LG-UNet Based Segmentation and Survival Prediction of Nasopharyngeal Carcinoma Using Multimodal MRI Imaging
Abstract
1. Introduction
2. Materials and Methods
2.1. Network Encoder
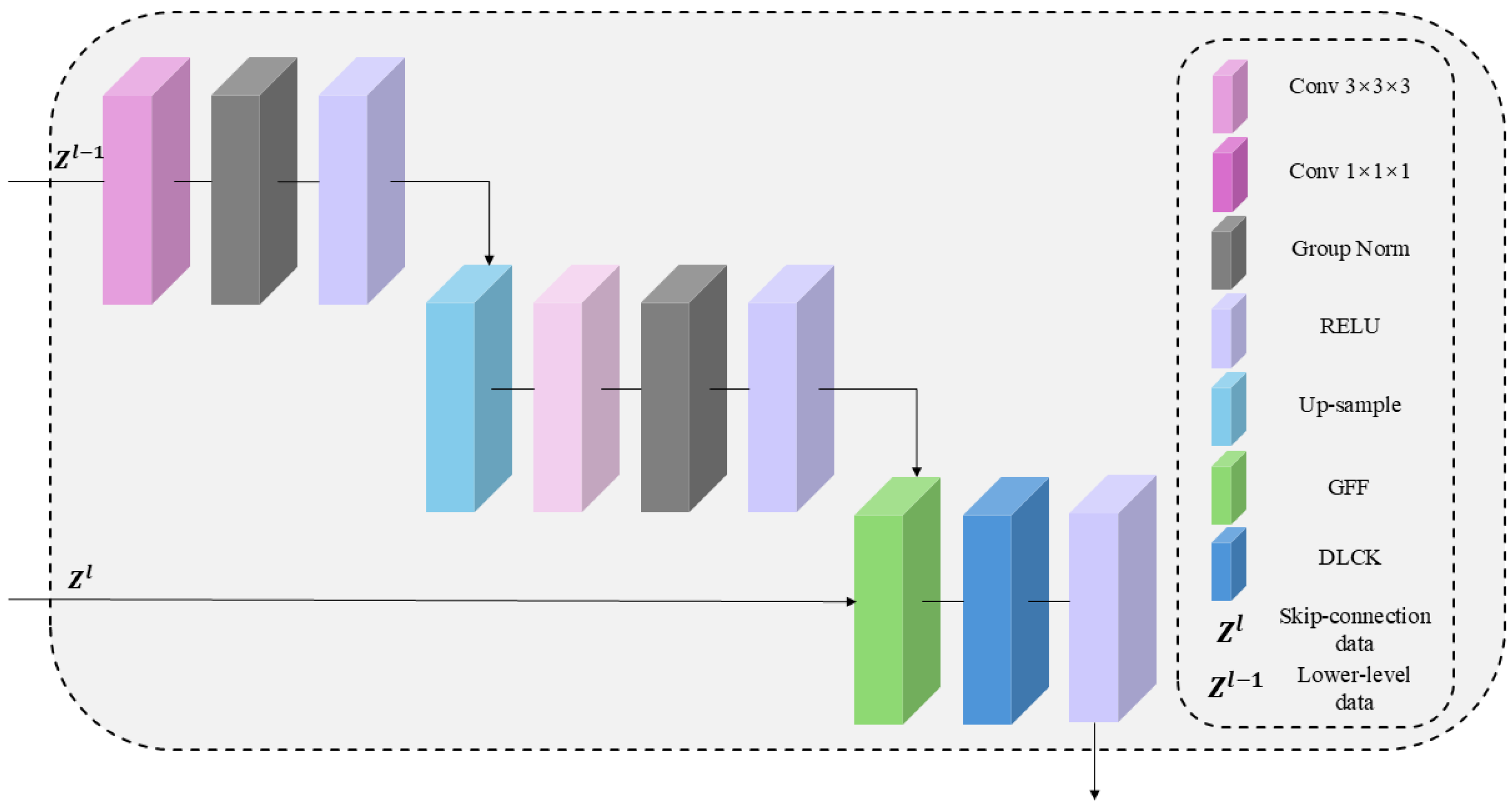
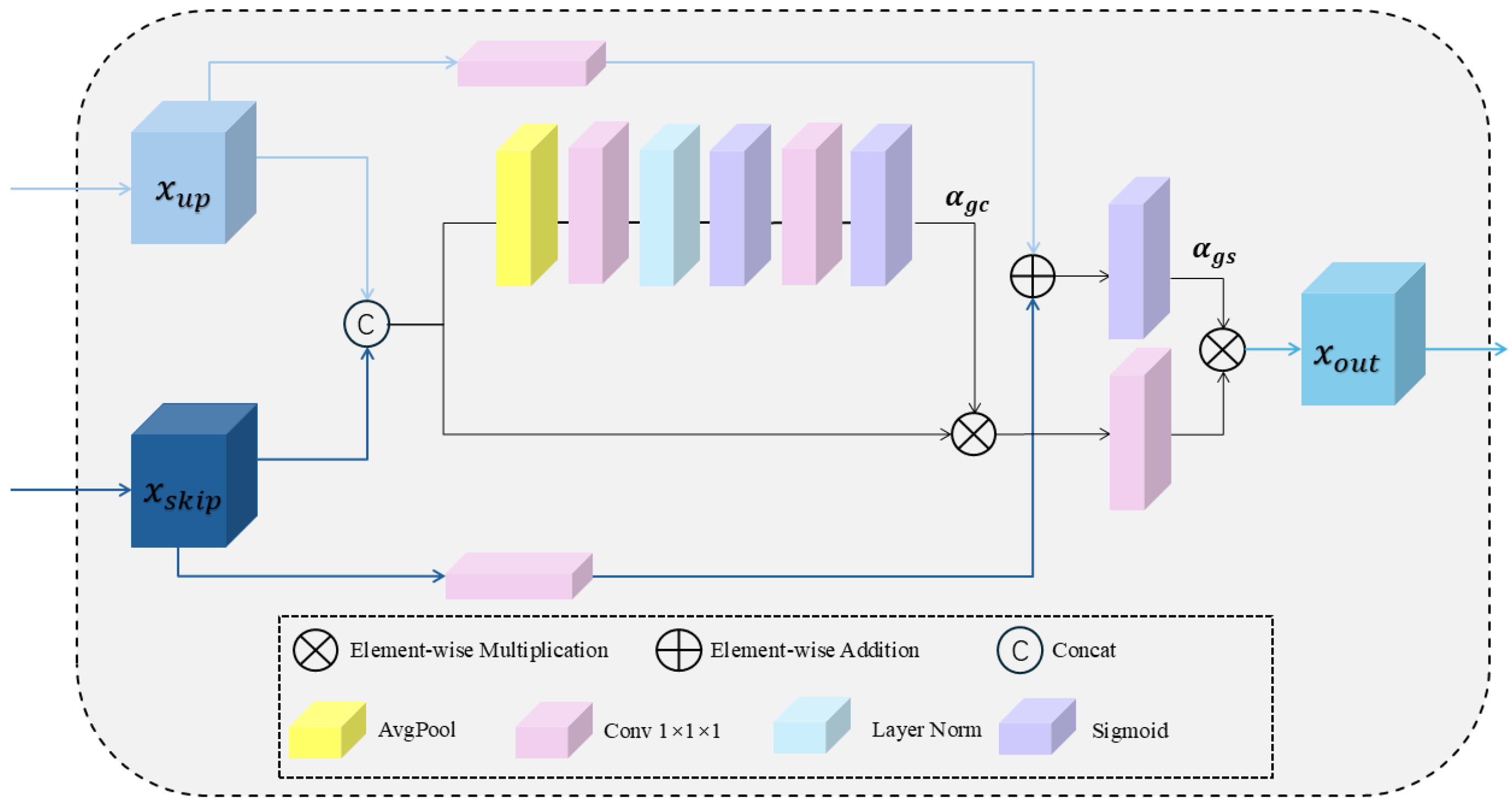
2.2. Network Decoder
2.3. The Architecture of the GDU Module
2.3.1. The Overview of GFF Module
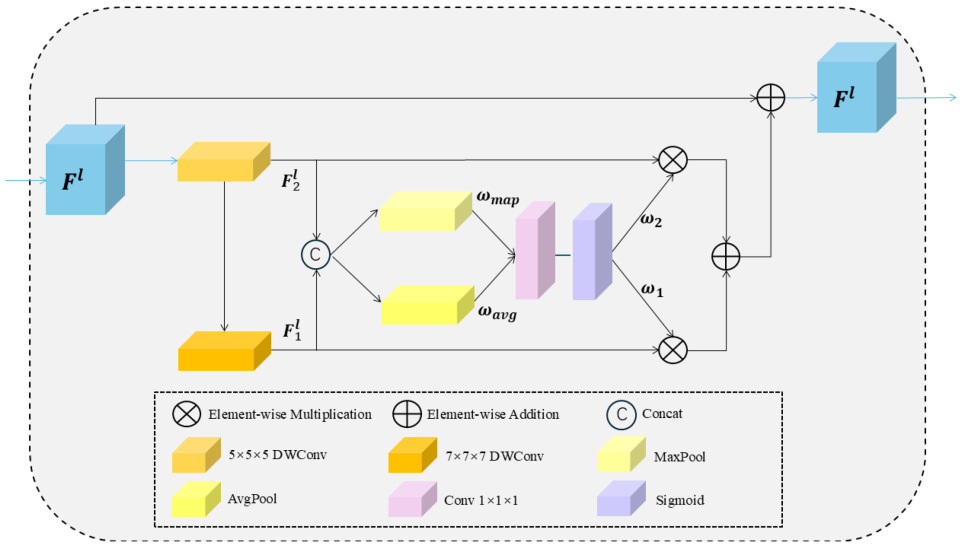
2.3.2. The Overview of DLCK Block
2.4. Survival Prediction Task
3. Data
3.1. Image Segmentation Task Data
3.2. Survival Prediction Task Data
Survival Prediction Data Preprocessing
3.3. Performancce Metrics
3.3.1. Segmentation Task Evaluation Metrics
3.3.2. Survival Prediction Task Evaluation Metrics
3.4. Implementation Details
4. Results
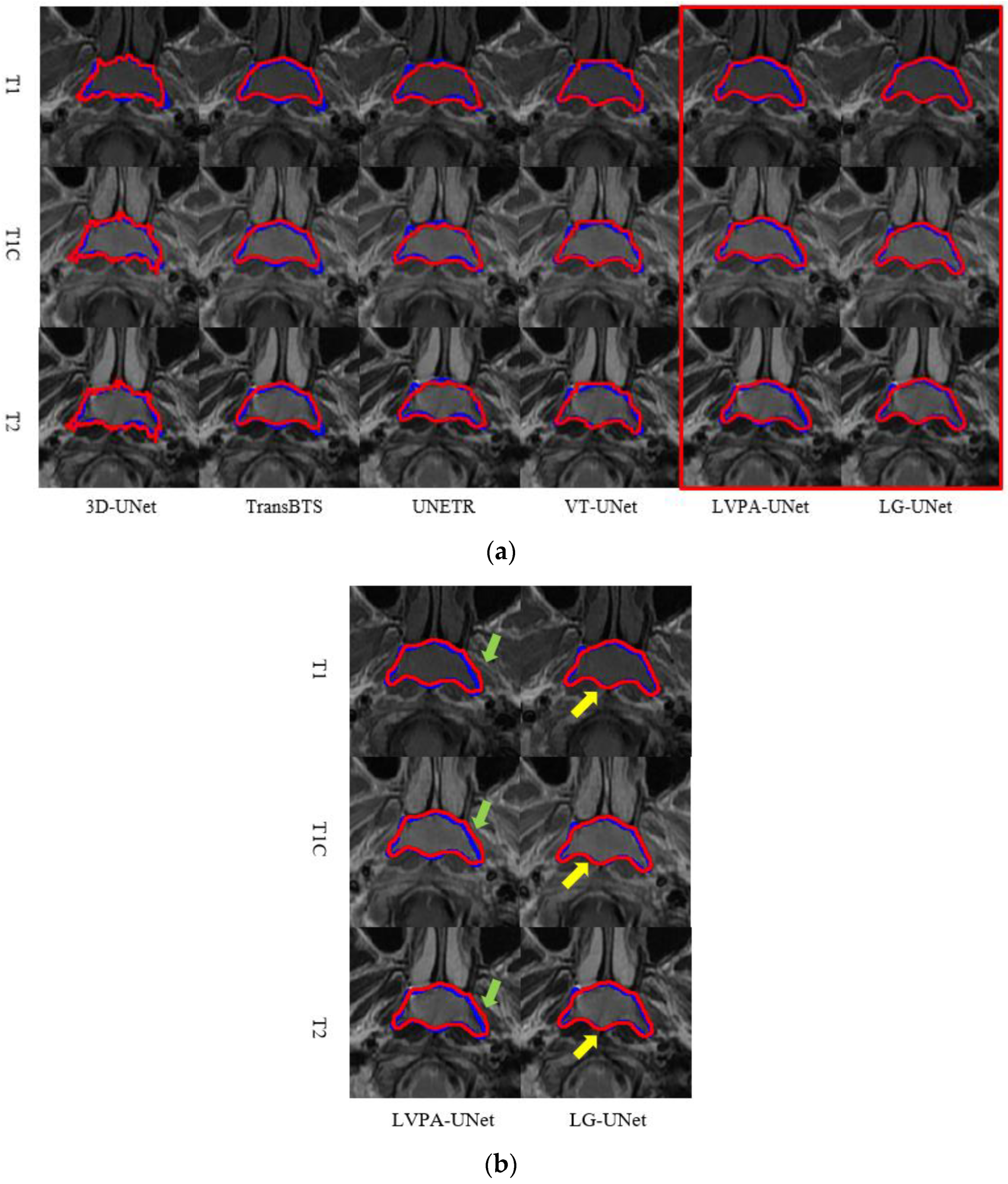
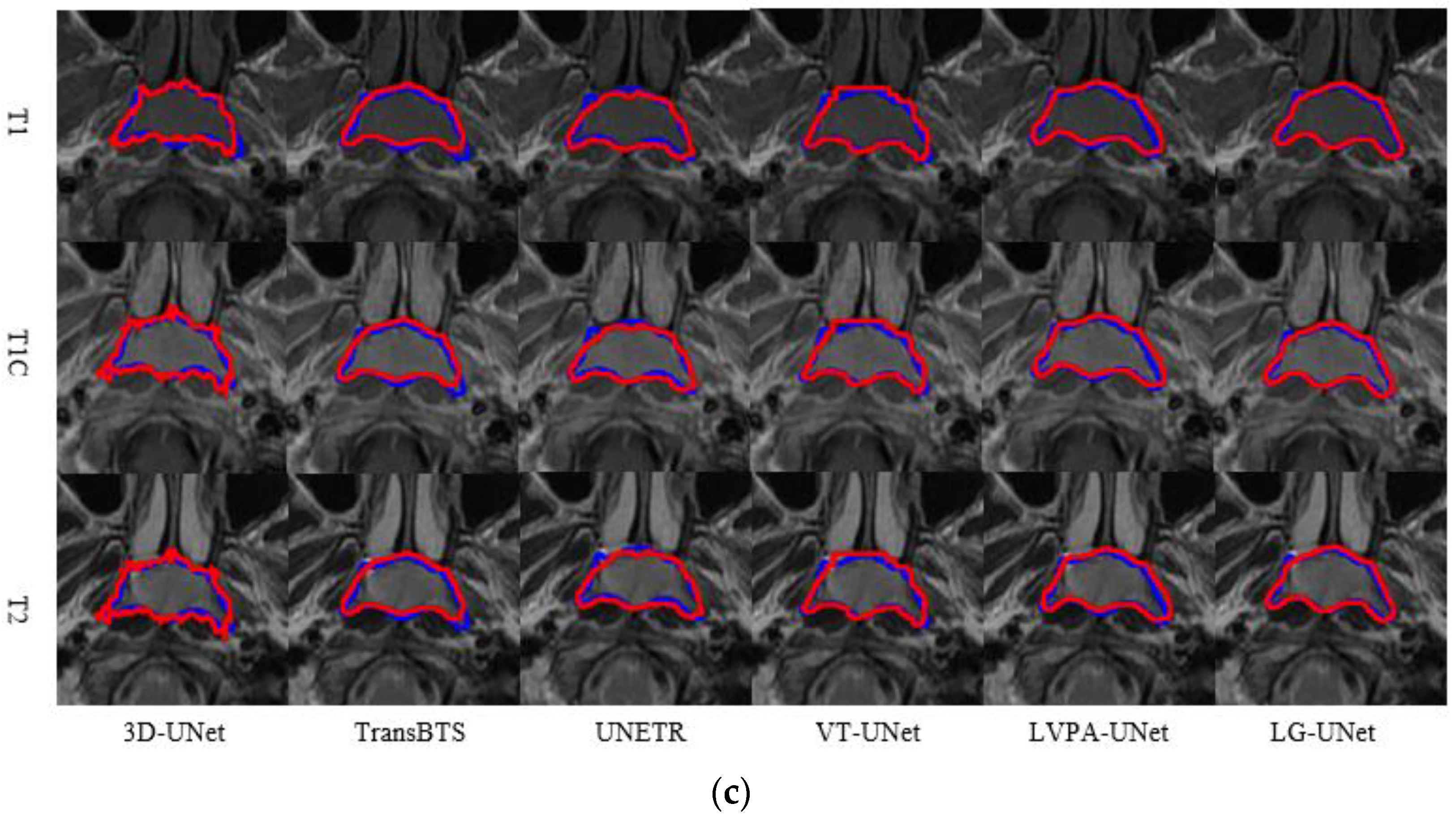
4.1. Analysis of Segmentation Experiment Results
4.1.1. Comparison of Model Experiments
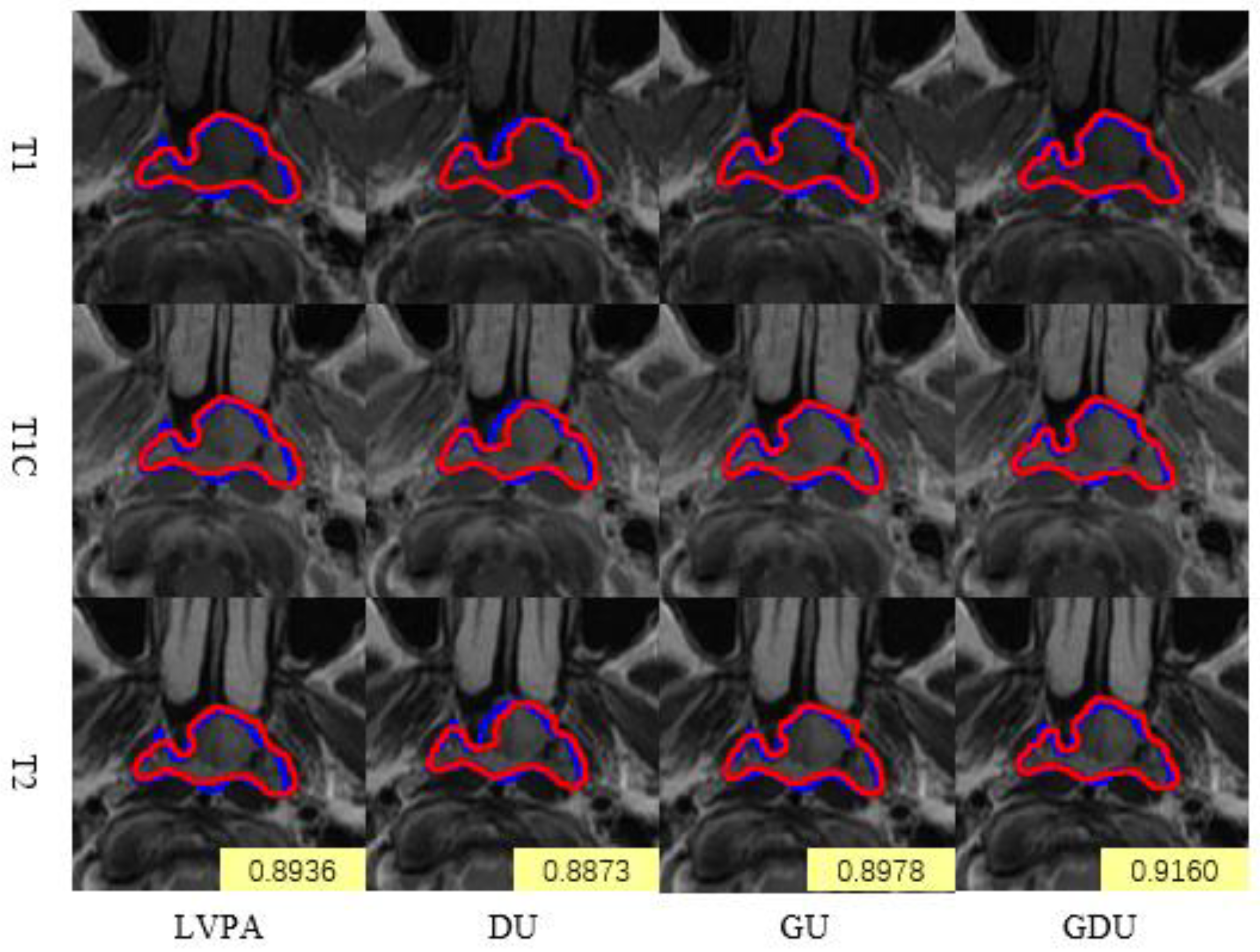
4.1.2. Comparison of Module Ablation Experiments
4.1.3. Comparison of Multimodal and Single-Modality Inputs
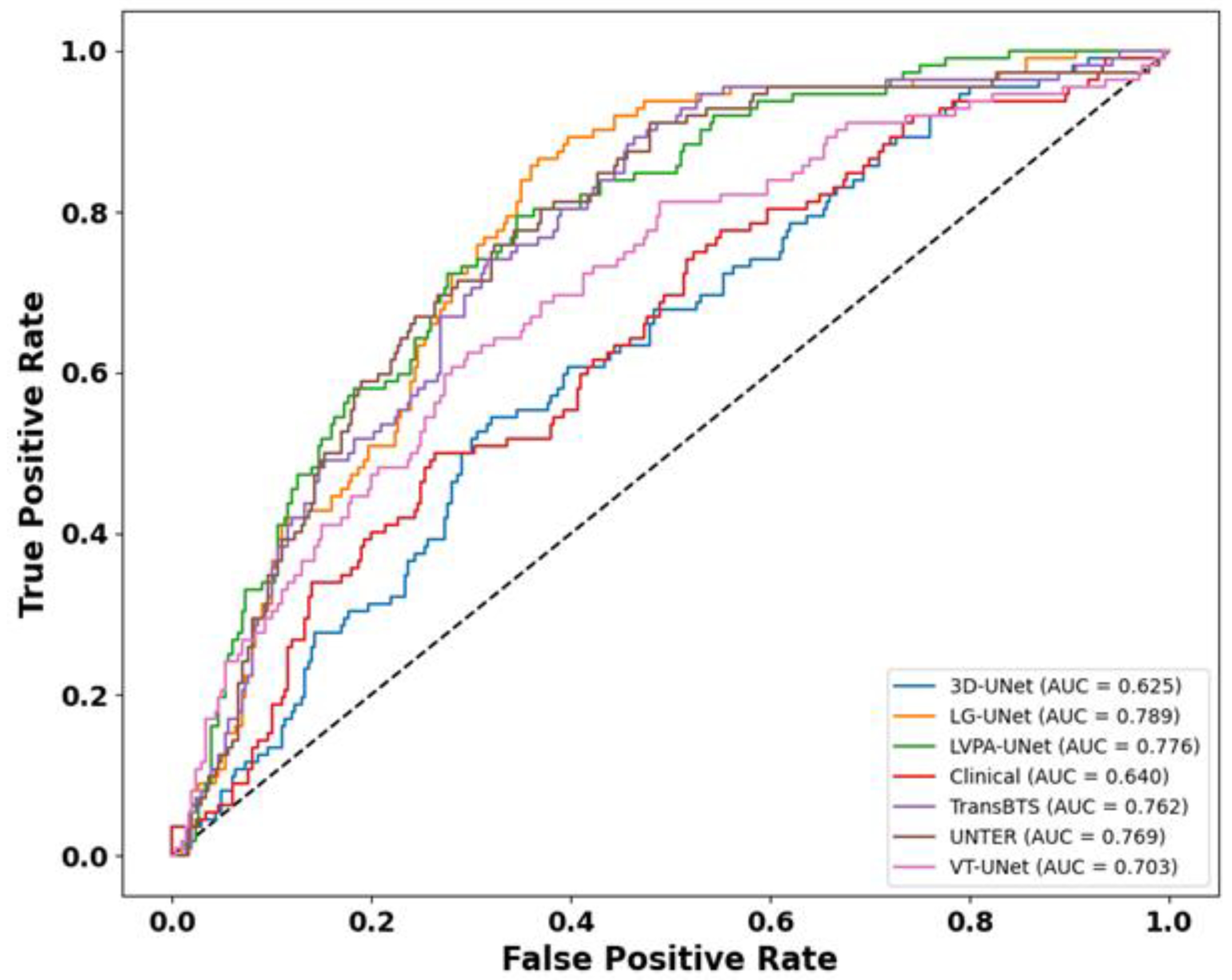
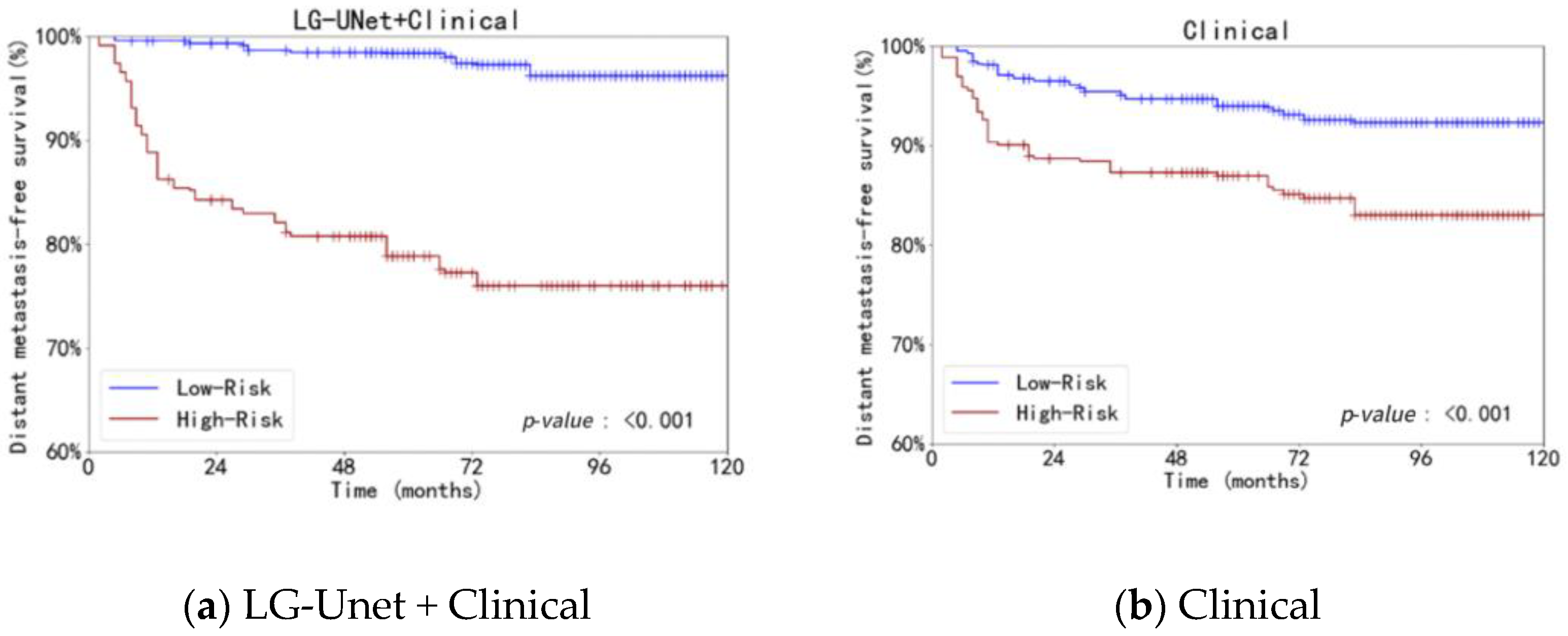
4.2. Analysis of Survival Prediction Experiment Results
5. Discussion
6. Conclusions
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| LVPA | Layer Volume Parallel Attention |
| CNN | Convolutional Neural Network |
| NPC | Nasopharyngeal Carcinoma |
| AJCC/UICC | American Joint Committee on Cancer/Union for International Cancer Control |
| MRI | Magnetic Resonance Imaging |
| DMFS | Distant Metastasis Free Survival |
| LDH | Lactate Dehydrogenase |
| EBV DNA | Epstein–Barr Virus Deoxyribonucleic Acid |
| DSC | Dice Similarity Coefficient |
| C-Index | Concordance Index |
| TD-ROC | Time-Dependent Receiver Operating Characteristic |
| DLCK | Dynamic Large Convolutional Kernel |
| GFF | Global Feature Fusion |
| GELU | Gaussian Error Linear Unit |
| DWConv | Depthwise Convolution |
Appendix A
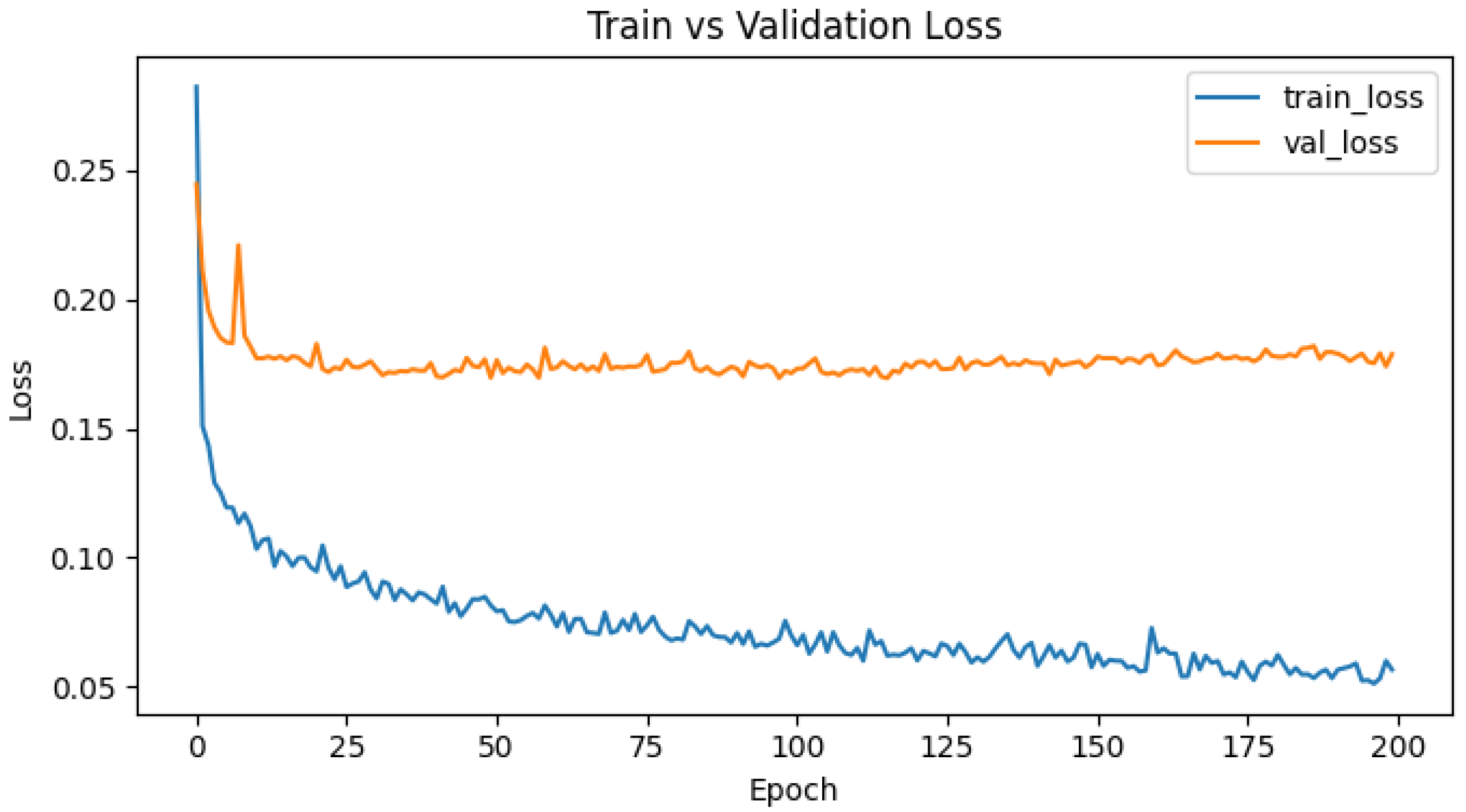
References
- De Martel, C.; Georges, D.; Bray, F.; Ferlay, J.; Clifford, G.M. Global burden of cancer attributable to infections in 2018: A worldwide incidence analysis. Lancet Glob. Health 2020, 8, e180–e190. [Google Scholar] [CrossRef]
- Chen, Y.-P.; Chan, A.T.C.; Le, Q.-T.; Blanchard, P.; Sun, Y.; Ma, J. Nasopharyngeal carcinoma. Lancet 2019, 394, 64–80. [Google Scholar] [CrossRef]
- Isensee, F.; Jaeger, P.F.; Kohl, S.A.A.; Petersen, J.; Maier-Hein, K.H. nnU-Net: A self-configuring method for deep learning-based biomedical image segmentation. Nat. Methods 2021, 18, 203–211. [Google Scholar] [CrossRef]
- Tang, P.; Zu, C.; Hong, M.; Yan, R.; Peng, X.; Xiao, J.; Wu, X.; Zhou, J.; Zhou, L.; Wang, Y. Da-dsunet: Dual attention-based dense su-net for automatic head-and-neck tumor segmentation in mri images. Neurocomputing 2021, 435, 103–113. [Google Scholar] [CrossRef]
- Huang, H.; Lin, L.; Tong, R.; Hu, H.; Zhang, Q.; Iwamoto, Y.; Han, X.; Chen, Y.-W.; Wu, J. UNet 3+: A Full-Scale Connected UNet for Medical Image Segmentation. In Proceedings of the ICASSP 2020—2020 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Barcelona, Spain, 4–8 May 2020; pp. 1055–1059. [Google Scholar] [CrossRef]
- Chen, L.-C.; Zhu, Y.; Papandreou, G.; Schroff, F.; Adam, H. Encoder-Decoder with Atrous Separable Convolution for Semantic Image Segmentation. arXiv 2018. [Google Scholar] [CrossRef]
- Qin, X.; Zhang, Z.; Huang, C.; Dehghan, M.; Zaiane, O.R.; Jagersand, M. U2-Net: Going deeper with nested U-structure for salient object detection. Pattern Recognit. 2020, 106, 107404. [Google Scholar] [CrossRef]
- Li, Y.; Dan, T.; Li, H.; Chen, J.; Peng, H.; Liu, L.; Cai, H. Npcnet: Jointly segment primary nasopharyngeal carcinoma tumors and metastatic lymph nodes in mr images. IEEE Trans. Med. Imaging 2022, 41, 1639–1650. [Google Scholar] [CrossRef] [PubMed]
- Hatamizadeh, A.; Nath, V.; Tang, Y.; Yang, D.; Roth, H.R.; Xu, D. Swin unetr: Swin transformers for semantic segmentation of brain tumors in mri images. In International MICCAI Brainlesion Workshop; Springer: Berlin/Heidelberg, Germany, 2021; pp. 272–284. [Google Scholar]
- Hatamizadeh, A.; Xu, Z.; Yang, D.; Li, W.; Roth, H.; Xu, D. UNetFormer: A Unified Vision Transformer Model and Pre-Training Framework for 3D Medical Image Segmentation. arXiv 2022. [Google Scholar] [CrossRef]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An Image is Worth 16x16 Words: Transformers for Image Recognition at Scale. arXiv 2021, arXiv:2010.11929. [Google Scholar] [CrossRef]
- Chen, J.; Lu, Y.; Yu, Q.; Luo, X.; Adeli, E.; Wang, Y.; Lu, L.; Yuille, A.L.; Zhou, Y. TransUNet: Transformers Make Strong Encoders for Medical Image Segmentation. arXiv 2021. [Google Scholar] [CrossRef]
- Wang, W.; Chen, C.; Ding, M.; Yu, H.; Zha, S.; Li, J. Transbts: Multimodal Brain Tumor Segmentation Using Transformer; Springer: Berlin/Heidelberg, Germany, 2021. [Google Scholar]
- Chen, S.; Luo, C.; Liu, S.; Li, H.; Liu, Y.; Zhou, H.; Liu, L.; Chen, H. Ld-unet: A long-distance perceptual model for segmentation of blurred boundaries in medical images. Comput. Biol. Med. 2024, 171, 108120. [Google Scholar] [CrossRef] [PubMed]
- Chowdary, G.J.A.Y. Diffusion transformer u-net for medical image segmentation. In Proceedings of the 26th International Conference, Vancouver, BC, Canada, 8–12 October 2023. [Google Scholar]
- Azad, R.; Niggemeier, L.; Hüttemann, M.; Kazerouni, A.; Aghdam, E.K.; Velichko, Y.; Bagci, U.; Merhof, D. Beyond self-attention: Deformable large kernel attention for medical image segmentation. In Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision, Waikoloa, HI, USA, 3–8 January 2024; pp. 1287–1297. [Google Scholar]
- Lee, H.H.; Bao, S.; Huo, Y.; Landman, B.A. 3d ux-net: A large kernel volumetric convnet modernizing hierarchical transformer for medical image segmentation. arXiv 2022, arXiv:2209.15076. [Google Scholar]
- Ding, X.; Zhang, X.; Zhou, Y.; Han, J.; Ding, G.; Sun, J. Scaling Up Your Kernels to 31x31: Revisiting Large Kernel Design in CNNs. arXiv 2022. [Google Scholar] [CrossRef]
- Chen, H.; Chu, X.; Ren, Y.; Zhao, X.; Huang, K. PeLK: Parameter-efficient Large Kernel ConvNets with Peripheral Convolution. arXiv 2024. [Google Scholar] [CrossRef]
- Ding, X. UniRepLKNet: A Universal Perception Large-Kernel ConvNet for Audio, Video, Point Cloud, Time-Series and Image Recognition. arXiv 2023, arXiv:2311.15599. [Google Scholar] [CrossRef]
- Saeed, N.; Sobirov, I.; Al Majzoub, R.; Yaqub, M. TMSS: An End-to-End Transformer-Based Multimodal Network for Segmentation and Survival Prediction. In Medical Image Computing and Computer Assisted Intervention—MICCAI 2022. MICCAI 2022; Wang, L., Dou, Q., Fletcher, P.T., Speidel, S., Li, S., Eds.; Lecture Notes in Computer Science; Springer: Cham, Switzerland, 2022; Volume 13437. [Google Scholar] [CrossRef]
- Liu, Z.; Mao, H.; Wu, C.Y.; Feichtenhofer, C.; Darrell, T.; Xie, S. A convnet for the 2020s. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, New Orleans, LA, USA, 18–24 June 2022; pp. 11976–11986. [Google Scholar]
- Wang, W.; Li, S.; Shao, J.; Jumahong, H. LKC-Net: Large kernel convolution object detection network. Sci. Rep. 2023, 13, 9535. [Google Scholar] [CrossRef]
- Cai, J.; Tang, Y.; Lu, L.; Harrison, A.P.; Yan, K.; Xiao, J.; Yang, L.; Summers, R.M. Accurate weakly-supervised deep lesion segmentation using large-scale clinical annotations: Slice-propagated 3d mask generation from 2d recist. In Proceedings of the Medical Image Computing and Computer Assisted Intervention–MICCAI 2018: 21st International Conference, Granada, Spain, 16–20 September 2018; Proceedings, Part IV 11. Springer: Berlin/Heidelberg, Germany, 2018; pp. 396–404. [Google Scholar]
- Poudel, R.P.K.; Lamata, P.; Montana, G. Recurrent fully convolutional neural networks for multi-slice MRI cardiac segmentation. In Proceedings of the Reconstruction, Segmentation, and Analysis of Medical Images: First International Workshops, RAMBO 2016 and HVSMR 2016, Held in Conjunction with MICCAI 2016, Athens, Greece, 17 October 2016; Revised Selected Papers 1. Springer: Berlin/Heidelberg, Germany, 2017; pp. 83–94. [Google Scholar]
- Li, J.; Chen, J.; Tang, Y.; Wang, C.; Landman, B.A.; Zhou, S.K. Transforming medical imaging with Transformers? A comparative review of key properties, current progresses, and future perspectives. Med. Image Anal. 2023, 85, 102762. [Google Scholar] [CrossRef]
- Bilic, P.; Christ, P.; Li, H.B.; Vorontsov, E.; Ben-Cohen, A.; Kaissis, G.; Szeskin, A.; Jacobs, C.; Mamani, G.E.H.; Chartrand, G.; et al. The liver tumor segmentation benchmark (lits). Med. Image Anal. 2023, 84, 102680. [Google Scholar] [CrossRef] [PubMed]
- Xia, F.; Peng, Y.; Wang, J.; Chen, X. A 2.5D multi-path fusion network framework with focusing on z-axis 3D joint for medical image segmentation. Biomed. Signal Process. Control. 2024, 91, 106049. [Google Scholar] [CrossRef]
- Zhao, J.; Xing, Z.; Chen, Z.; Wan, L.; Han, T.; Fu, H.; Zhu, L. Uncertainty-Aware Multi-Dimensional Mutual Learning for Brain and Brain Tumor Segmentation. IEEE J. Biomed. Health Inform. 2023, 27, 43624372. [Google Scholar] [CrossRef]
- Zhang, Y.; Xu, H.R.; Wen, J.H.; Hu, Y.J.; Diao, Y.L.; Chen, J.L.; Xia, Y.F. A novel LVPA-UNet network for target volume automatic delineation: An MRI case study of nasopharyngeal carcinoma. Heliyon 2024, 10, e30763. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Yang, J.; Qiu, P.; Zhang, Y.; Marcus, D.S.; Sotiras, A. D-Net: Dynamic Large Kernel with Dynamic Feature Fusion for Volumetric Medical Image Segmentation. arXiv 2024, arXiv:2403.10674. [Google Scholar] [CrossRef]
- Zhang, L.; Wu, X.; Liu, J.; Zhang, B.; Mo, X.; Chen, Q.; Fang, J.; Wang, F.; Li, M.; Chen, Z.; et al. Mri-based deep-learning model for distant metastasis-free survival in locoregionally advanced nasopharyngeal carcinoma. J. Magn. Reson. Imaging 2021, 53, 167–178. [Google Scholar] [CrossRef]
- Huang, S.H.; O’Sullivan, B. Overview of the 8th edition TNM classification for head and neck cancer. Curr. Treat. Options Oncol. 2017, 18, 40. [Google Scholar] [CrossRef]
- Hui, E.; Li, W.; Ma, B.; Lam, W.; Chan, K.; Mo, F.; Ai, Q.; King, A.; Wong, C.; Guo, R.; et al. Integrating postradiotherapy plasma epstein-barr virus DNA and TNM stage for risk stratification of nasopharyngeal carcinoma to adjuvant therapy. Ann. Oncol. 2020, 31, 769–779. [Google Scholar] [CrossRef]
- Demirjian, N.L.; Varghese, B.A.; Cen, S.Y.; Hwang, D.H.; Aron, M.; Siddiqui, I.; Fields, B.K.K.; Lei, X.; Yap, F.Y.; Rivas, M.; et al. CT-based radiomics stratification of tumor grade and TNM stage of clear cell renal cell carcinoma. Eur. Radiol. 2022, 32, 2552–2563. [Google Scholar] [CrossRef] [PubMed]
- Gu, B.; Meng, M.; Xu, M.; Feng, D.D.; Bi, L.; Kim, J.; Song, S. Multi-task deep learning-based radiomic nomogram for prognostic prediction in locoregionally advanced nasopharyngeal carcinoma. Eur. J. Nucl. Med. Mol. Imaging 2023, 50, 3996–4009. [Google Scholar] [CrossRef]
- Delfan, N.; Abrishami Moghaddam, H.; Modaresi, M.; Afshari, K.; Nezamabadi, K.; Pak, N.; Ghaemi, O.; Forouzanfar, M. CT-LungNet: A Deep Learning Framework for Precise Lung Tissue Segmentation in 3D Thoracic CT Scans. arXiv 2022, arXiv:2212.13971. [Google Scholar] [CrossRef]
- Tibshirani, R. The lasso method for variable selection in the cox model. Stat. Med. 1997, 16, 38595. [Google Scholar] [CrossRef]
- Gu, A.; Dao, T. Mamba: Linear-time sequence modeling with selective state spaces. arXiv 2023, arXiv:2312.00752. [Google Scholar] [CrossRef]
- Zhang, H.; Zhu, Y.; Wang, D.; Zhang, L.; Chen, T.; Wang, Z.; Ye, Z. A survey on visual mamba. Appl. Sci. 2024, 14, 5683. [Google Scholar] [CrossRef]
- Xu, R.; Yang, S.; Wang, Y.; Du, B.; Chen, H. A survey on vision mamba: Models, applications and challenges. arXiv 2024, arXiv:2404.18861. [Google Scholar] [CrossRef]
- Liu, X.; Zhang, C.; Zhang, L. Vision mamba: A comprehensive survey and taxonomy. arXiv 2024, arXiv:2405.04404. [Google Scholar] [CrossRef]
- Li, J.; Wang, W.; Chen, C.; Zhang, T.; Zha, S.; Wang, J.; Yu, H. Transbtsv2: Towards better and more efficient volumetric segmentation of medical images. arXiv 2022, arXiv:2201.12785. [Google Scholar] [CrossRef]
- Zhang, Y.; Liao, Q.; Ding, L.; Zhang, J. Bridging 2d and 3d segmentation networks for computation-efficient volumetric medical image segmentation: An empirical study of 2.5 d solutions. Comput. Med. Imaging Graph. 2022, 99, 102088. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Lin, Z.; Zhou, Q.; Peng, X.; Xiao, J.; Zu, C.; Jiao, Z.; Wang, Y. Multi-Transsp: Multimodal Transformer for Survival Prediction of Nasopharyngeal Carcinoma Patients; Springer: Berlin/Heidelberg, Germany, 2022. [Google Scholar]


| Characteristics | Dataset (n = 388) |
|---|---|
| Age (years), median (IQR) 6 | 44 (38.51) |
| Sex, No (%) | |
| Male | 271 (69.9) |
| Female | 117 (30.1) |
| Histological type (WHO) 6, No (%) | |
| WHO I | 0 |
| WHO II | 20 (5.2) |
| WHO III | 368 (94.8) |
| T a, No (%) | |
| T1 | 197 (50.8) |
| T2 | 191 (49.2) |
| N a, No (%) | |
| N0 | 48 (12.4) |
| N1 | 340 (87.6) |
| LDH b 6, No (%) | |
| Normal | 369 (95.1) |
| Abnormal | 19 (4.9) |
| Unknown | 0 |
| EBV DNA c 6, No (%) | |
| Undetectable | 208 (53.6) |
| Detectable | 161 (41.5) |
| Unknown | 19 (4.9) |
| DMFS 6, No (%) | |
| Metastasis-free | 353 (91.0) |
| Metastasis | 35 (9.0) |
| Models | DSC (%) ↑ | HD95 (mm) ↓ | Precision (%) ↑ | Recall (%) ↑ |
|---|---|---|---|---|
| 3D UNet | 76.31 ± 7.29 | 2.26 ± 1.41 | 76.88 ± 11.05 | 77.68 ± 10.74 |
| TransBTS | 77.85 ± 6.40 | 2.45 ± 1.92 | 78.45 ± 9.01 | 78.65 ± 10.10 |
| UNETR | 78.76 ± 6.87 | 2.19 ± 1.54 | 77.94 ± 8.84 | 79.71 ± 8.43 |
| VT-UNet | 78.67 ± 7.83 | 2.04 ± 2.13 | 78.36 ± 10.48 | 80.52 ± 10.49 |
| LVPA-UNet | 80.09 ± 7.25 | 1.76 ± 1.02 | 79.06 ± 10.46 | 82.29 ± 8.73 |
| LG-UNet | 82.22 ± 6.47 | 1.68 ± 0.97 | 82.36 ± 7.96 | 82.97 ± 9.08 |
| Models | DSC (%) ↑ | HD95 (mm) ↓ | Precision (%) ↑ | Recall (%) ↑ |
|---|---|---|---|---|
| LVPA-UNet (U) | 80.09 ± 7.25 | 1.76 ± 1.02 | 79.06 ± 10.46 | 82.29 ± 8.73 |
| DU | 79.31 ± 6.83 | 2.11 ± 2.55 | 79.02 ± 10.78 | 80.65 ± 10.51 |
| GU | 80.42 ± 6.89 | 1.75 ± 0.96 | 81.04 ± 9.81 | 81.01 ± 9.35 |
| GDU | 82.22 ± 6.47 | 1.68 ± 0.97 | 82.36 ± 7.96 | 82.97 ± 9.08 |
| Input Channels | DSC (%) ↑ | HD95 (mm) ↓ | Precision (%) ↑ | Recall (%) ↑ |
|---|---|---|---|---|
| T1 | 77.18 ± 6.31 | 1.89 ± 1.02 | 79.05 ± 7.10 | 80.21 ± 6.55 |
| T1C | 80.05 ± 6.55 | 1.81 ± 0.96 | 80.10 ± 7.51 | 81.33 ± 7.48 |
| T2 | 78.76 ± 6.71 | 1.93 ± 1.13 | 78.60 ± 6.68 | 79.94 ± 6.61 |
| T1 + T1C + T2 | 82.22 ± 6.47 | 1.68 ± 0.97 | 82.36 ± 7.96 | 82.97 ± 9.08 |
| Models | AUC (95%CI) | C-Index (95%CI) |
|---|---|---|
| CoxPH (3D UNet + Clinical) | 0.625 (0.571–0.679) | 0.584 (0.528–0.641) |
| CoxPH (TransBTS + Clinical) | 0.762 (0.715–0.807) | 0.726 (0.678–0.775) |
| CoxPH (UNETR + Clinical) | 0.769 (0.725–0.812) | 0.716 (0.667–0.784) |
| CoxPH (VT-UNet + Clinical) | 0.703 (0.652–0.753) | 0.694 (0.673–0.762) |
| CoxPH (LVPA-UNet + Clinical) | 0.776 (0.728–0.820) | 0.734 (0.687–0.781) |
| CoxPH (LG-UNet + Clinical) | 0.789 (0.746–0.829) | 0.756 (0.699–0.804) |
| CoxPH (Clinical) | 0.640 (0.589–0.694) | 0.636 (0.577–0.684) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, Y.; Wen, J.; Wu, T.; Dong, J.; Xia, Y.; Zhang, Y. LG-UNet Based Segmentation and Survival Prediction of Nasopharyngeal Carcinoma Using Multimodal MRI Imaging. Bioengineering 2025, 12, 1051. https://doi.org/10.3390/bioengineering12101051
Yang Y, Wen J, Wu T, Dong J, Xia Y, Zhang Y. LG-UNet Based Segmentation and Survival Prediction of Nasopharyngeal Carcinoma Using Multimodal MRI Imaging. Bioengineering. 2025; 12(10):1051. https://doi.org/10.3390/bioengineering12101051
Chicago/Turabian StyleYang, Yuhao, Junhao Wen, Tianyi Wu, Jinrang Dong, Yunfei Xia, and Yu Zhang. 2025. "LG-UNet Based Segmentation and Survival Prediction of Nasopharyngeal Carcinoma Using Multimodal MRI Imaging" Bioengineering 12, no. 10: 1051. https://doi.org/10.3390/bioengineering12101051
APA StyleYang, Y., Wen, J., Wu, T., Dong, J., Xia, Y., & Zhang, Y. (2025). LG-UNet Based Segmentation and Survival Prediction of Nasopharyngeal Carcinoma Using Multimodal MRI Imaging. Bioengineering, 12(10), 1051. https://doi.org/10.3390/bioengineering12101051





