A Deep Learning Approach for Improving Two-Photon Vascular Imaging Speeds
Abstract
1. Introduction
2. Methods
2.1. In Vivo Imaging
2.1.1. Animal Preparation
2.1.2. Image Acquisition
2.2. Image Processing
2.2.1. Image Preprocessing
2.2.2. Stitching
2.3. Semi-Synthetic Image Generation
2.3.1. Single-Frame Images
2.3.2. Multi-Frame Images
2.4. Neural Networks and Training
2.4.1. Training/Test Images
2.4.2. Hardware
2.5. Image Quality Evaluation
2.6. Vectorization
2.7. Statistical Analysis
3. Results
3.1. Structure and Analysis Pipeline Overview

3.2. Transfer Learning, Creation and Evaluation of Semi-Synthetic Training Data
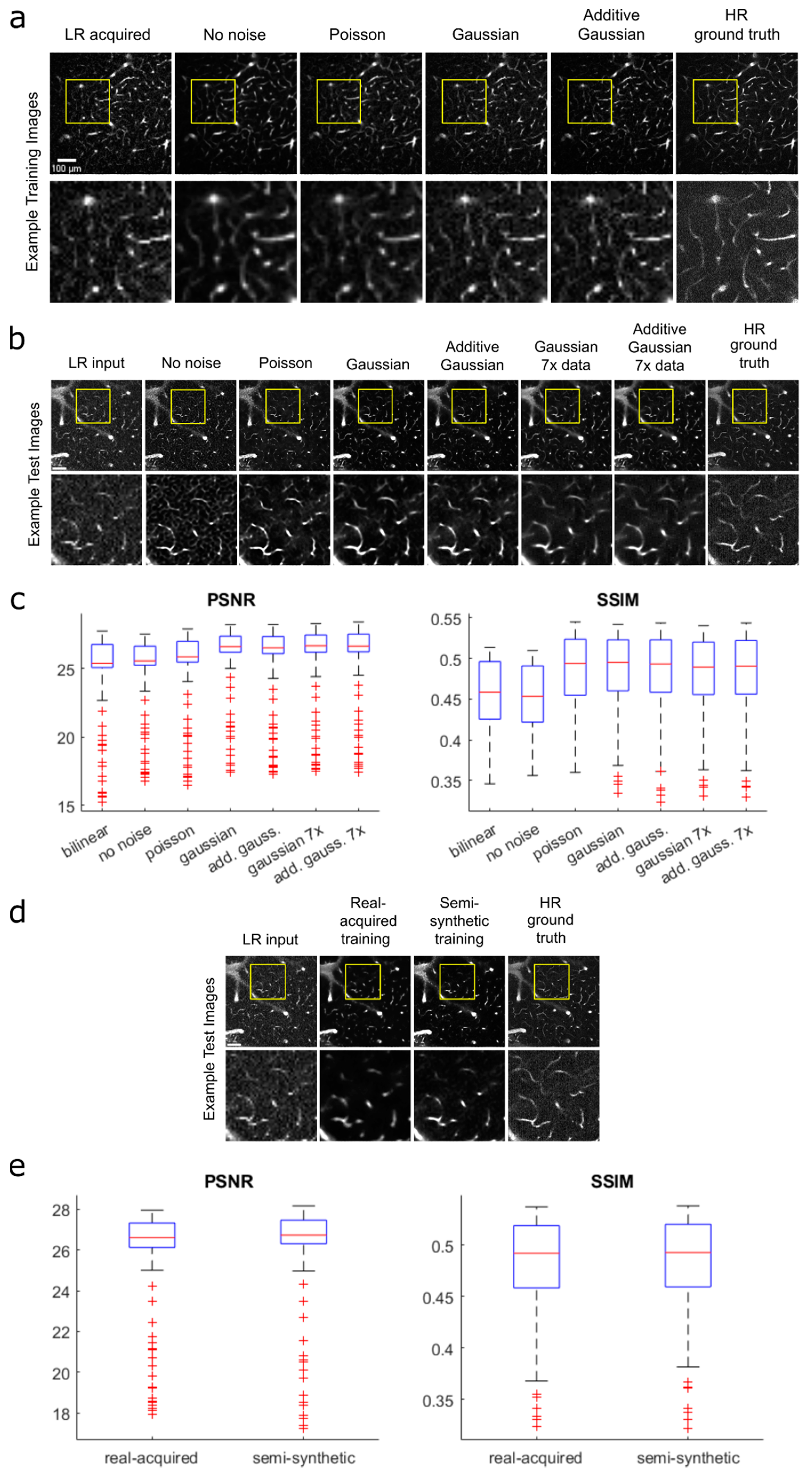
3.3. Single-Frame vs. Multi-Frame Training
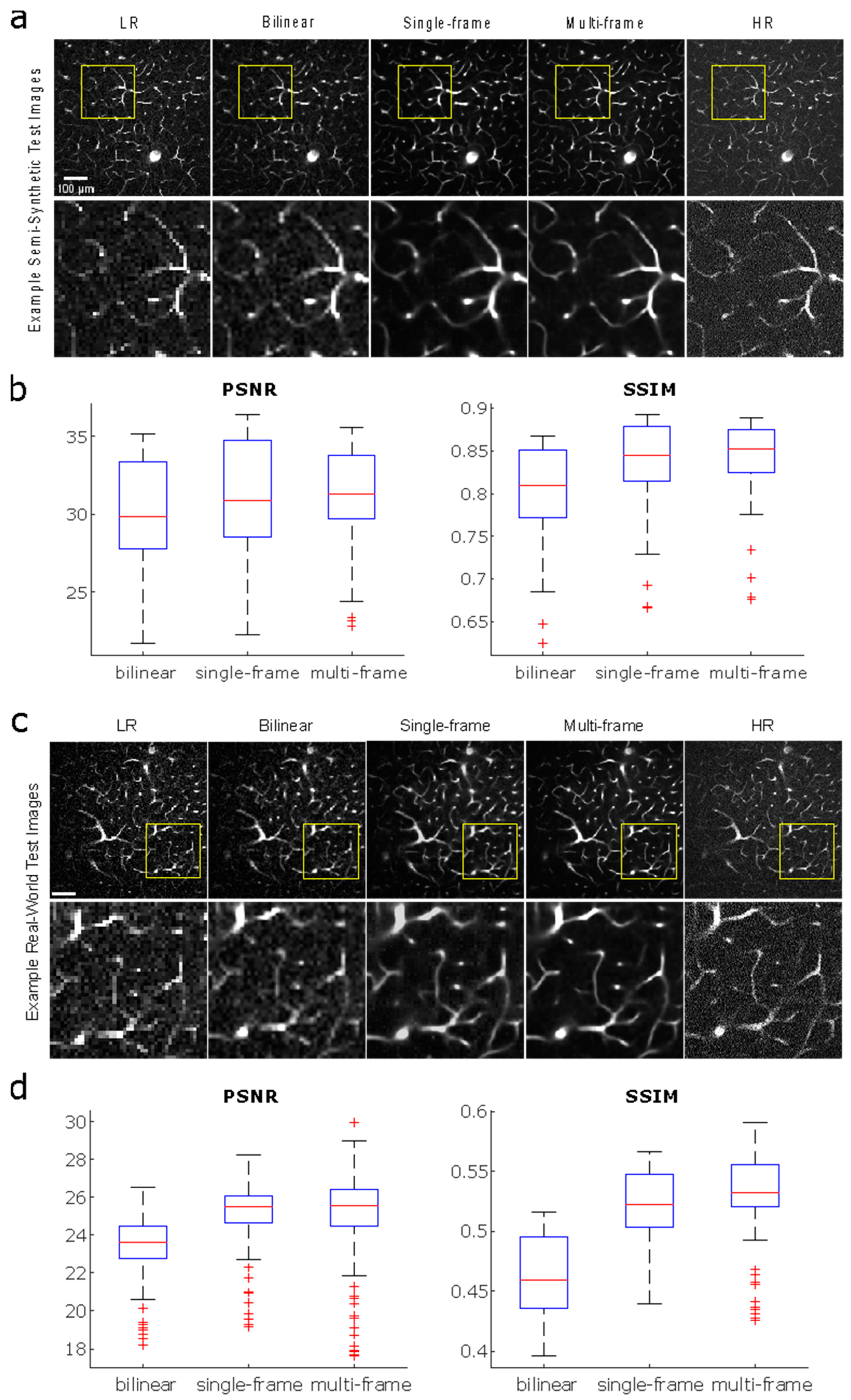
3.4. Reconstruction and Stitching of Infarct Images

3.5. Vectorization
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lecrux, C.; Hamel, E. The neurovascular unit in brain function and disease. Acta Physiol. 2011, 203, 47–59. [Google Scholar] [CrossRef] [PubMed]
- Girouard, H.; Iadecola, C. Neurovascular coupling in the normal brain and in hypertension, stroke, and Alzheimer disease. J. Appl. Physiol. 2006, 100, 328–335. [Google Scholar] [CrossRef] [PubMed]
- Meng, G.; Zhong, J.; Zhang, Q.; Wong, J.S.; Wu, J.; Tsia, K.K.; Ji, N. Ultrafast two-photon fluorescence imaging of cerebral blood circulation in the mouse brain in vivo. Proc. Natl. Acad. Sci. USA 2022, 119, e2117346119. [Google Scholar] [CrossRef] [PubMed]
- Hatakeyama, M.; Ninomiya, I.; Kanazawa, M. Angiogenesis and neuronal remodeling after ischemic stroke. Neural Regen. Res. 2020, 15, 16. [Google Scholar] [PubMed]
- Roundtable, Stroke Therapy Academic Industry. Recommendations for Standards Regarding Preclinical Neuroprotective and Restorative Drug Development. Stroke 1999, 30, 2752–2758. [Google Scholar] [CrossRef] [PubMed]
- Shih, A.Y.; Driscoll, J.D.; Drew, P.J.; Nishimura, N.; Schaffer, C.B.; Kleinfeld, D. Two-photon microscopy as a tool to study blood flow and neurovascular coupling in the rodent brain. J. Cereb. Blood Flow. Metab. 2012, 32, 1277–1309. [Google Scholar] [CrossRef]
- Voleti, V.; Patel, K.B.; Li, W.; Perez Campos, C.; Bharadwaj, S.; Yu, H.; Ford, C.; Casper, M.J.; Yan, R.W.; Liang, W.; et al. Real-time volumetric microscopy of in vivo dynamics and large-scale samples with SCAPE 2.0. Nat. Methods 2019, 16, 1054–1062. [Google Scholar] [CrossRef]
- Beaulieu, D.R.; Davison, I.G.; Kılıç, K.; Bifano, T.G.; Mertz, J. Simultaneous multiplane imaging with reverberation two-photon microscopy. Nat. Methods 2020, 17, 283–286. [Google Scholar] [CrossRef]
- Mahou, P.; Vermot, J.; Beaurepaire, E.; Supatto, W. Multicolor two-photon light-sheet microscopy. Nat. Methods 2014, 11, 600–601. [Google Scholar] [CrossRef]
- Wu, J.; Liang, Y.; Chen, S.; Hsu, C.L.; Chavarha, M.; Evans, S.W.; Shi, D.; Lin, M.Z.; Tsia, K.K.; Ji, N. Kilohertz two-photon fluorescence microscopy imaging of neural activity in vivo. Nat. Methods 2020, 17, 287–290. [Google Scholar] [CrossRef]
- Sità, L.; Brondi, M.; Lagomarsino de Leon Roig, P.; Curreli, S.; Panniello, M.; Vecchia, D.; Fellin, T. A deep-learning approach for online cell identification and trace extraction in functional two-photon calcium imaging. Nat. Commun. 2022, 13, 1529. [Google Scholar] [CrossRef] [PubMed]
- Soltanian-Zadeh, S.; Sahingur, K.; Blau, S.; Gong, Y.; Farsiu, S. Fast and robust active neuron segmentation in two-photon calcium imaging using spatiotemporal deep learning. Proc. Natl. Acad. Sci. USA 2019, 116, 8554–8563. [Google Scholar] [CrossRef] [PubMed]
- Luo, L.; Xu, Y.; Pan, J.; Wang, M.; Guan, J.; Liang, S.; Li, Y.; Jia, H.; Chen, X.; Li, X.; et al. Restoration of Two-Photon Ca2+ Imaging Data Through Model Blind Spatiotemporal Filtering. Front. Neurosci. 2021, 15, 630250. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Negishi, M.; Urakubo, H.; Kasai, H.; Ishii, S. Mu-net: Multi-scale U-net for two-photon microscopy image denoising and restoration. Neural Netw. 2020, 125, 92–103. [Google Scholar] [CrossRef] [PubMed]
- Tahir, W.; BoasDavid, A. Anatomical Modeling of Brain Vasculature in Two-Photon Microscopy by Generalizable Deep Learning. BME Front. 2021, 2021, 8620932. [Google Scholar] [CrossRef]
- Guan, H.; Li, D.; Park, H.C.; Li, A.; Yue, Y.; Gau, Y.T.A.; Li, M.J.; Bergles, D.E.; Lu, H.; Li, X. Deep-learning two-photon fiberscopy for video-rate brain imaging in freely-behaving mice. Nat. Commun. 2022, 13, 1534. [Google Scholar] [CrossRef]
- Schrandt, C.J.; Kazmi, S.M.S.; Jones, T.A.; Dunn, A.K. Chronic Monitoring of Vascular Progression after Ischemic Stroke Using Multiexposure Speckle Imaging and Two-Photon Fluorescence Microscopy. J. Cereb. Blood Flow. Metab. 2015, 35, 933–942. [Google Scholar] [CrossRef]
- Zhou, A.; Engelmann, S.A.; Mihelic, S.A.; Tomar, A.; Hassan, A.M.; Dunn, A.K. Evaluation of resonant scanning as a high-speed imaging technique for two-photon imaging of cortical vasculature. Biomed. Opt. Express 2022, 13, 1374. [Google Scholar] [CrossRef]
- Williamson, M.R.; Engelmann, S.A.; Zhou, A.; Hassan, A.M.; Jarrett, J.W.; Perillo, E.P.; Tomar, A.; Spence, D.J.; Jones, T.A.; Dunn, A.K. Diamond Raman laser and Yb fiber amplifier for in vivo multiphoton fluorescence microscopy. Biomed. Opt. Express 2022, 13, 1888–1898. [Google Scholar]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef]
- Preibisch, S.; Saalfeld, S.; Tomancak, P. Globally optimal stitching of tiled 3D microscopic image acquisitions. Bioinformatics 2009, 25, 1463–1465. [Google Scholar] [CrossRef] [PubMed]
- Fang, L.; Monroe, F.; Novak, S.W.; Kirk, L.; Schiavon, C.R.; Yu, S.B.; Zhang, T.; Wu, M.; Kastner, K.; Latif, A.A.; et al. Deep learning-based point-scanning super-resolution imaging. Nat. Methods 2021, 18, 406–416. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Bovik, A.C.; Sheikh, H.R.; Simoncelli, E.P. Image quality assessment: From error visibility to structural similarity. IEEE Trans. Image Process. 2004, 13, 600–612. [Google Scholar] [CrossRef] [PubMed]
- Mihelic, S.A.; Sikora, W.A.; Hassan, A.M.; Williamson, M.R.; Jones, T.A.; Dunn, A.K. Segmentation-Less, Automated, Vascular Vectorization. PLoS Comput. Biol. 2021, 17, e1009451. [Google Scholar] [CrossRef] [PubMed]
- Ting, K.M. Confusion Matrix. Encycl. Mach. Learn. 2011, 2011, 209. [Google Scholar] [CrossRef]
- Abdellah, M.; Guerrero, N.R.; Lapere, S.; Coggan, J.S.; Keller, D.; Coste, B.; Dagar, S.; Courcol, J.D.; Markram, H.; Schürmann, F. Interactive visualization and analysis of morphological skeletons of brain vasculature networks with VessMorphoVis. Bioinformatics 2020, 36, i534–i541. [Google Scholar] [CrossRef]
- Linninger, A.; Hartung, G.; Badr, S.; Morley, R. Mathematical synthesis of the cortical circulation for the whole mouse brain-part I. Theory and image integration. Comput. Biol. Med. 2019, 110, 265–275. [Google Scholar] [CrossRef] [PubMed]
- Hartung, G.; Badr, S.; Mihelic, S.; Dunn, A.; Cheng, X.; Kura, S.; Boas, D.A.; Kleinfeld, D.; Alaraj, A.; Linninger, A.A. Mathematical synthesis of the cortical circulation for the whole mouse brain—Part II: Microcirculatory closure. Microcirculation 2021, 28, e12687. [Google Scholar] [CrossRef]
- Hopt, A.; Neher, E. Highly Nonlinear Photodamage in Two-Photon Fluorescence Microscopy. Biophys. J. 2001, 80, 2029–2036. [Google Scholar] [CrossRef]
- Wang, T.; Wu, C.; Ouzounov, D.G.; Gu, W.; Xia, F.; Kim, M.; Yang, X.; Warden, M.R.; Xu, C. Quantitative analysis of 1300-nm three-photon calcium imaging in the mouse brain. eLife 2020, 9, e53205. [Google Scholar] [CrossRef]
- Cao, R.; Li, J.; Ning, B.; Sun, N.; Wang, T.; Zuo, Z.; Hu, S. Functional and oxygen-metabolic photoacoustic microscopy of the awake mouse brain. Neuroimage 2017, 150, 77–87. [Google Scholar] [CrossRef] [PubMed]
- Jafari, C.Z.; Mihelic, S.A.; Engelmann, S.; Dunn, A.K. High-resolution three-dimensional blood flow tomography in the subdiffuse regime using laser speckle contrast imaging. J. Biomed. Opt. 2022, 27, 083011. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, A.; Mihelic, S.A.; Engelmann, S.A.; Tomar, A.; Dunn, A.K.; Narasimhan, V.M. A Deep Learning Approach for Improving Two-Photon Vascular Imaging Speeds. Bioengineering 2024, 11, 111. https://doi.org/10.3390/bioengineering11020111
Zhou A, Mihelic SA, Engelmann SA, Tomar A, Dunn AK, Narasimhan VM. A Deep Learning Approach for Improving Two-Photon Vascular Imaging Speeds. Bioengineering. 2024; 11(2):111. https://doi.org/10.3390/bioengineering11020111
Chicago/Turabian StyleZhou, Annie, Samuel A. Mihelic, Shaun A. Engelmann, Alankrit Tomar, Andrew K. Dunn, and Vagheesh M. Narasimhan. 2024. "A Deep Learning Approach for Improving Two-Photon Vascular Imaging Speeds" Bioengineering 11, no. 2: 111. https://doi.org/10.3390/bioengineering11020111
APA StyleZhou, A., Mihelic, S. A., Engelmann, S. A., Tomar, A., Dunn, A. K., & Narasimhan, V. M. (2024). A Deep Learning Approach for Improving Two-Photon Vascular Imaging Speeds. Bioengineering, 11(2), 111. https://doi.org/10.3390/bioengineering11020111






