Ultrawidefield-to-Conventional Fundus Image Translation with Scaled Feature Registration and Distorted Vessel Correction
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Cohort
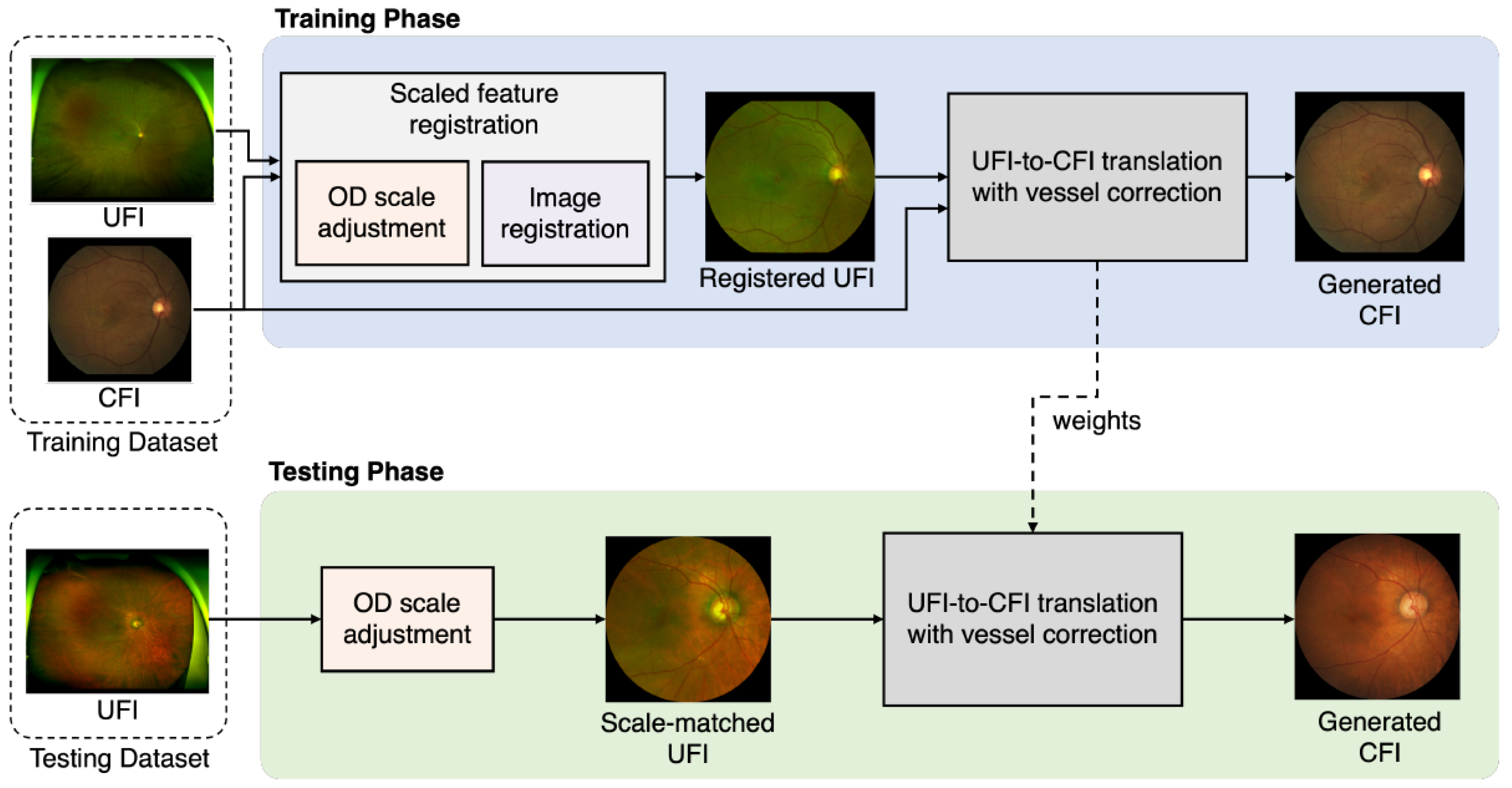
2.2. Overall Structure of the Proposed Method
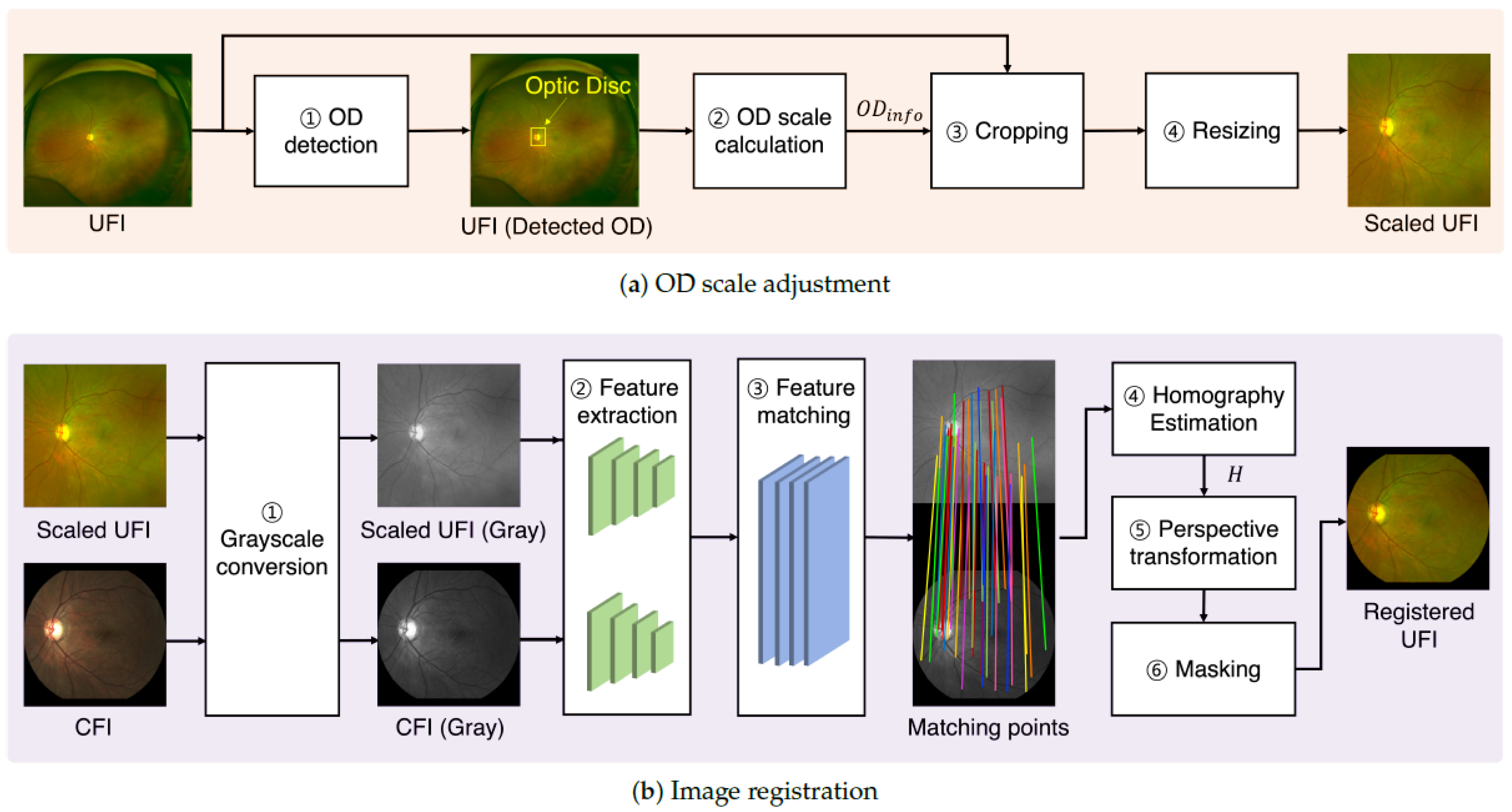
2.3. Scaled Feature Registration
2.4. UFI-to-CFI Translation with Distorted Vessel Correction Branch
2.5. Loss Functions
2.6. Training Models
3. Results
3.1. Evaluation of UFI-to-CFI Registration
3.2. Evaluation of UFI-to-CFI Translation
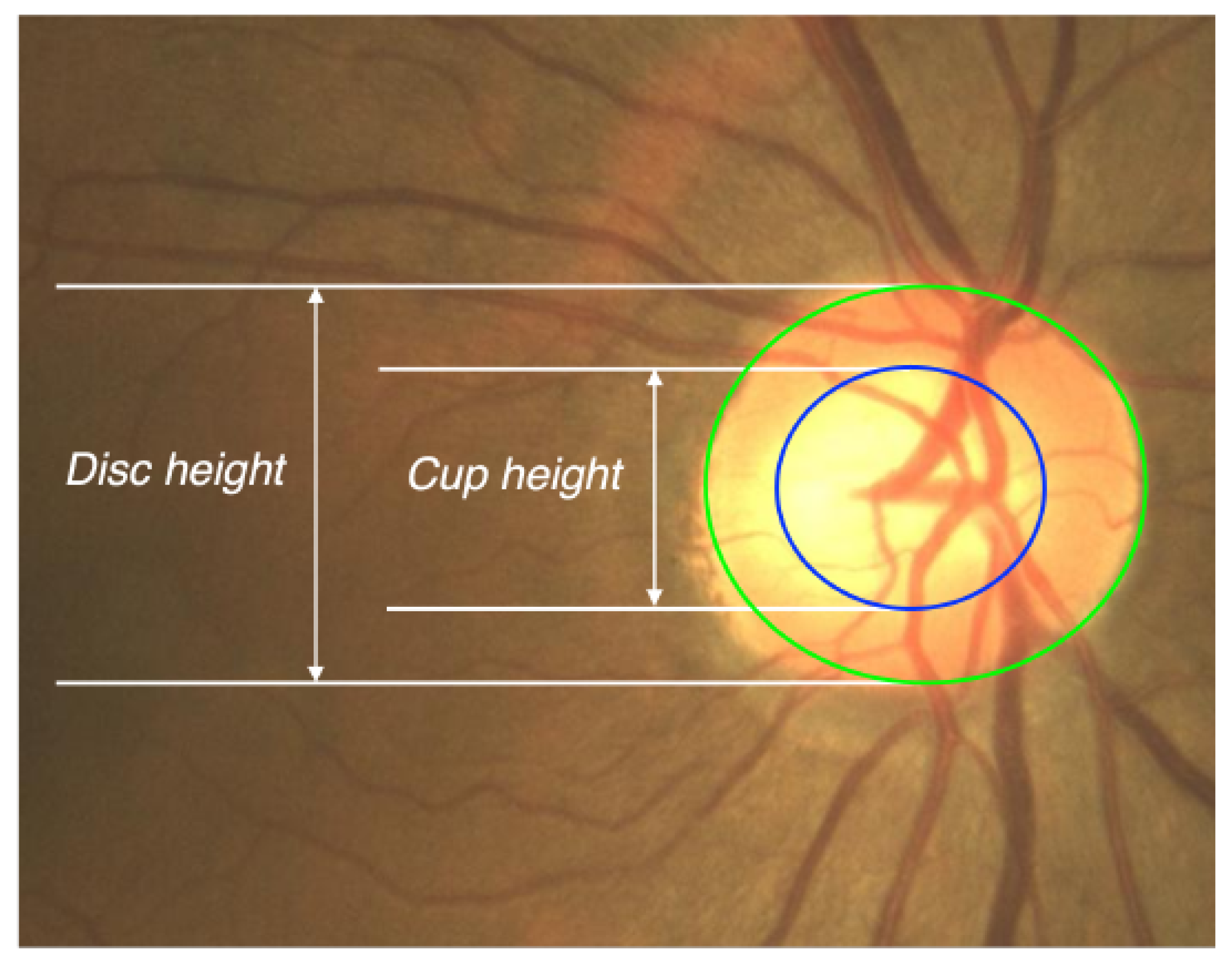
3.3. Qualitative Evaluation for Clinical Usefulness
3.4. Further Experiments
- Computational Performance. The model was trained on dual NVIDIA RTX A6000 GPUs, reaching a peak memory usage of approximately 72 GB (36 GB per GPU) with a batch size of 64. During inference, the translation step averaged 32 ms per image. The complete pipeline, including both scale adjustment and translation, averaged 52 ms per image, corresponding to a processing speed of approximately 19 frames per second (FPS). This performance demonstrates significant computational efficiency, supporting the model’s feasibility for near-real-time clinical applications.
- External Validataion. To address the limitations of relying on a single-center dataset, we conducted external validation experiments to assess the generalizability of our model. Specifically, we evaluated its zero-shot generalization capability using public UFI datasets (DeepDRiD, MSHF, and UWF-IQA [40,41,42]) and compared the Fréchet Inception Distance (FID) [43] between real CFIs and the generated CFIs. FID measures the distance between the feature distributions of real and generated images in the latent space of a pre-trained Inception network; lower values indicate closer alignment in terms of both image quality and diversity.
3.5. Ablation Study
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| CFI | Conventional Fundus Image |
| UFI | Ultrawidefield Fundus Image |
| OD | Optic Disc |
| AMD | Age-related Macular Degeneration |
| ERM | Epiretinal Membranes |
| CNN | Convolutional Neural Network |
| GNN | Graph-base Neural Network |
| cGAN | conditional Generative Adversarial Networks |
| CBAM | Convolutional Block Attention Module |
| PCK | Percentage of Correct Key points |
| MSE | Mean Squared Error |
| PSNR | Peak Signal-to-Noise Ratio |
| SSIM | Structural Similarity Index |
| MS-SSIM | Multiscale Structural Similarity Index |
References
- Yoo, T.K.; Ryu, I.H.; Kim, J.K.; Lee, I.S.; Kim, J.S.; Kim, H.K.; Choi, J.Y. Deep learning can generate traditional retinal fundus photographs using ultrawidefield images via generative adversarial networks. Comput. Methods Programs Biomed. 2020, 197, 105761. [Google Scholar] [CrossRef]
- Peng, Y.; Dharssi, S.; Chen, Q.; Keenan, T.D.; Agrón, E.; Wong, W.T.; Chew, E.Y.; Lu, Z. DeepSeeNet: A deep learning model for automated classification of patient-based age-related macular degeneration severity from color fundus photographs. Ophthalmology 2019, 126, 565–575. [Google Scholar] [CrossRef] [PubMed]
- Patel, S.N.; Shi, A.; Wibbelsman, T.D.; Klufas, M.A. Ultra-widefield retinal imaging: An update on recent advances. Ther. Adv. Ophthalmol. 2020, 12, 2515841419899495. [Google Scholar] [CrossRef] [PubMed]
- Nagiel, A.; Lalane, R.A.; Sadda, S.R.; Schwartz, S.D. Ultra-widefield fundus imaging: A review of clinical applications and future trends. Retina 2016, 36, 660–678. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.A.; Liu, P.; Cheng, J.; Fu, H. A deep step pattern representation for multimodal retinal image registration. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Seoul, Republic of Korea, 27 October–2 November 2019; pp. 5076–5085. [Google Scholar]
- Wang, T.C.; Liu, M.Y.; Zhu, J.Y.; Tao, A.; Kautz, J.; Catanzaro, B. High-resolution image synthesis and semantic manipulation with conditional GANs. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 8798–8807. [Google Scholar]
- Zhu, J.Y.; Park, T.; Isola, P.; Efros, A.A. Unpaired Image-to-Image Translation using Cycle-Consistent Adversarial Networks. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 2223–2232. [Google Scholar]
- Perez-Rovira, A.; Trucco, E.; Wilson, P.; Liu, J. Deformable registration of retinal fluorescein angiogram sequences using vasculature structures. In Proceedings of the 2010 Annual International Conference of the IEEE Engineering in Medicine and Biology, Buenos Aires, Argentina, 31 August–4 September 2010; pp. 4383–4386. [Google Scholar]
- Pham, V.-N.; Le, D.T.; Bum, J.; Lee, E.J.; Han, J.C.; Choo, H. Attention-aided generative learning for multi-scale multi-modal fundus image translation. IEEE Access 2023, 11, 51701–51711. [Google Scholar] [CrossRef]
- Zhang, Q.; Nie, Y.; Zheng, W.S. Dual illumination estimation for robust exposure correction. Comput. Graph. Forum 2019, 38, 243–252. [Google Scholar] [CrossRef]
- Woo, S.; Park, J.; Lee, J.Y.; Kweon, I.S. CBAM: Convolutional block attention module. In Proceedings of the European Conference on Computer Vision (ECCV), Munich, Germany, 8–14 September 2018; pp. 3–19. [Google Scholar]
- Ren, S.; He, K.; Girshick, R.; Sun, J. Faster R-CNN: Towards real-time object detection with region proposal networks. IEEE Trans. Pattern Anal. Mach. Intell. 2016, 39, 1137–1149. [Google Scholar] [CrossRef]
- Tu, H.; Wang, Z.; Zhao, Y. Unpaired image-to-image translation with diffusion adversarial network. Mathematics 2024, 12, 3178. [Google Scholar] [CrossRef]
- Go, S.; Ji, Y.; Park, S.J.; Lee, S. Generation of structurally realistic retinal fundus images with diffusion models. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR) Workshops, Seattle, WA, USA, 17–18 June 2024. [Google Scholar]
- Zhang, L.; Rao, A.; Agrawala, M. Adding conditional control to text-to-image diffusion models. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Paris, France, 1–6 October 2023. [Google Scholar]
- Fang, Z.; Chen, Z.; Wei, P.; Li, W.; Zhang, S.; Elazab, A.; Jia, G.; Ge, R.; Wang, C. UWAT-GAN: Fundus fluorescein angiography synthesis via ultra-wide-angle transformation multi-scale GAN. In Proceedings of the 26th International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI), Vancouver, BC, Canada, 8–12 October 2023. [Google Scholar]
- Ge, R.; Fang, Z.; Wei, P.; Chen, Z.; Jiang, H.; Elazab, A.; Li, W.; Wan, X.; Zhang, S.; Wang, C. UWAFA-GAN: Ultra-wide-angle fluorescein angiography transformation via multi-scale generation and registration enhancement. IEEE J. Biomed. Health Inform. 2024, 28, 4820–4829. [Google Scholar] [CrossRef]
- Wang, D.; Lian, J.; Jiao, W. Multi-label classification of retinal disease via a novel vision transformer model. Front. Neurosci. 2024, 17, 1290803. [Google Scholar] [CrossRef]
- Malik, S.M.F.; Nafis, M.T.; Ahad, M.A.; Tanweer, S. Grading and Anomaly Detection for Automated Retinal Image Analysis using Deep Learning. arXiv 2024, arXiv:2409.16721. [Google Scholar]
- Wang, Y. Research on the application of vision transformer model in 14-visual-field classification of fundus images. In Proceedings of the International Conference on Computer Application and Information Security (ICCAIS 2024), Wuhan, China, 20–22 December 2024. [Google Scholar]
- Huang, C.; Jiang, Y.; Yang, X.; Wei, C.; Chen, H.; Xiong, W.; Lin, H.; Wang, X.; Tian, T.; Tan, H. Enhancing Retinal Fundus Image Quality Assessment with Swin-Transformer–Based Learning Across Multiple Color-Spaces. Transl. Vis. Sci. Technol. 2024, 13, 8. [Google Scholar] [CrossRef] [PubMed]
- Isola, P.; Zhu, J.Y.; Zhou, T.; Efros, A.A. Image-to-image translation with conditional adversarial networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 1125–1134. [Google Scholar]
- Lee, M.E.; Kim, S.H.; Seo, I.H. Intensity-based registration of medical images. In Proceedings of the 2009 International Conference on Test and Measurement, Hong Kong, China, 5–6 December 2009; pp. 239–242. [Google Scholar]
- Luo, G.; Chen, X.; Shi, F.; Peng, Y.; Xiang, D.; Chen, Q.; Xu, X.; Zhu, W.; Fan, Y. Multimodal affine registration for ICGA and MCSL fundus images of high myopia. Biomed. Opt. Express 2020, 11, 4443–4457. [Google Scholar] [CrossRef] [PubMed]
- Hernandez, M.; Medioni, G.; Hu, Z.; Sadda, S. Multimodal registration of multiple retinal images based on line structures. In Proceedings of the 2015 IEEE Winter Conference on Applications of Computer Vision, Waikoloa, HI, USA, 5–9 January 2015; pp. 907–914. [Google Scholar]
- Li, Z.; Huang, F.; Zhang, J.; Dashtbozorg, B.; Abbasi-Sureshjani, S.; Sun, Y.; Long, X.; Yu, Q.; Romeny, B.t.H.; Tan, T. Multi-modal and multi-vendor retina image registration. Biomed. Opt. Express 2018, 9, 410–422. [Google Scholar] [CrossRef] [PubMed]
- Hervella, Á.S.; Rouco, J.; Novo, J.; Ortega, M. Multimodal registration of retinal images using domain-specific landmarks and vessel enhancement. Proc. Comput. Sci. 2018, 126, 97–104. [Google Scholar] [CrossRef]
- Ghassabi, Z.; Shanbehzadeh, J.; Sedaghat, A.; Fatemizadeh, E. An efficient approach for robust multimodal retinal image registration based on UR-SIFT Features and PIIFD descriptors. EURASIP J. Image Video Process. 2013, 2013, 25. [Google Scholar] [CrossRef]
- Wang, G.; Wang, Z.; Chen, Y.; Zhao, W. Robust point matching method for multimodal retinal image registration. Biomed. Signal Process. Control 2015, 19, 68–76. [Google Scholar] [CrossRef]
- Chen, J.; Tian, J.; Lee, N.; Zheng, J.; Smith, R.T.; Laine, A.F. A partial intensity invariant feature descriptor for multimodal retinal image registration. IEEE Trans. Biomed. Eng. 2010, 57, 1707–1718. [Google Scholar] [CrossRef]
- Lee, J.A.; Cheng, J.; Lee, B.H.; Ong, E.P.; Xu, G.; Wong, D.W.K.; Liu, J.; Laude, A.; Lim, T.H. A low-dimensional step pattern analysis algorithm with application to multimodal retinal image registration. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Boston, MA, USA, 7–12 June 2015; pp. 1046–1053. [Google Scholar]
- Rocco, I.; Arandjelović, R.; Sivic, J. Convolutional neural network architecture for geometric matching. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017. [Google Scholar]
- Ballé, J.; Chou, P.A.; Minnen, D.; Singh, S.; Johnston, N.; Agustsson, E.; Hwang, S.J.; Toderici, G. Nonlinear transform coding. IEEE J. Sel. Top. Signal Process. 2020, 14, 1414–1425. [Google Scholar] [CrossRef]
- Li, H.; Liu, H.; Hu, Y.; Higashita, R.; Zhao, Y.; Qi, H.; Liu, J. Restoration of cataract fundus images via unsupervised domain adaptation. In Proceedings of the 2021 IEEE 18th International Symposium on Biomedical Imaging (ISBI), Nice, France, 13–16 April 2021; pp. 516–520. [Google Scholar]
- Sarlin, P.E.; DeTone, D.; Malisiewicz, T.; Rabinovich, A. SuperGlue: Learning feature matching with graph neural networks. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Seattle, WA, USA, 13–19 June 2020; pp. 4938–4947. [Google Scholar]
- Lyu, W.; Chen, L.; Zhou, Z.; Wu, W. Weakly supervised object-aware convolutional neural networks for semantic feature matching. Neurocomputing 2021, 447, 257–271. [Google Scholar] [CrossRef]
- Lowe, D.G. Distinctive image features from scale-invariant keypoints. Int. J. Comput. Vis. 2004, 60, 91–110. [Google Scholar] [CrossRef]
- Rublee, E.; Rabaud, V.; Konolige, K.; Bradski, G. ORB: An efficient alternative to SIFT or SURF. In Proceedings of the 2011 International Conference on Computer Vision, Barcelona, Spain, 6–13 November 2011; pp. 2564–2571. [Google Scholar]
- Sara, U.; Akter, M.; Uddin, M.S. Image quality assessment through FSIM, SSIM, MSE and PSNR—A Comparative Study. J. Comput. Commun. 2019, 7, 19–32. [Google Scholar] [CrossRef]
- Liu, R.; Wang, X.; Wu, Q.; Dai, L.; Fang, X.; Yan, T.; Son, J.; Tang, S.; Li, J.; Gao, Z.; et al. DeepDRiD: Diabetic retinopathy—grading and image quality estimation challenge. Patterns 2022, 3, 100512. [Google Scholar] [CrossRef]
- He, S.; Ye, X.; Xie, W.; Shen, Y.; Yang, S.; Zhong, X.; Guan, H.; Zhou, X.; Wu, J.; Shen, L. Open ultrawidefield fundus image dataset with disease diagnosis and clinical image quality assessment. Sci. Data 2024, 11, 1251. [Google Scholar] [CrossRef]
- Jin, K.; Gao, Z.; Jiang, X.; Wang, Y.; Ma, X.; Li, Y.; Ye, J. MSHF: A Multi-Source Heterogeneous Fundus (MSHF) Dataset for Image Quality Assessment. Sci. Data 2023, 10, 286. [Google Scholar] [CrossRef]
- Heusel, M.; Ramsauer, H.; Unterthiner, T.; Nessler, B.; Hochreiter, S. GANs Trained by a Two Time-Scale Update Rule Converge to a Local Nash Equilibrium. Adv. Neural Inf. Process. Syst. 2017, 30, 6626–6637. [Google Scholar]






| Methods | Mean PCK (%) | ||
|---|---|---|---|
| SIFT [37] | 31.6 | 46.7 | 69.0 |
| ORB [38] | 27.8 | 41.9 | 57.4 |
| SuperGlue [35] | 46.9 | 78.6 | 89.2 |
| Ours | 57.3 | 83.1 | 94.8 |
| Methods | MSE ↓ | PSNR ↑ | SSIM (%) ↑ | MS-SSIM (%) ↑ | |
|---|---|---|---|---|---|
| Unpaired Learning | CycleGAN [7] | 90.42 | 28.60 | 43.41 | 46.03 |
| Yoo et al. [1] | 75.78 | 29.44 | 75.46 | 74.87 | |
| Pham et al. [9] | 70.19 | 28.71 | 78.79 | 78.74 | |
| Paired Learning | Pix2Pix [22] | 70.71 | 29.71 | 84.49 | 84.30 |
| Pix2PixHD [6] | 60.64 | 30.49 | 87.71 | 88.02 | |
| Ours | 57.38 | 30.74 | 88.73 | 89.71 | |
| Evaluation Criteria | Items | Score (Mean ± SD) | |
|---|---|---|---|
| Registered UFI (Regi. Only) | Generated CFI (After Trans.) | ||
| Optic nerve structures | Cup–to–disc ratio | 1.53 ± 0.59 | 2.68 ± 0.49 |
| Color of the disc | 1.23 ± 0.45 | 3.00 ± 0.00 | |
| Vascular distribution | Overall morphology | 2.43 ± 0.50 | 2.83 ± 0.38 |
| Vessel contrast | 1.87 ± 0.63 | 2.66 ± 0.50 | |
| Drusen | Drusen pattern | 2.18 ± 0.41 | 2.53 ± 0.50 |
| Drusen number | 2.24 ± 0.43 | 2.53 ± 0.50 | |
| Dataset | Comparison | FID ↓ |
|---|---|---|
| UWF-IQA [42] | generated CFI vs. SMC real CFI | 51.72 |
| MSHF [41] | generated CFI vs. real CFI | 104.32 |
| DeepDRiD [40] | generated CFI vs. real CFI | 132.24 |
| Method Components | Performance Metrics | ||||
|---|---|---|---|---|---|
| Scaled Feature Registration | Vessel Correction Registration | MSE | PSNR | SSIM (%) | MS-SSIM (%) |
| ↓ | ↑ | ↑ | ↑ | ||
| ✗ | ✗ | 70.14 | 28.67 | 78.74 | 79.63 |
| ✗ | ✓ | 78.96 | 28.04 | 74.33 | 73.91 |
| ✓ | ✗ | 60.64 | 30.49 | 87.71 | 88.02 |
| ✓ | ✓ | 57.38 | 30.74 | 88.73 | 89.71 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, J.; Bum, J.; Le, D.-T.; Son, C.-H.; Lee, E.J.; Han, J.C.; Choo, H. Ultrawidefield-to-Conventional Fundus Image Translation with Scaled Feature Registration and Distorted Vessel Correction. Bioengineering 2025, 12, 1046. https://doi.org/10.3390/bioengineering12101046
Kim J, Bum J, Le D-T, Son C-H, Lee EJ, Han JC, Choo H. Ultrawidefield-to-Conventional Fundus Image Translation with Scaled Feature Registration and Distorted Vessel Correction. Bioengineering. 2025; 12(10):1046. https://doi.org/10.3390/bioengineering12101046
Chicago/Turabian StyleKim, JuChan, Junghyun Bum, Duc-Tai Le, Chang-Hwan Son, Eun Jung Lee, Jong Chul Han, and Hyunseung Choo. 2025. "Ultrawidefield-to-Conventional Fundus Image Translation with Scaled Feature Registration and Distorted Vessel Correction" Bioengineering 12, no. 10: 1046. https://doi.org/10.3390/bioengineering12101046
APA StyleKim, J., Bum, J., Le, D.-T., Son, C.-H., Lee, E. J., Han, J. C., & Choo, H. (2025). Ultrawidefield-to-Conventional Fundus Image Translation with Scaled Feature Registration and Distorted Vessel Correction. Bioengineering, 12(10), 1046. https://doi.org/10.3390/bioengineering12101046






