AGSAM: Agent-Guided Segment Anything Model for Automatic Segmentation in Few-Shot Scenarios
Abstract
1. Introduction
- Introducing a novel fully automatic segmentation approach: AGSAM. This method leverages SAM-Med2D’s pre-trained feature extraction and generalized decoding capabilities. The guided model seamlessly replaces prompt generation and embedding, making it adaptable to any segmentation method.
- Introduction of a feature augmentation convolution module (FACM) in AGSAM, a parameter-free and computationally efficient module that enhances model accuracy. FACM ensures more stable feature representations, minimizing image noise impact on segmentation.
- Experimental comparisons demonstrate that AGSAM consistently outperforms comparative methods across various metrics, showcasing its effectiveness in few-shot scenarios.
2. Related Work
2.1. SAM and SAM-Med2D
2.2. SAM-Based Method
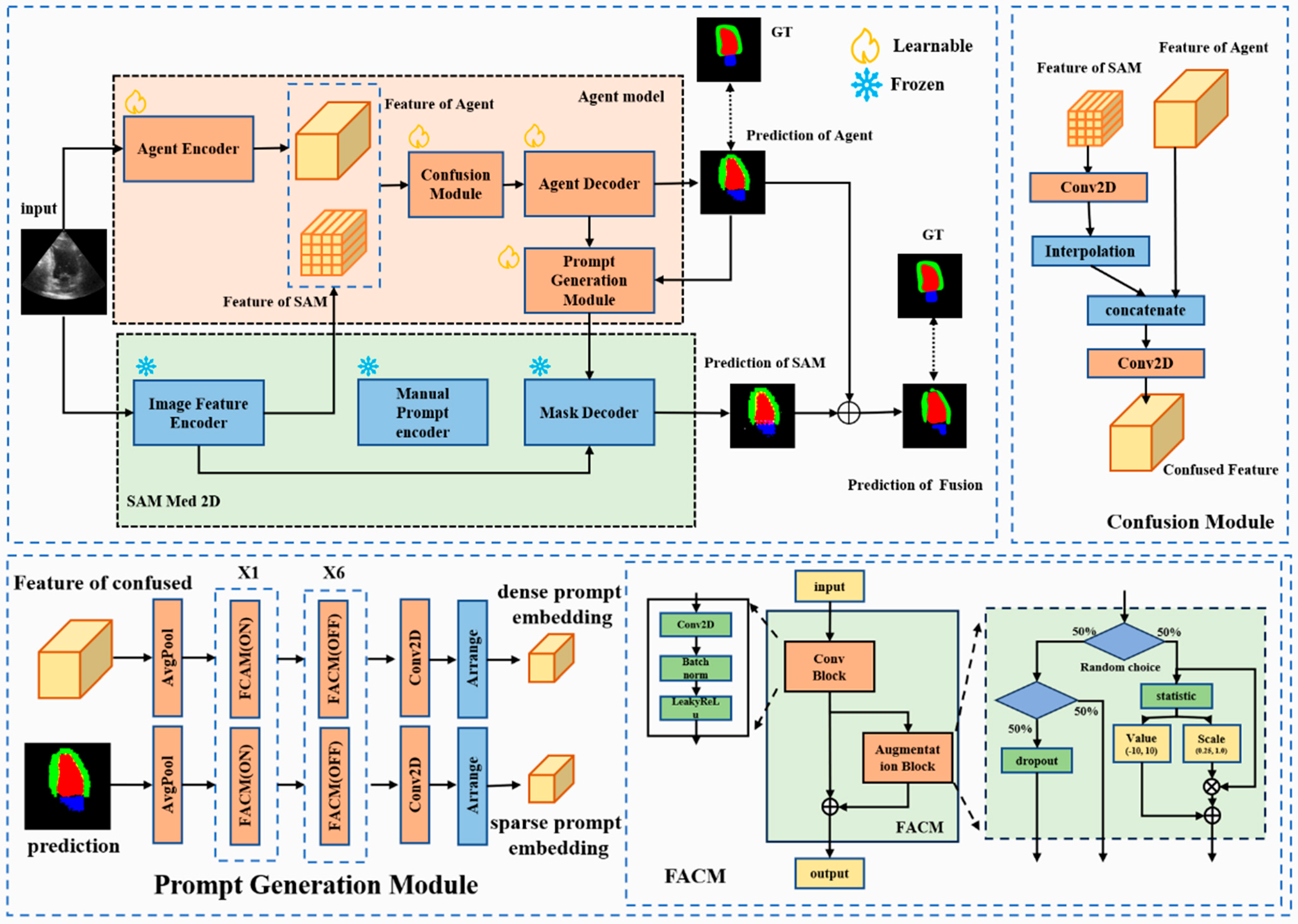
3. Methods
3.1. Architecture Overview
3.2. Agent-Guided SAM
3.3. Feature Augmentation Convolution Module
4. Experiments
4.1. Datasets
4.2. Data Pre-Processing
4.3. Implementation Details and Evaluation Metrics
4.3.1. Architecture Description
4.3.2. Setting of Training
4.3.3. Evaluation Metrics
4.4. Experimental Settings
4.4.1. Comparative Study
4.4.2. Ablation Study
5. Results and Discussion
5.1. Comparison in Few-Shot Scenario with CAMUS
5.2. Comparison in Few-Shot Scenario with REFUGE
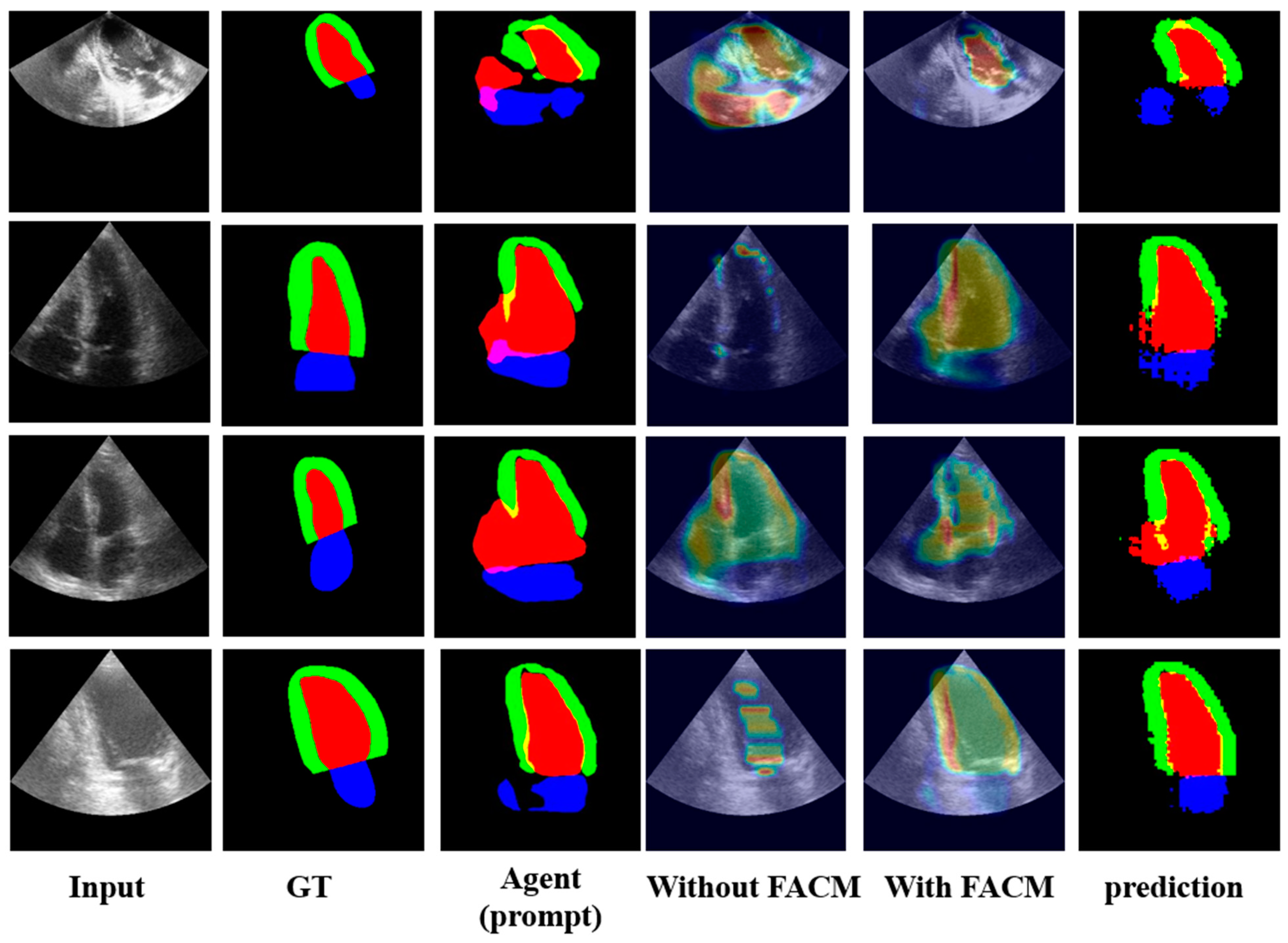
5.3. Ablation Studies of AGSAM
5.4. Limitations of AGSAM
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sheikh, T.S.; Cho, M. Segmentation of Variants of Nuclei on Whole Slide Images by Using Radiomic Features. Bioengineering 2024, 11, 252. [Google Scholar] [CrossRef]
- Karn, P.K.; Abdulla, W.H. Advancing Ocular Imaging: A Hybrid Attention Mechanism-Based U-Net Model for Precise Segmentation of Sub-Retinal Layers in OCT Images. Bioengineering 2024, 11, 240. [Google Scholar] [CrossRef]
- Deng, J.; Dong, W.; Socher, R.; Li, L.-J.; Li, K.; Fei-Fei, L. Imagenet: A large-scale hierarchical image database. In Proceedings of the Computer Vision and Pattern Recognition, Miami, FL, USA, 20–25 June 2009; pp. 248–255. [Google Scholar]
- Li, Y.; Jing, B.; Feng, X.; Li, Z.; He, Y.; Wang, J.; Zhang, Y. nnSAM: Plug-and-play segment anything model improves nnunet performance. arXiv 2023, arXiv:2309.16967. [Google Scholar]
- Shamrat, F.J.M.; Akter, S.; Azam, S.; Karim, A.; Ghosh, P.; Tasnim, Z.; Hasib, K.M.; De Boer, F.; Ahmed, K. AlzheimerNet: An effective deep learning based proposition for alzheimer’s disease stages classification from functional brain changes in magnetic resonance images. IEEE Access 2023, 11, 16376–16395. [Google Scholar] [CrossRef]
- Shamrat, F.J.M.; Azam, S.; Karim, A.; Ahmed, K.; Bui, F.M.; De Boer, F. High-precision multiclass classification of lung disease through customized MobileNetV2 from chest X-ray images. Comput. Biol. Med. 2023, 155, 106646. [Google Scholar] [CrossRef]
- Lang, C.; Cheng, G.; Tu, B.; Han, J. Learning what not to segment: A new perspective on few-shot segmentation. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, New Orleans, LA, USA, 18–24 June 2022; pp. 8057–8067. [Google Scholar]
- Floridi, L.; Chiriatti, M. GPT-3: Its nature, scope, limits, and consequences. Minds Mach. 2020, 30, 681–694. [Google Scholar] [CrossRef]
- Luo, R.; Sun, L.; Xia, Y.; Qin, T.; Zhang, S.; Poon, H.; Liu, T.-Y. BioGPT: Generative pre-trained transformer for biomedical text generation and mining. Brief. Bioinform. 2022, 23, bbac409. [Google Scholar] [CrossRef]
- Sufi, F. Generative Pre-Trained Transformer (GPT) in Research: A Systematic Review on Data Augmentation. Information 2024, 15, 99. [Google Scholar] [CrossRef]
- Kirillov, A.; Mintun, E.; Ravi, N.; Mao, H.; Rolland, C.; Gustafson, L.; Xiao, T.; Whitehead, S.; Berg, A.C.; Lo, W.-Y. Segment anything. arXiv 2023, arXiv:2304.02643. [Google Scholar]
- Cheng, J.; Ye, J.; Deng, Z.; Chen, J.; Li, T.; Wang, H.; Su, Y.; Huang, Z.; Chen, J.; Jiang, L. Sam-med2d. arXiv 2023, arXiv:2308.16184. [Google Scholar]
- Hu, X.; Xu, X.; Shi, Y. How to Efficiently Adapt Large Segmentation Model (SAM) to Medical Images. arXiv 2023, arXiv:2306.13731. [Google Scholar]
- Villa, M.; Dardenne, G.; Nasan, M.; Letissier, H.; Hamitouche, C.; Stindel, E. FCN-based approach for the automatic segmentation of bone surfaces in ultrasound images. Int. J. Comput. Assist. Radiol. Surg. 2018, 13, 1707–1716. [Google Scholar] [CrossRef]
- Leclerc, S.; Smistad, E.; Pedrosa, J.; Ostvik, A.; Cervenansky, F.; Espinosa, F.; Espeland, T.; Berg, E.A.R.; Jodoin, P.M.; Grenier, T.; et al. Deep Learning for Segmentation Using an Open Large-Scale Dataset in 2D Echocardiography. IEEE Trans. Med. Imaging 2019, 38, 2198–2210. [Google Scholar] [CrossRef]
- Orlando, J.I.; Fu, H.; Barbosa Breda, J.; van Keer, K.; Bathula, D.R.; Diaz-Pinto, A.; Fang, R.; Heng, P.A.; Kim, J.; Lee, J.; et al. REFUGE Challenge: A unified framework for evaluating automated methods for glaucoma assessment from fundus photographs. Med. Image Anal. 2020, 59, 101570. [Google Scholar] [CrossRef]
- Triyani, Y.; Nugroho, H.A.; Rahmawaty, M.; Ardiyanto, I.; Choridah, L. Performance analysis of image segmentation for breast ultrasound images. In Proceedings of the 2016 8th International Conference on Information Technology and Electrical Engineering (ICITEE), Yogyakarta, Indonesia, 5–6 October 2016; pp. 1–6. [Google Scholar]
- Aydin, O.U.; Taha, A.A.; Hilbert, A.; Khalil, A.A.; Galinovic, I.; Fiebach, J.B.; Frey, D.; Madai, V.I. On the usage of average hausdorff distance for segmentation performance assessment: Hidden bias when used for ranking. arXiv 2020, arXiv:2009.00215. [Google Scholar] [CrossRef]
- Yurtkulu, S.C.; Şahin, Y.H.; Unal, G. Semantic segmentation with extended DeepLabv3 architecture. In Proceedings of the 2019 27th (SIU), Sivas, Turkey, 24–26 April 2019; pp. 1–4. [Google Scholar]
- Zhao, H.; Shi, J.; Qi, X.; Wang, X.; Jia, J. Pyramid scene parsing network. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 2881–2890. [Google Scholar]
- Poudel, R.P.; Liwicki, S.; Cipolla, R. Fast-scnn: Fast semantic segmentation network. arXiv 2019, arXiv:1902.04502. [Google Scholar]
- Tomar, N.K.; Jha, D.; Bagci, U.; Ali, S. TGANet: Text-guided attention for improved polyp segmentation. Med. Image Comput. Comput. Assist. Interv. 2022, 13433, 151–160. [Google Scholar] [CrossRef]
- Xie, E.; Wang, W.; Yu, Z.; Anandkumar, A.; Alvarez, J.M.; Luo, P. SegFormer: Simple and efficient design for semantic segmentation with transformers. Adv. Neural Inf. Process. Syst. 2021, 34, 12077–12090. [Google Scholar]
- Zhou, Z.; Siddiquee, M.M.R.; Tajbakhsh, N.; Liang, J. UNet++: Redesigning Skip Connections to Exploit Multiscale Features in Image Segmentation. IEEE Trans. Med. Imaging 2020, 39, 1856–1867. [Google Scholar] [CrossRef]
- Wang, Z.; Zheng, J.-Q.; Zhang, Y.; Cui, G.; Li, L. Mamba-unet: Unet-like pure visual mamba for medical image segmentation. arXiv 2024, arXiv:2402.05079. [Google Scholar]
- Ma, J.; He, Y.; Li, F.; Han, L.; You, C.; Wang, B. Segment anything in medical images. Nat. Commun. 2024, 15, 654. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Han, D.; Qiao, Y.; Kim, J.U.; Bae, S.-H.; Lee, S.; Hong, C.S. Faster Segment Anything: Towards Lightweight SAM for Mobile Applications. arXiv 2023, arXiv:2306.14289. [Google Scholar]
- Selvaraju, R.R.; Cogswell, M.; Das, A.; Vedantam, R.; Parikh, D.; Batra, D. Grad-cam: Visual explanations from deep networks via gradient-based localization. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 618–626. [Google Scholar]

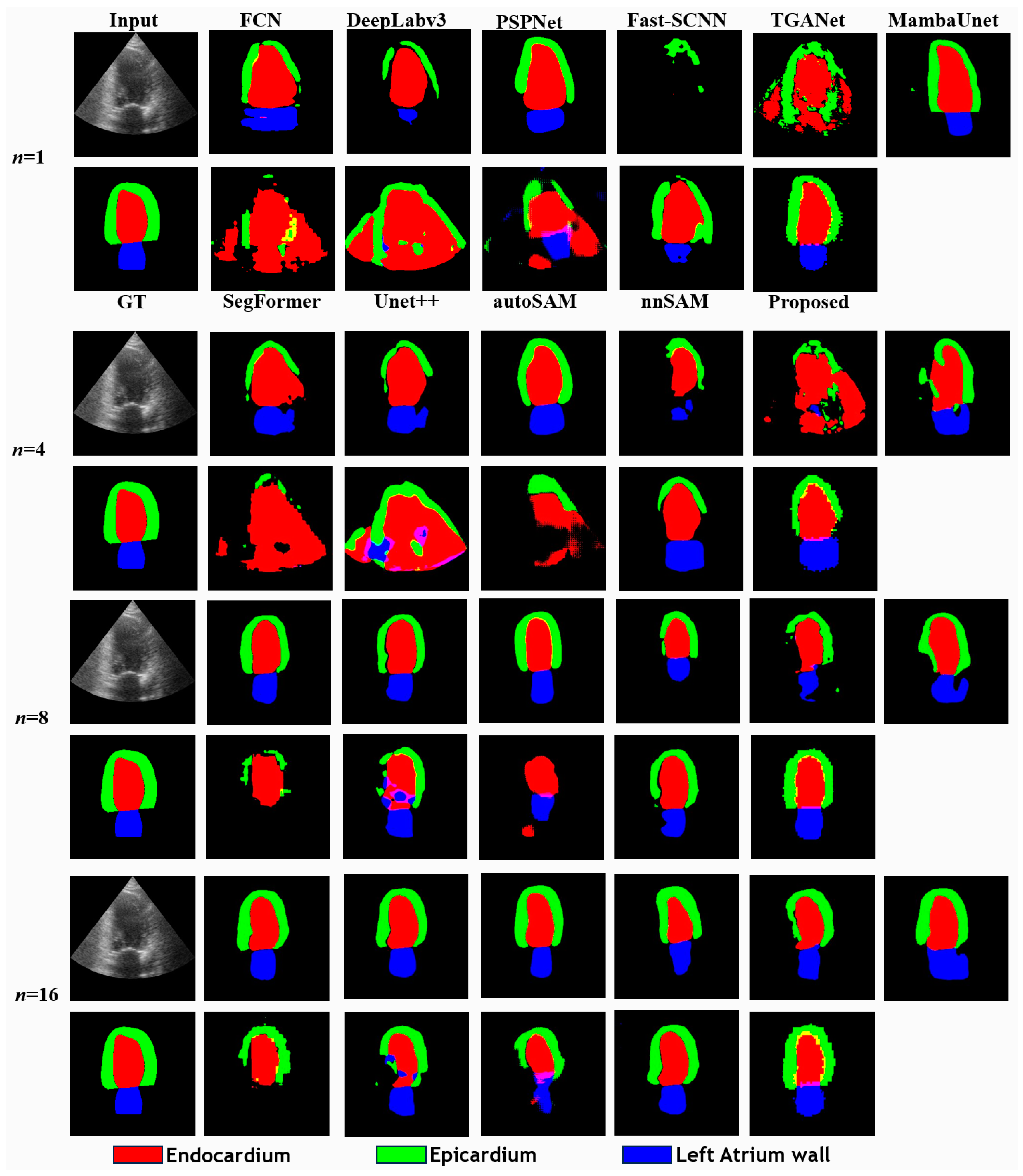
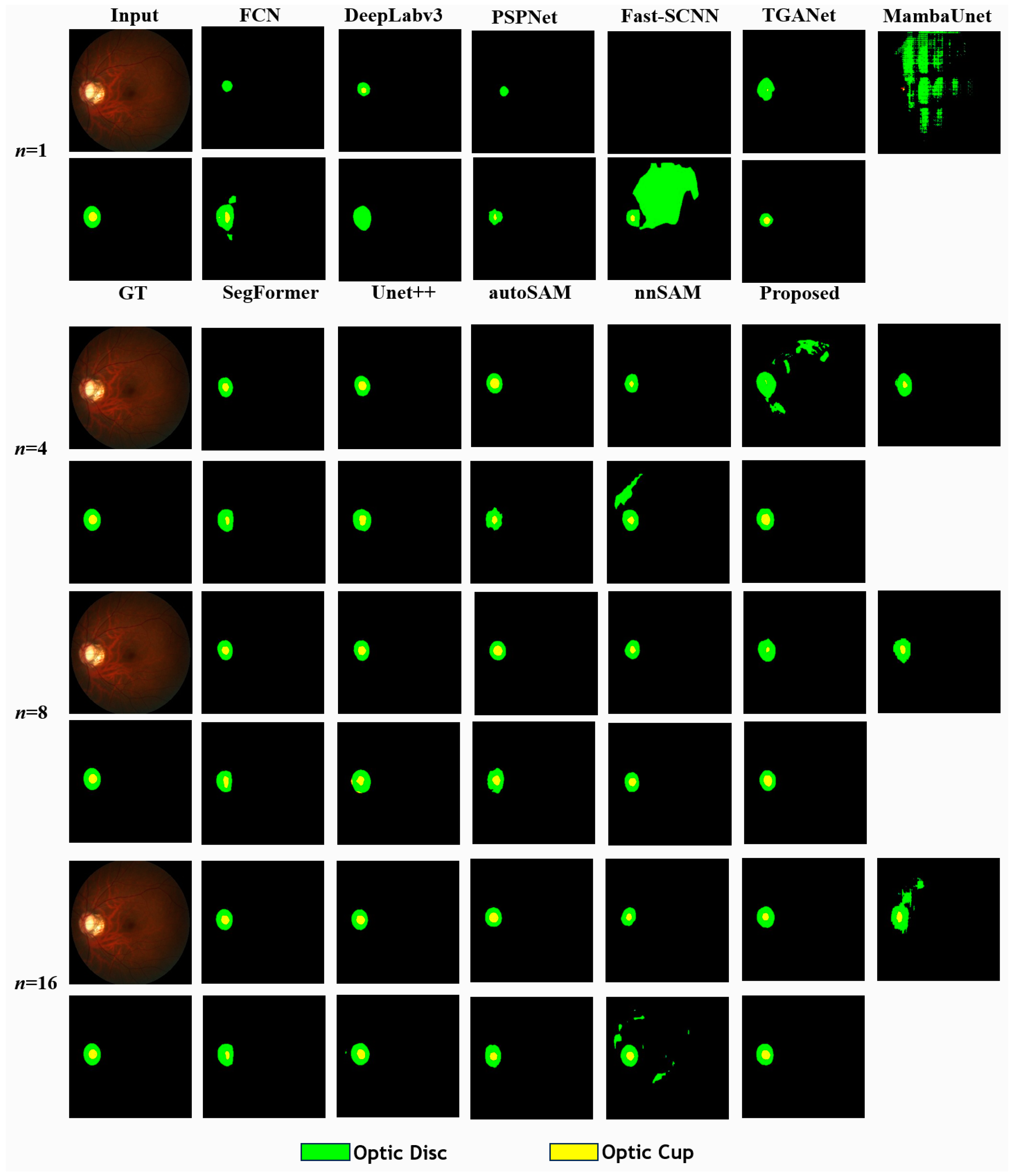
| Method | Metrics | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DICE | HD | |||||||||||||||
| Training Sample Size (n) | ||||||||||||||||
| 1 | 2 | 4 | 6 | 8 | 12 | 16 | 20 | 1 | 2 | 4 | 6 | 8 | 12 | 16 | 20 | |
| FCN | 0.4819 *** | 0.6044 *** | 0.5962 *** | 0.7507 * | 0.7607 *** | 0.7954 ** | 0.8058 ns | 0.8036 ns | 30.7356 *** | 307356 *** | 25.0324 *** | 12.4143 ns | 10.7692 *** | 8.7511 * | 7.9985 ** | 8.1526 ns |
| DeepLabV3 | 0.5157 *** | 0.6312 *** | 0.6382 * | 0.7417 *** | 0.7512 *** | 0.7983 ns | 0.8029 * | 0.8118 ns | 26.2979 *** | 24.4466 *** | 20.1742 *** | 12.1354 ns | 11.6624 *** | 8.3925 *** | 8.5411 *** | 7.3718 ns |
| PSPNet | 0.5308 *** | 0.6198 *** | 0.6059 *** | 0.7002 *** | 0.7206 *** | 0.7400 *** | 0.7592 *** | 0.7681 *** | 23.5507 *** | 17.9434 *** | 18.7494 *** | 12.3391 *** | 11.1914 *** | 11.0718 *** | 9.4149 *** | 9.7201 *** |
| Fast-SCNN | 0.2311 *** | 0.3133 *** | 0.4271 *** | 0.5263 *** | 0.5418 *** | 0.6374 *** | 0.6328 *** | 0.6501 *** | 55.0956 *** | 36.9229 *** | 36.3592 *** | 24.4900 *** | 23.6173 *** | 16.7035 *** | 17.6839 *** | 17.6589 *** |
| TGANet | 0.3387 *** | 0.3503 *** | 0.3886 *** | 0.6406 *** | 0.6435 *** | 0.7145 *** | 0.7216 *** | 0.7069 *** | 57.5892 *** | 56.7400 *** | 44.2247 *** | 19.1582 *** | 19.0647 *** | 14.6703 *** | 14.8420 *** | 15.9357 *** |
| SegFormer | 0.2637 *** | 0.4378 *** | 0.2495 *** | 0.6084 *** | 0.4136 *** | 0.6591 *** | 0.4737 *** | 0.6593 *** | 65.4658 *** | 50.6095 *** | 67.2951 *** | 30.3880 *** | 47.6450 *** | 18.7880 *** | 42.6189 *** | 19.2131 *** |
| Unet++ | 0.2486 *** | 0.2915 *** | 0.3562 *** | 0.6098 *** | 0.6779 *** | 0.7119 *** | 0.7063 *** | 0.7405 *** | 66.9296 *** | 67.5001 *** | 68.7728 *** | 29.8018 *** | 22.7979 *** | 19.4106 *** | 19.3498 *** | 16.7502 *** |
| autoSAM | 0.4482 *** | 0.4911 *** | 0.4465 *** | 0.5844 *** | 0.5807 *** | 0.6671 *** | 0.6642 *** | 0.6682 *** | 61.9208 *** | 51.3325 *** | 47.9954 *** | 20.5984 *** | 20.8936 *** | 17.8528 *** | 17.9705 *** | 18.9544 *** |
| Mamba-Unet | 0.5040 *** | 0.5982 *** | 0.6089 *** | 0.6290 *** | 0.6534 *** | 0.6528 *** | 0.6674 *** | 0.7067 *** | 23.3985 *** | 18.8779 *** | 19.3776 *** | 18.5073 *** | 16.8112 *** | 16.9241 *** | 15.9930 *** | 14.8776 *** |
| nnSAM (FCN) | 0.5087 *** | 0.5906 *** | 0.5882 *** | 0.7564 ns | 0.7786 ns | 0.8021 ns | 0.8069 *** | 0.8010 ns | 32.6712 *** | 31.6343 *** | 23.6663 *** | 11.1222 * | 9.2595 ns | 8.1799 ns | 7.3214 *** | 8.2214 ns |
| Proposed (FCN) | 0.5419 *** | 0.6164 *** | 0.6103 *** | 0.7570 ns | 0.7818 | 0.8060 | 0.8091 | 0.8052 | 25.5238 *** | 23.3281 *** | 19.4167 *** | 10.5465 | 8.7999 | 7.9875 | 7.1839 | 7.8991 |
| nnSAM (deep) | 0.5323 *** | 0.6417 *** | 0.6435 ns | 0.7530 ns | 0.7588 *** | 0.7915 *** | 0.8031 ns | 0.8058 ns | 24.2617 *** | 19.6827 *** | 17.5183 ns | 12.6661 ns | 10.7260 *** | 9.3173 *** | 8.4043 ** | 8.0490 ns |
| Proposed (deep) | 0.5758 | 0.6584 | 0.6519 | 0.7599 | 0.7672 ** | 0.7973 * | 0.8091 ns | 0.8104 ns | 20.7766 | 17.5505 | 16.7683 | 11.8957 *** | 10.3071 *** | 8.7514 * | 7.8906 * | 7.6765 ns |
| Method | Metrics | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DICE | HD | |||||||||||||||
| Training Sample Size (n) | ||||||||||||||||
| 1 | 2 | 4 | 6 | 8 | 12 | 16 | 20 | 1 | 2 | 4 | 6 | 8 | 12 | 16 | 20 | |
| FCN | 0.4790 *** | 0.6759 *** | 0.7762 *** | 0.8210 *** | 0.8130 *** | 0.8726 ns | 0.8718 ns | 0.8751 ns | 20.0650 *** | 10.7355 *** | 5.3205 *** | 3.0799 ** | 3.3850 *** | 1.7938 ns | 1.9800 ns | 1.6347 ns |
| DeepLabV3 | 0.5657 *** | 0.7034 *** | 0.8229 *** | 0.8145 *** | 0.8350 ns | 0.8736 ns | 0.8754 ns | 0.8763 ns | 19.1857 *** | 8.0980 *** | 3.3823 *** | 4.2733 *** | 2.8859 * | 1.7468 ns | 1.7494 ns | 1.4995 ns |
| PSPNet | 0.2952 *** | 0.5397 *** | 0.6876 *** | 0.7643 *** | 0.7935 *** | 0.8342 *** | 0.8432 *** | 0.8349 *** | 36.6751 *** | 8.7706 *** | 6.3812 *** | 8.7565 *** | 7.6729 *** | 5.8643 *** | 3.9266 *** | 3.8622 *** |
| Fast-SCNN | 0.3473 *** | 0.5133 *** | 0.6423 *** | 0.6507 *** | 0.6662 *** | 0.7837 *** | 0.7746 *** | 0.7987 *** | 45.4614 *** | 31.5551 *** | 15.1610 *** | 15.2604 *** | 15.1658 *** | 6.2228 *** | 6.7428 *** | 6.1936 *** |
| TGANet | 0.5750 *** | 0.6537 *** | 0.6491 *** | 0.6970 *** | 0.7471 *** | 0.8157 *** | 0.8091 *** | 0.7976 *** | 26.2602 *** | 26.0183 *** | 31.2509 *** | 26.2529 *** | 15.8280 *** | 8.5710 *** | 9.9134 *** | 10.0348 *** |
| SegFormer | 0.6014 *** | 0.6518 *** | 0.7065 *** | 0.7574 *** | 0.7638 *** | 0.7764 *** | 0.7559 *** | 0.7578 *** | 30.1322 *** | 16.0105 *** | 14.2888 *** | 7.9404 *** | 8.1005 *** | 9.7448 *** | 9.4702 *** | 9.1201 *** |
| Unet++ | 0.5241 *** | 0.4020 *** | 0.7922 *** | 0.8215 *** | 0.7961 *** | 0.8426 *** | 0.8463 *** | 0.8563 *** | 41.3638 *** | 82.2642 *** | 9.9493 *** | 8.1613 *** | 10.7172 *** | 10.3751 *** | 9.2046 *** | 7.2565 *** |
| autoSAM | 0.4723 *** | 0.4456 *** | 0.7208 *** | 0.7627 *** | 0.7789 *** | 0.8042 *** | 0.7972 *** | 0.8076 *** | 22.5677 *** | 35.1402 *** | 12.9685 *** | 8.2359 *** | 8.5600 *** | 6.2316 *** | 5.2601 *** | 5.3802 *** |
| Mamba-Unet | 0.2567 *** | 0.3326 *** | 0.3225 *** | 0.6176 *** | 0.6935 *** | 0.7320 *** | 0.6709 *** | 0.7605 *** | 41.4738 *** | 33.2122 *** | 15.8653 *** | 13.8365 *** | 10.6301 *** | 13.8848 *** | 16.3959 *** | 8.5436 *** |
| nnSAM (FCN) | 0.6049 *** | 0.7886 ns | 0.7994 *** | 0.8172 *** | 0.8412 *** | 0.8737 ns | 0.8668 *** | 0.8720 ** | 13.0598 *** | 3.8768 ns | 4.5948 *** | 3.3072 *** | 2.6651 *** | 1.7309 ns | 1.9841 ns | 2.0956 * |
| Proposed (FCN) | 0.7141 | 0.7898 | 0.8427 | 0.8449 | 0.8432 | 0.8743 | 0.8773 | 0.8800 | 7.5007 | 4.6304 | 2.3837 | 2.3026 | 2.3543 | 1.7615 | 1.8195 | 1.5801 |
| nnSAM (deep) | 0.6347 *** | 0.7007 *** | 0.8028 *** | 0.8176 *** | 0.8395 ns | 0.8721 ns | 0.8681 ** | 0.8748 ns | 10.5907 *** | 9.2379 *** | 3.9076 *** | 3.6697 *** | 2.8225 ns | 2.0105 ns | 1.9409 ns | 1.7629 ns |
| Proposed (deep) | 0.6725 *** | 0.7282 *** | 0.8075 *** | 0.8229 *** | 0.8329 * | 0.8674 * | 0.8741 ns | 0.8784 ns | 11.8223 *** | 7.5185 *** | 4.2160 *** | 3.3738 *** | 2.6646 ns | 2.1048 ns | 2.2357 ns | 1.5954 ns |
| Ablation study (FCN) n = 1 | Modules | Metrics | |||
| FE | MD | FACM | DICE | HD | |
| × | × | × | 0.4819 | 33.53 | |
| √ | × | × | 0.5087 | 32.67 | |
| √ | √ | × | 0.5324 | 25.57 | |
| √ | √ | √ | 0.5419 | 25.52 | |
| Ablation study (DeeplabV3) n = 1 | Modules | Metrics | |||
| FE | MD | FACM | DICE | HD | |
| × | × | × | 0.5157 | 26.30 | |
| √ | × | × | 0.5323 | 24.26 | |
| √ | √ | × | 0.5689 | 20.67 | |
| √ | √ | √ | 0.5758 | 20.78 | |
| Ablation study (Unet++) n = 1 | Modules | Metrics | |||
| FE | MD | FACM | DICE | HD | |
| × | × | × | 0.2486 | 66.93 | |
| √ | × | × | 0.2430 | 68.49 | |
| √ | √ | × | 0.2592 | 66.88 | |
| √ | √ | √ | 0.2615 | 66.46 | |
| Fusion Weight of Agent and SAM | Training Sample Size (n) | |||||
|---|---|---|---|---|---|---|
| Agent | SAM | Metrics | 1 | 4 | 8 | 16 |
| 0.1 | 0.9 | DICE | 0.5758 | 0.6428 | 0.7683 | 0.7986 |
| 0.25 | 0.75 | DICE | 0.5683 | 0.6427 | 0.7672 | 0.8091 |
| 0.5 | 0.5 | DICE | 0.5581 | 0.6385 | 0.7618 | 0.8063 |
| 0.75 | 0.25 | DICE | 0.5536 | 0.6357 | 0.7591 | 0.8023 |
| 0.9 | 0.1 | DICE | 0.5518 | 0.6345 | 0.7580 | 0.8005 |
| 0.1 | 0.9 | HD | 20.7766 | 18.2607 | 10.1163 | 8.0842 |
| 0.25 | 0.75 | HD | 20.6299 | 18.4923 | 10.3071 | 7.8906 |
| 0.5 | 0.5 | HD | 20.5974 | 18.9707 | 10.6805 | 9.6143 |
| 0.75 | 0.25 | HD | 20.6733 | 19.2346 | 10.8251 | 11.4226 |
| 0.9 | 0.1 | HD | 20.6801 | 19.3462 | 10.8708 | 12.2420 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, H.; He, Y.; Cui, X.; Xie, Z. AGSAM: Agent-Guided Segment Anything Model for Automatic Segmentation in Few-Shot Scenarios. Bioengineering 2024, 11, 447. https://doi.org/10.3390/bioengineering11050447
Zhou H, He Y, Cui X, Xie Z. AGSAM: Agent-Guided Segment Anything Model for Automatic Segmentation in Few-Shot Scenarios. Bioengineering. 2024; 11(5):447. https://doi.org/10.3390/bioengineering11050447
Chicago/Turabian StyleZhou, Hao, Yao He, Xiaoxiao Cui, and Zhi Xie. 2024. "AGSAM: Agent-Guided Segment Anything Model for Automatic Segmentation in Few-Shot Scenarios" Bioengineering 11, no. 5: 447. https://doi.org/10.3390/bioengineering11050447
APA StyleZhou, H., He, Y., Cui, X., & Xie, Z. (2024). AGSAM: Agent-Guided Segment Anything Model for Automatic Segmentation in Few-Shot Scenarios. Bioengineering, 11(5), 447. https://doi.org/10.3390/bioengineering11050447






