Enhancing Oral Squamous Cell Carcinoma Detection Using Histopathological Images: A Deep Feature Fusion and Improved Haris Hawks Optimization-Based Framework
Abstract
1. Introduction
2. Materials and Methods
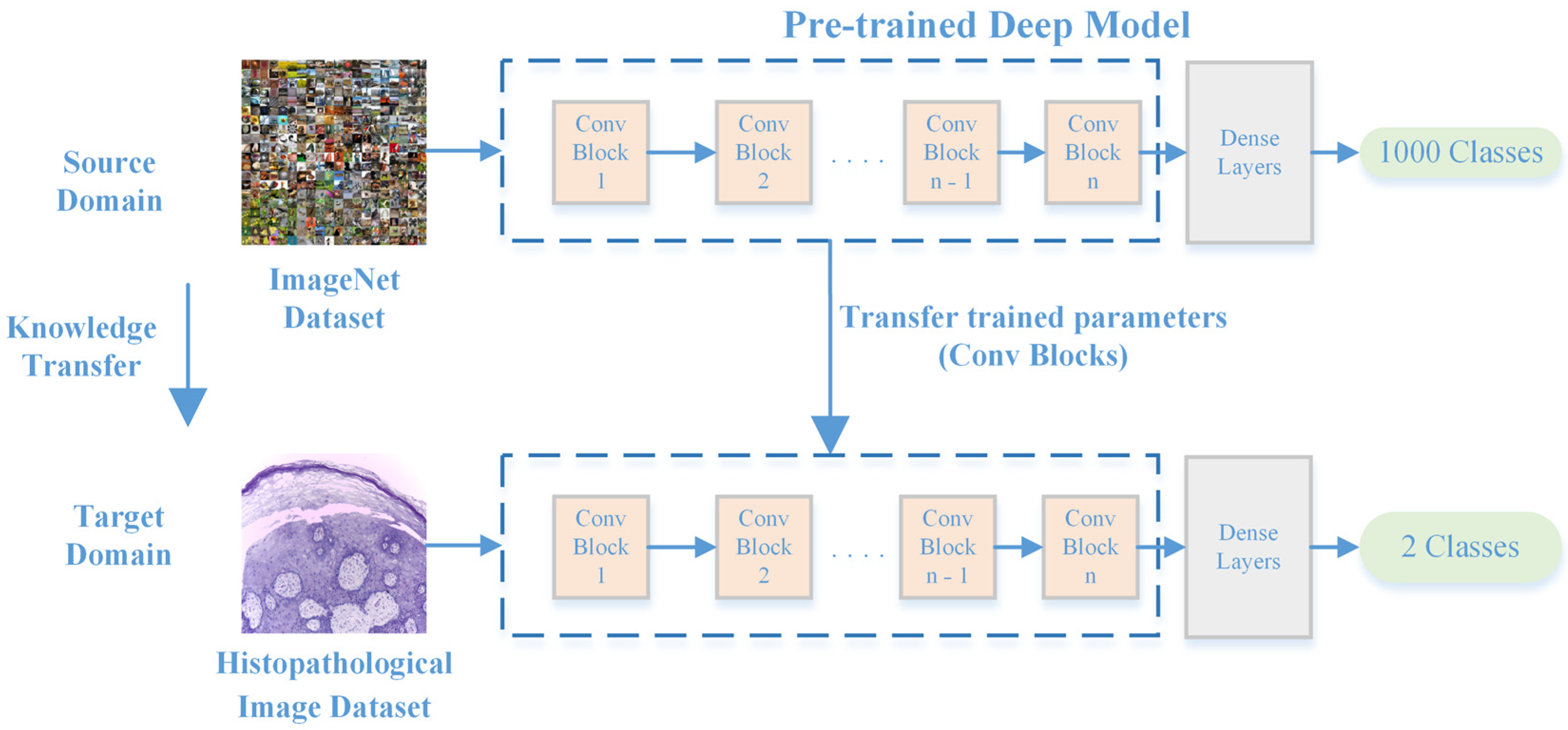
2.1. Proposed OSCC Detection Framework
2.2. Histopathological Images Dataset
2.3. Feature Extraction from Histopathological Images
2.3.1. CNNs
2.3.2. Deep Feature Extraction Using CNNs
2.3.3. Feature Fusion Using Canonical Correlation Analysis
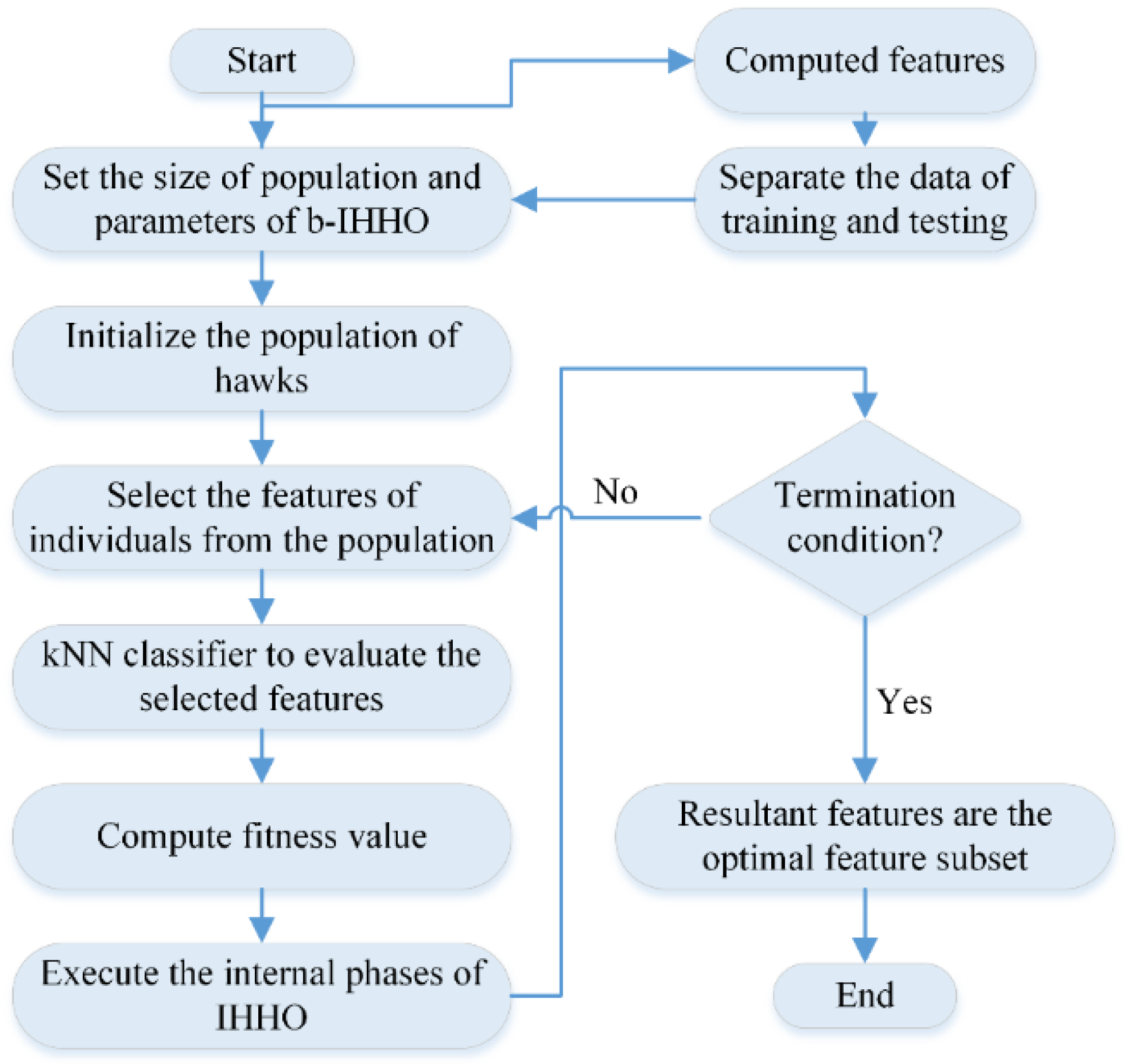
2.3.4. HHO
2.3.5. IHHO
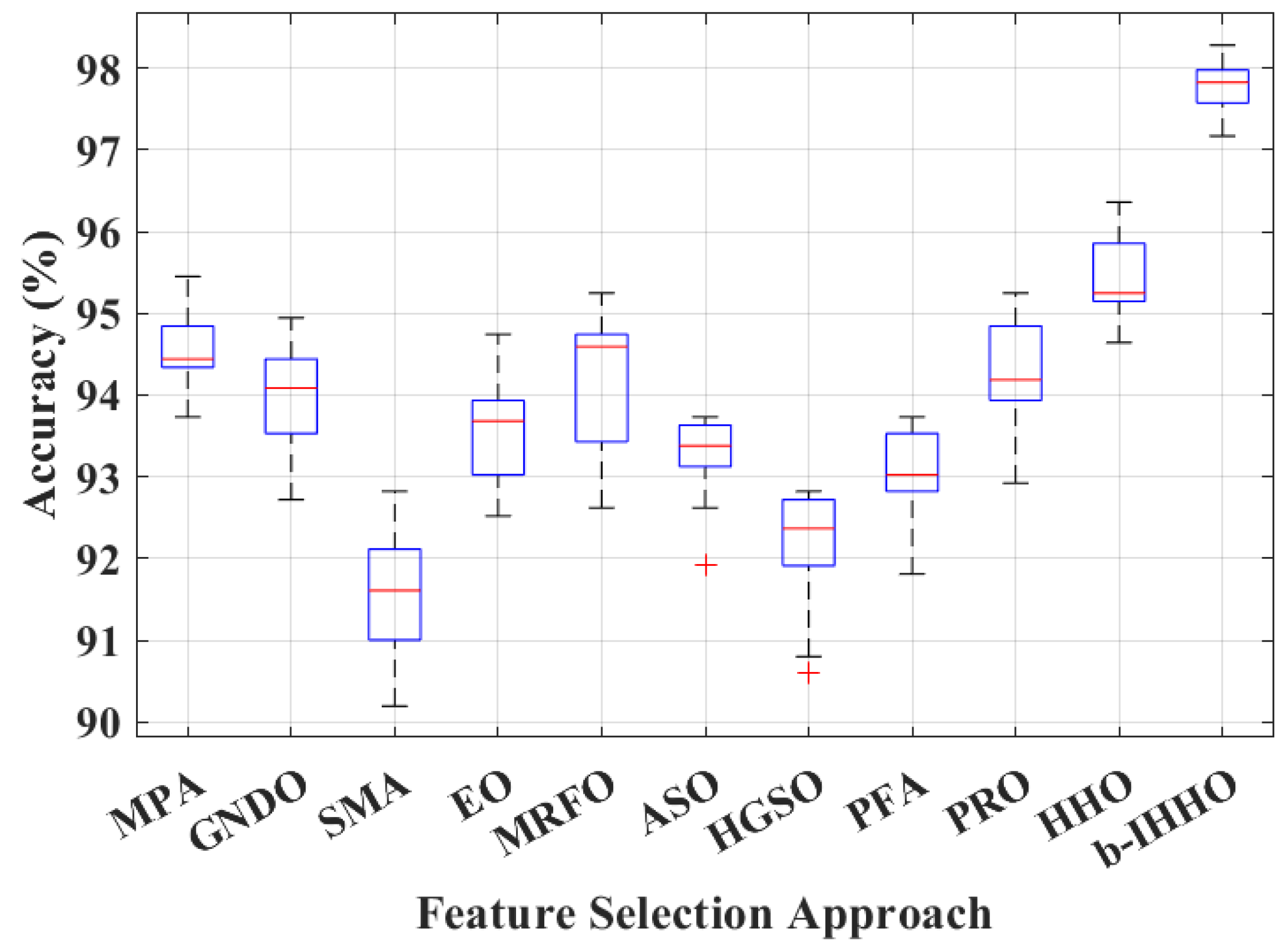
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ghantous, Y.; Abu Elnaaj, I. Global incidence and risk factors of oral cancer. Harefuah 2017, 156, 645–649. [Google Scholar] [PubMed]
- Zygogianni, A.G.; Kyrgias, G.; Karakitsos, P.; Psyrri, A.; Kouvaris, J.; Kelekis, N.; Kouloulias, V. Oral squamous cell cancer: Early detection and the role of alcohol and smoking. Head Neck Oncol. 2011, 3, 2. [Google Scholar] [CrossRef] [PubMed]
- Khijmatgar, S.; Yong, J.; Rübsamen, N.; Lorusso, F.; Rai, P.; Cenzato, N.; Gaffuri, F.; Del Fabbro, M.; Tartaglia, G.M. Salivary biomarkers for early detection of oral squamous cell carcinoma (OSCC) and head/neck squamous cell carcinoma (HNSCC): A systematic review and network meta-analysis. Jpn. Dent. Sci. Rev. 2024, 60, 32–39. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Boccia, S.; Hashibe, M.; Gallì, P.; De Feo, E.; Asakage, T.; Hashimoto, T.; Hiraki, A.; Katoh, T.; Nomura, T.; Yokoyama, A.; et al. Aldehyde dehydrogenase 2 and head and neck cancer: A meta-analysis implementing a Mendelian randomization approach. Cancer Epidemiol. Biomark. Prev. 2009, 18, 248–254. [Google Scholar] [CrossRef]
- Patel, S.C.; Carpenter, W.R.; Tyree, S.; Couch, M.E.; Weissler, M.; Hackman, T.; Hayes, D.N.; Shores, C.; Chera, B.S. Increasing incidence of oral tongue squamous cell carcinoma in young white women, age 18 to 44 years. J. Clin. Oncol. 2011, 29, 1488–1494. [Google Scholar] [CrossRef]
- Müller, S.; Pan, Y.; Li, R.; Chi, A.C. Changing trends in oral squamous cell carcinoma with particular reference to young patients: 1971–2006. The Emory University experience. Head Neck Pathol. 2008, 2, 60–66. [Google Scholar] [CrossRef]
- Ferreira E Costa, R.; Leão, M.L.B.; Sant’Ana, M.S.P.; Mesquita, R.A.; Gomez, R.S.; Santos-Silva, A.R.; Khurram, S.A.; Tailor, A.; Schouwstra, C.-M.; Robinson, L.; et al. Oral squamous cell carcinoma frequency in young patients from referral centers around the world. Head Neck Pathol. 2022, 16, 755–762. [Google Scholar] [CrossRef]
- Faeli Ghadikolaei, R.; Ghorbani, H.; Seyedmajidi, M.; Ebrahimnejad Gorji, K.; Moudi, E.; Seyedmajidi, S. Genotoxicity and cytotoxicity effects of X-rays on the oral mucosa epithelium at different fields of view: A cone beam computed tomography technique. Casp. J. Intern. Med. 2023, 14, 121–127. [Google Scholar] [CrossRef]
- Nien, H.-H.; Wang, L.-Y.; Liao, L.-J.; Lin, P.-Y.; Wu, C.-Y.; Shueng, P.-W.; Chung, C.-S.; Lo, W.-C.; Lin, S.-C.; Hsieh, C.-H. Advances in image-guided radiotherapy in the treatment of oral cavity cancer. Cancers 2022, 14, 4630. [Google Scholar] [CrossRef]
- Marcus, C.; Subramaniam, R.M. PET imaging of oral cavity and oropharyngeal cancers. PET Clin. 2022, 17, 223–234. [Google Scholar] [CrossRef]
- Maraghelli, D.; Pietragalla, M.; Calistri, L.; Barbato, L.; Locatello, L.G.; Orlandi, M.; Landini, N.; Lo Casto, A.; Nardi, C. Techniques, tricks, and stratagems of oral cavity computed tomography and magnetic resonance imaging. Appl. Sci. 2022, 12, 1473. [Google Scholar] [CrossRef]
- Azam, M.A.; Sampieri, C.; Ioppi, A.; Benzi, P.; Giordano, G.G.; De Vecchi, M.; Campagnari, V.; Li, S.; Guastini, L.; Paderno, A.; et al. Videomics of the upper aero-digestive tract cancer: Deep learning applied to white light and narrow band imaging for automatic segmentation of endoscopic images. Front. Oncol. 2022, 12, 900451. [Google Scholar] [CrossRef]
- Esteva, A.; Kuprel, B.; Novoa, R.A.; Ko, J.; Swetter, S.M.; Blau, H.M.; Thrun, S. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017, 542, 115–118. [Google Scholar] [CrossRef]
- Tshering Vogel, D.W.T.; Zbaeren, P.; Thoeny, H.C. Cancer of the oral cavity and oropharynx. Cancer Imaging 2010, 10, 62–72. [Google Scholar] [CrossRef]
- de Chauveron, J.; Unger, M.; Lescaille, G.; Wendling, L.; Kurtz, C.; Rochefort, J. Artificial intelligence for oral squamous cell carcinoma detection based on oral photographs: A comprehensive literature review. Cancer Med. 2024, 13, e6822. [Google Scholar] [CrossRef]
- Amin, I.; Zamir, H.; Khan, F.F. Histopathological image analysis for oral squamous cell carcinoma classification using concatenated deep learning models. medRxiv 2021. [Google Scholar] [CrossRef]
- Das, N.; Hussain, E.; Mahanta, L.B. Automated classification of cells into multiple classes in epithelial tissue of oral squamous cell carcinoma using transfer learning and convolutional neural network. Neural Netw. 2020, 128, 47–60. [Google Scholar] [CrossRef]
- Das, M.; Dash, R.; Mishra, S.K. Automatic detection of oral squamous cell carcinoma from histopathological images of oral mucosa using deep convolutional neural network. Int. J. Environ. Res. Public Health 2023, 20, 2131. [Google Scholar] [CrossRef]
- Khan, M.A.; Mir, M.; Ullah, M.S.; Hamza, A.; Jabeen, K.; Gupta, D. A fusion framework of pre-trained deep learning models for oral squamous cell carcinoma classification. In Proceedings of the Third International Conference on Computing and Communication Networks, Manchester, UK, 17–18 November 2023; Springer: Singapore, 2024; pp. 769–782. [Google Scholar] [CrossRef]
- Akram, M.W.; Li, G.; Jin, Y.; Chen, X.; Zhu, C.; Ahmad, A. Automatic detection of photovoltaic module defects in infrared images with isolated and develop-model transfer deep learning. Sol. Energy 2020, 198, 175–186. [Google Scholar] [CrossRef]
- Oyetade, I.S.; Ayeni, J.O.; Ogunde, A.O.; Oguntunde, B.O.; Olowookere, T.A. Hybridized deep convolutional neural network and fuzzy support vector machines for breast cancer detection. SN Comput. Sci. 2022, 3, 581. [Google Scholar] [CrossRef]
- Fatima, M.; Khan, M.A.; Shaheen, S.; Almujally, N.A.; Wang, S.-H. B2C3NetF2: Breast cancer classification using an end-to-end deep learning feature fusion and satin bowerbird optimization controlled Newton Raphson feature selection. CAAI Trans. Intell. Technol. 2023, 8, 1374–1390. [Google Scholar] [CrossRef]
- Zahoor, S.; Shoaib, U.; Lali, I.U. Breast cancer mammograms classification using deep neural network and entropy-controlled whale optimization algorithm. Diagnostics 2022, 12, 557. [Google Scholar] [CrossRef]
- Baltruschat, I.M.; Nickisch, H.; Grass, M.; Knopp, T.; Saalbach, A. Comparison of deep learning approaches for multi-label chest X-ray classification. Sci. Rep. 2019, 9, 6381. [Google Scholar] [CrossRef]
- Kang, J.; Gwak, J. Ensemble of instance segmentation models for polyp segmentation in colonoscopy images. IEEE Access 2019, 7, 26440–26447. [Google Scholar] [CrossRef]
- Heidari, A.A.; Mirjalili, S.; Faris, H.; Aljarah, I.; Mafarja, M.; Chen, H. Harris hawks optimization: Algorithm and applications. Future Gener. Comput. Syst. 2019, 97, 849–872. [Google Scholar] [CrossRef]
- Peng, L.; Cai, Z.; Heidari, A.A.; Zhang, L.; Chen, H. Hierarchical Harris hawks optimizer for feature selection. J. Adv. Res. 2023, 53, 261–278. [Google Scholar] [CrossRef]
- Agrawal, P.; Abutarboush, H.F.; Ganesh, T.; Mohamed, A.W. Metaheuristic algorithms on feature selection: A survey of one decade of research (2009–2019). IEEE Access 2021, 9, 26766–26791. [Google Scholar] [CrossRef]
- Sukegawa, S.; Ono, S.; Tanaka, F.; Inoue, Y.; Hara, T.; Yoshii, K.; Nakano, K.; Takabatake, K.; Kawai, H.; Katsumitsu, S.; et al. Effectiveness of deep learning classifiers in histopathological diagnosis of oral squamous cell carcinoma by pathologists. Sci. Rep. 2023, 13, 11676. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Ding, J.; Liu, W.; Tang, X.; Xia, J.; Liang, S.; Jing, R.; Zhu, L.; Zhang, T. Deep multi-feature fusion residual network for oral squamous cell carcinoma classification and its intelligent system using Raman spectroscopy. Biomed. Signal Process Control 2023, 86, 105339. [Google Scholar] [CrossRef]
- Chang, X.; Yu, M.; Liu, R.; Jing, R.; Ding, J.; Xia, J.; Zhu, Z.; Li, X.; Yao, Q.; Zhu, L.; et al. Deep learning methods for oral cancer detection using Raman spectroscopy. Vib. Spectrosc. 2023, 126, 103522. [Google Scholar] [CrossRef]
- Panigrahi, S.; Nanda, B.S.; Bhuyan, R.; Kumar, K.; Ghosh, S.; Swarnkar, T. Classifying histopathological images of oral squamous cell carcinoma using deep transfer learning. Heliyon 2023, 9, e13444. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Pan, H.; Shang, J.; Zhang, J.; Liang, Y. Deep-learning-based automated identification and visualization of oral cancer in optical coherence tomography images. Biomedicines 2023, 11, 802. [Google Scholar] [CrossRef] [PubMed]



| Normal | Sick (OSCC) | |
|---|---|---|
| Histopathological images |  | 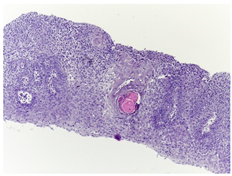 |
| Images per class | 2435 | 2511 |
| Model | Feature Vector Size | Accuracy (%) |
|---|---|---|
| Xception | 2048 | 87.77 |
| SqueezeNet | 1000 | 82.91 |
| ShuffleNet | 544 | 84.93 |
| ResNet-18 | 512 | 86.05 |
| ResNet-50 | 2048 | 89.69 |
| ResNet-101 | 2048 | 91.51 |
| NASNet-Mobile | 1056 | 84.73 |
| MobileNet-v2 | 1280 | 84.53 |
| Inception-v3 | 2048 | 86.86 |
| Inception-ResNet-v2 | 1536 | 89.69 |
| GoogLeNet | 1024 | 81.60 |
| GoogLeNet365 | 1024 | 85.04 |
| EfficientNet-b0 | 1280 | 91.61 |
| DenseNet-201 | 1920 | 87.87 |
| DarkNet-53 | 1024 | 86.96 |
| DarkNet-19 | 1000 | 87.36 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zafar, A.; Khalid, M.; Farrash, M.; Qadah, T.M.; Lahza, H.F.M.; Kim, S.-H. Enhancing Oral Squamous Cell Carcinoma Detection Using Histopathological Images: A Deep Feature Fusion and Improved Haris Hawks Optimization-Based Framework. Bioengineering 2024, 11, 913. https://doi.org/10.3390/bioengineering11090913
Zafar A, Khalid M, Farrash M, Qadah TM, Lahza HFM, Kim S-H. Enhancing Oral Squamous Cell Carcinoma Detection Using Histopathological Images: A Deep Feature Fusion and Improved Haris Hawks Optimization-Based Framework. Bioengineering. 2024; 11(9):913. https://doi.org/10.3390/bioengineering11090913
Chicago/Turabian StyleZafar, Amad, Majdi Khalid, Majed Farrash, Thamir M. Qadah, Hassan Fareed M. Lahza, and Seong-Han Kim. 2024. "Enhancing Oral Squamous Cell Carcinoma Detection Using Histopathological Images: A Deep Feature Fusion and Improved Haris Hawks Optimization-Based Framework" Bioengineering 11, no. 9: 913. https://doi.org/10.3390/bioengineering11090913
APA StyleZafar, A., Khalid, M., Farrash, M., Qadah, T. M., Lahza, H. F. M., & Kim, S.-H. (2024). Enhancing Oral Squamous Cell Carcinoma Detection Using Histopathological Images: A Deep Feature Fusion and Improved Haris Hawks Optimization-Based Framework. Bioengineering, 11(9), 913. https://doi.org/10.3390/bioengineering11090913







