic clinical settings. In the future, we plan to validate our proposed AF-DETR method in clinical practice at hospitals, and expand datasets with diverse arrhythmia data collected from different environments for training and evaluation to achieve higher accuracy in AF heartbeat detection. Additionally, we aim to investigate its interpretability.