A Unified Multi-Task Learning Model with Joint Reverse Optimization for Simultaneous Skin Lesion Segmentation and Diagnosis
Abstract
1. Introduction
2. Related Works
2.1. AI-Based Segmentation Methods
2.2. AI-Based Classification Methods
2.3. Integrated AI-Based Methods
| Reference | Task | Technique | Dataset | TNR | TPR | Accuracy | F1 Score | IOU | DSC |
|---|---|---|---|---|---|---|---|---|---|
| Yuan et al., 2017 [4] | AI-based Segmentation Methods | Deep FCN with Jaccard distance | ISIC 2016 | 96.7 | 90.4 | 95.3 | - | 83.6 | 90.3 |
| Li et al., 2019 [14] | Dense deconvolutional network | ISIC 2016 | 96.0 | 95.1 | 95.9 | - | 87.0 | 93.1 | |
| ISIC 2017 | 98.4 | 82.5 | 93.9 | 76.5 | 86.6 | ||||
| Al-masni et al., 2018 [15] | Deep full-resolution CNN | ISIC 2017 | 96.69 | 85.40 | 94.03 | - | 77.11 | 87.08 | |
| Xie et al., 2020 [16] | High-resolution CNN | ISIC 2016 | 96.4 | 87.0 | 93.8 | - | 85.8 | 91.8 | |
| ISIC 2017 | 96.4 | 87.0 | 93.8 | 78.3 | 86.2 | ||||
| PH2 | 94.2 | 96.3 | 94.9 | 85.7 | 91.9 | ||||
| Wu et al., 2021 [18] | CNN with adaptive dual attention module | ISIC 2017 | 96.28 | 90.61 | 95.70 | - | 82.55 | 89.69 | |
| ISIC 2018 | 94.10 | 94.2 | 94.70 | 84.4 | 90.8 | ||||
| Cao et al., 2023 [19] | Global and local inter-pixel correlations learning network | ISIC 2018 | 92.9 | 94.1 | 94.4 | - | 83.9 | 90.3 | |
| Al-masni et al., 2021 [20] | Contextual multi-scale multi-level network | ISIC 2017 | 96.23 | 87.69 | 93.93 | - | 77.65 | 85.78 | |
| Wu et al., 2022 [21] | Dual encoder with CNNs and Transformer | ISIC 2016 | 96.02 | 92.59 | 96.04 | - | - | 91.59 | |
| ISIC 2017 | 97.25 | 83.92 | 93.26 | - | - | 85.00 | |||
| ISIC 2018 | 96.99 | 91.00 | 95.78 | - | - | 89.03 | |||
| PH2 | 97.41 | 94.41 | 97.03 | - | - | 94.40 | |||
| Zhu et al., 2024 [23] | Multi-spatial-shift MLP-based U-Net | ISIC 2017 | 98.28 | 91.31 | - | - | - | 92.08 | |
| ISIC 2018 | 97.71 | 90.15 | - | - | - | 91.03 | |||
| PH2 | 97.97 | 96.50 | - | - | - | 96.40 | |||
| Li et al., 2024 [27] | Uncertainty self-learning network | ISIC 2017 | 93.7 | 88.6 | 90.5 | - | 68.5 | 80.5 | |
| ISIC 2018 | 87.8 | 90.9 | 88.5 | - | 68.3 | 80.8 | |||
| PH2 | 93.1 | 93.6 | 92.4 | - | 80.1 | 88.9 | |||
| Cheong et al., 2021 [28] | AI-based Classification Methods | SVM with the radial basis function | ISIC 2016 | 98.49 | 96.68 | 97.58 | - | - | - |
| Xie et al., 2017 [32] | Back propagation and fuzzy NN | Xanthous | 93.75 | 95.00 | 94.17 | - | - | - | |
| Caucasians | 95.00 | 83.33 | 91.11 | - | - | - | |||
| Abbes et al., 2021 [33] | KNN and Feature extraction | Collected Dataset | 89.00 | 96.00 | 92.00 | - | - | - | |
| Patil and Bellary, 2022 [36] | CNN with similarity measure | UCO AYRNA | 96.33 | 96.03 | 96.0 | 95.96 | - | - | |
| Yu et al., 2017 [5] | Integrated AI-based Methods (non-end-to-end approaches) | Very deep residual networks | ISIC 2016 | 94.1 | 50.7 | 85.5 | - | - | - |
| Al-masni et al., 2020 [8] | Full-resolution convolutional network | ISIC 2016 | 71.40 | 81.80 | 81.79 | 82.59 | - | - | |
| ISIC 2017 | 80.62 | 75.33 | 81.57 | 75.75 | - | - | |||
| ISIC 2018 | 87.16 | 81.00 | 89.28 | 81.28 | - | - | |||
| Dhivyaa et al., 2020 [39] | Decision trees and random forest | ISIC 2017 | 99.0 | 87.7 | 97.3 | - | - | - | |
| Balaji et al., 2020 [40] | Dynamic graph cut and Naive Bayes classifier | ISIC 2017 | 70.1 | 91.7 | 72.7 | - | - | - | |
| Kadirappa et al., 2023 [43] | SASegNet and EfficientNet B1 | ISIC 2017 | 97.3 | 95.6 | 95.60 | 95.4 | - | - | |
| ISIC 2018 | 95.4 | 92.5 | 92.73 | 92.8 | - | - | |||
| ISIC 2019 | 97.7 | 92.4 | 91.73 | 92.5 | - | - | |||
| ISIC 2020 | 92.4 | 90.6 | 91.19 | 90.7 | - | - | |||
| Xie et al., 2020 [44] | Mutual bootstrapping DCNN | ISIC 2017 | 93.0 | 78.6 | 90.4 | - | - | - | |
| PH2 | 93.8 | 95.0 | 94.0 | - | - | - | |||
| Al-masni and Al-Shamiri, 2023 [47] | Integrated AI-based Methods (end-to-end approaches) | nnU-Net and FC-NN | Segmentation | ||||||
| ISIC 2016 | - | - | - | - | - | 89.03 | |||
| Classification | |||||||||
| ISIC 2016 | 89.47 | 80.47 | - | 79.94 | - | - | |||
| Jin et al., 2021 [48] | Cascade knowledge diffusion | Segmentation | |||||||
| ISIC 2017 | 96.1 | 88.7 | 94.6 | - | 80.0 | 87.7 | |||
| ISIC 2018 | 90.4 | 96.7 | 93.4 | - | 79.4 | 87.7 | |||
| Classification | |||||||||
| ISIC 2017 | 92.5 | 70.0 | 88.1 | - | - | - | |||
| ISIC 2018 | 97.6 | 80.2 | 96.3 | - | - | - | |||
| Song et al., 2020 [49] | End-to-end multi-task deep learning | Segmentation | |||||||
| ISIC 2017 | 98.5 | 88.8 | 95.6 | - | 84.9 | 91.1 | |||
| Classification | |||||||||
| ISIC 2016 | 72.3 | 99.6 | 89.1 | - | - | - | |||
| ISIC 2017 | 73.1 | 97.7 | 81.3 | - | - | - | |||
| He et al., 2023 [50] | MTL-CNN | Segmentation | |||||||
| ISIC 2016 | 97.5 | 93.8 | 97.2 | - | 87.9 | 93.4 | |||
| ISIC 2017 | 98.3 | 88.6 | 95.5 | - | 81.5 | 88.7 | |||
| Xiangya-Clinic | 97.8 | 93.1 | 96.9 | - | 86.4 | 92.4 | |||
| Classification | |||||||||
| ISIC 2016 | 96.3 | 67.3 | 88.5 | - | - | - | |||
| ISIC 2017 | 93.0 | 76.8 | 90.7 | - | - | - | |||
| Xiangya-Clinic | 86.6 | 97.0 | 95.9 | - | - | - | |||
| Yang et al., 2017 [9] | Multi-task deep learning | Segmentation | |||||||
| ISIC 2017 | 98.5 | 84.9 | 95.6 | - | 76.0 | 84.6 | |||
| PH2 | 96.0 | 97.3 | 96.5 | - | 88.2 | 93.1 | |||
| Classification | |||||||||
| ISIC 2017 | 92.5 | 67.8 | 85.0 | - | - | - | |||
| PH2 | 93.6 | 94.3 | 93.3 | - | - | - | |||
| Chen et al., 2018 [10] | MTL with feature passing module | Segmentation | |||||||
| ISIC 2017 | - | - | 94.4 | - | 78.7 | 86.8 | |||
| Classification | |||||||||
| ISIC 2017 | - | - | 80.1 | - | - | - |
3. Materials and Methods
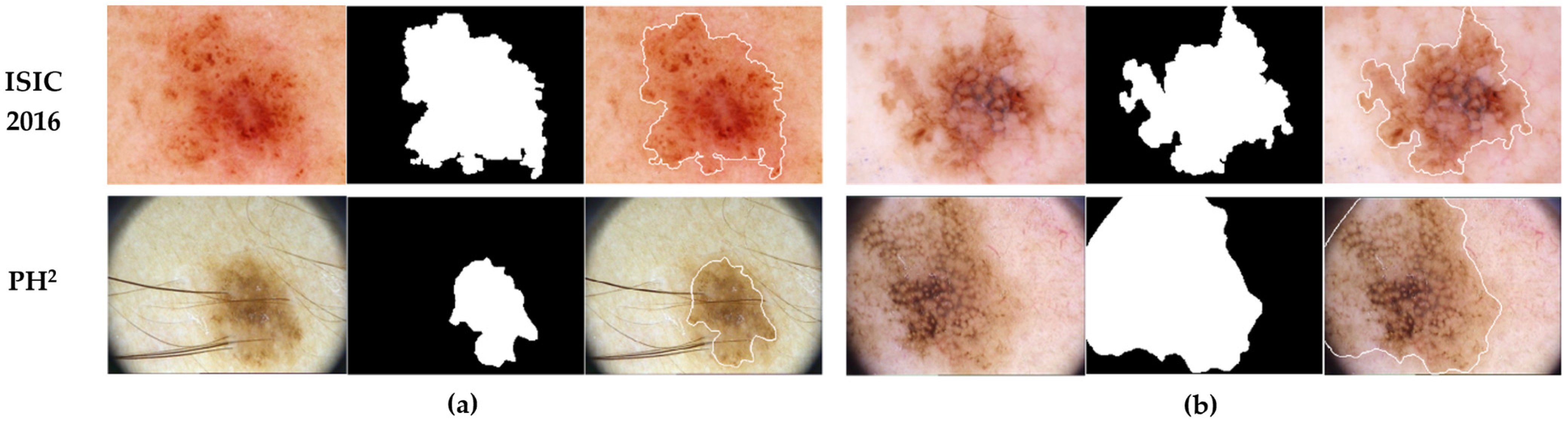
3.1. Dataset
3.2. Data Preparation
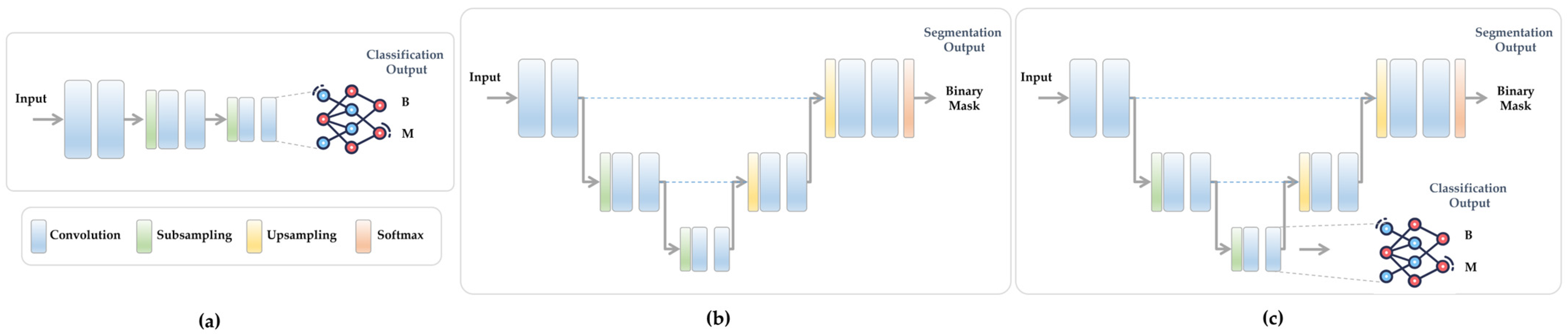
3.3. What Is Multi-Task Learning (MTL)?
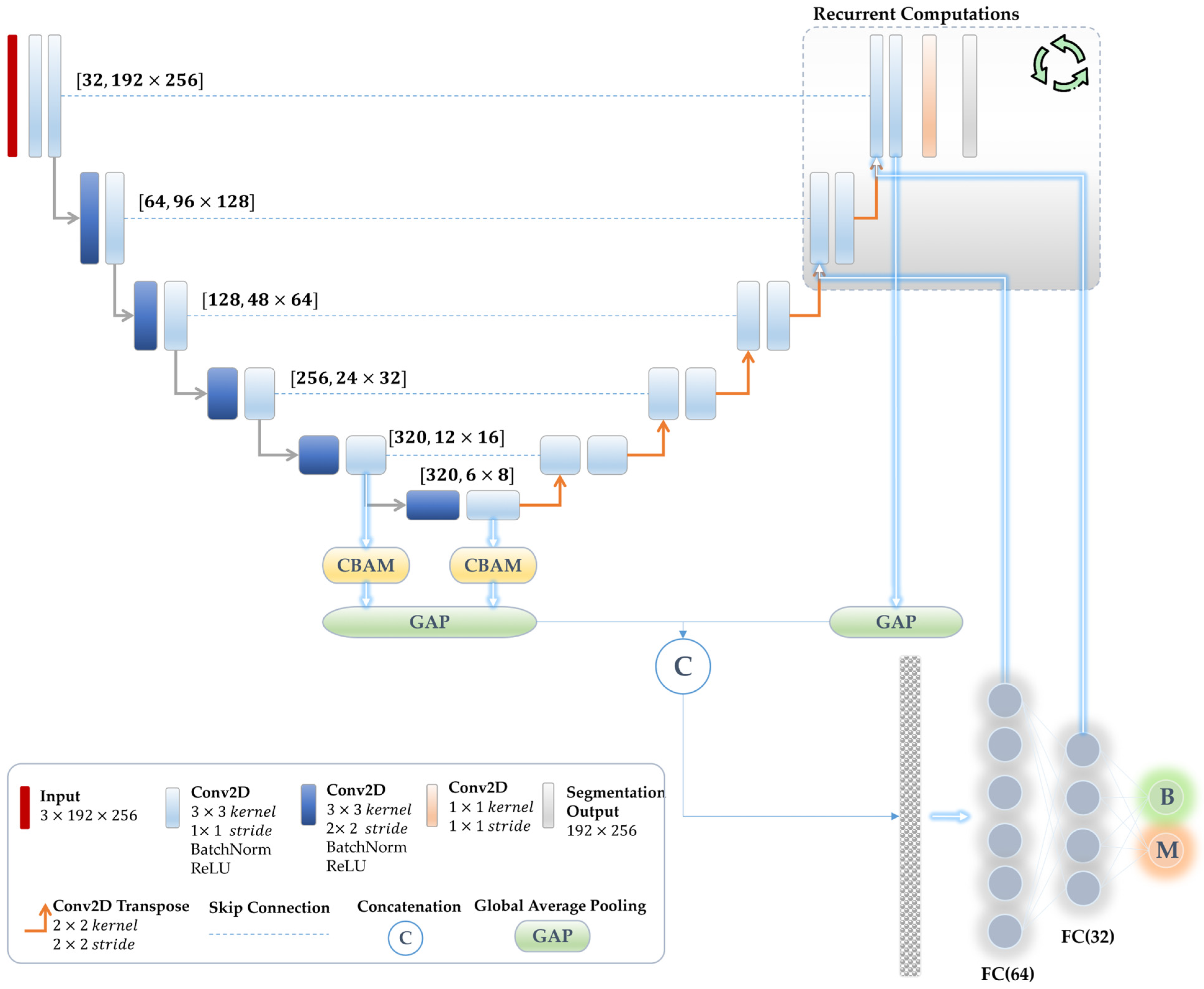
3.4. Proposed Multi-Task Learning Model
3.4.1. Network Configuration
3.4.2. Joint Reverse Learning
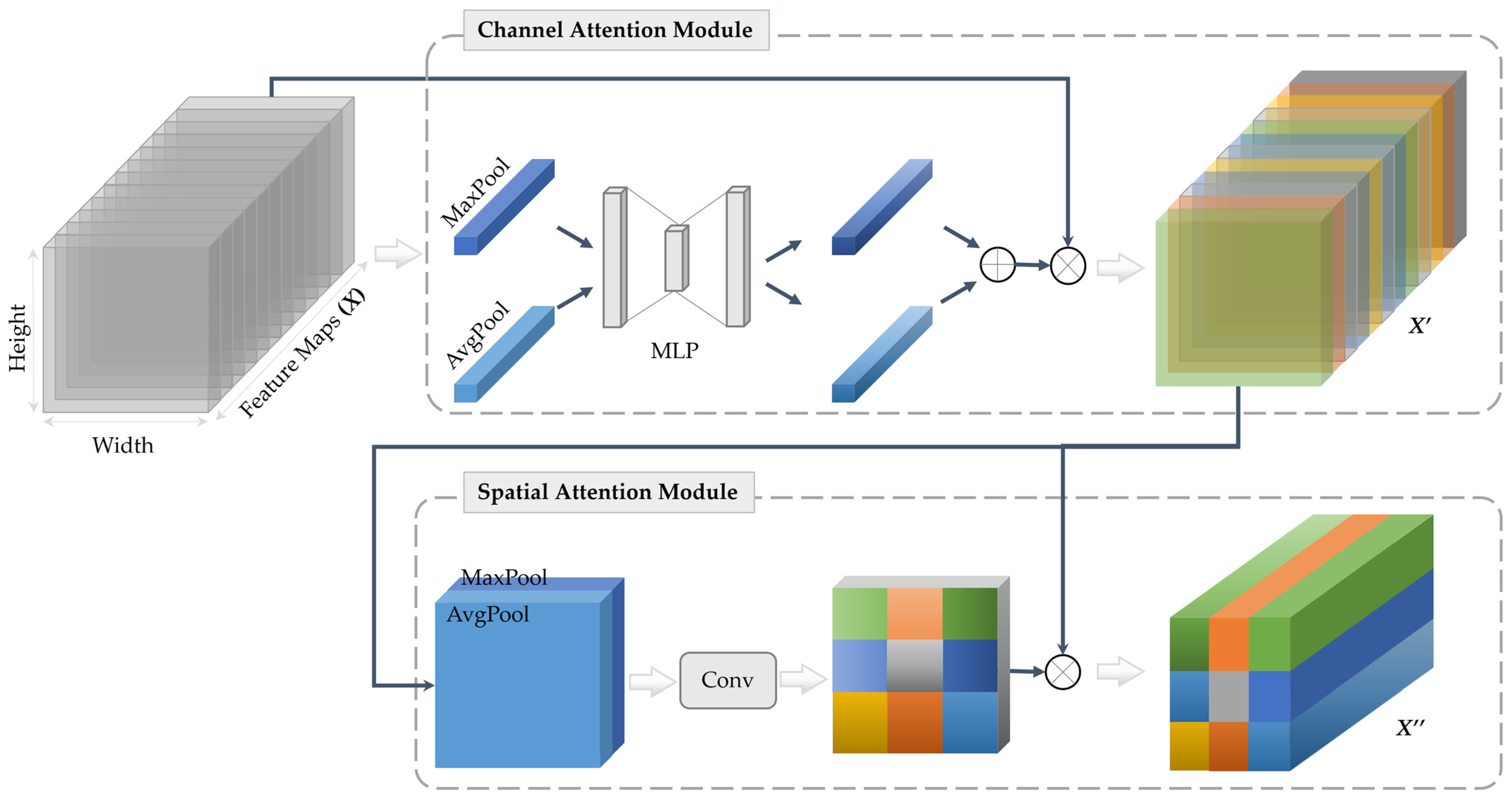
3.4.3. Attention Mechanism
3.4.4. Implementation Details
3.5. Evaluation Measures
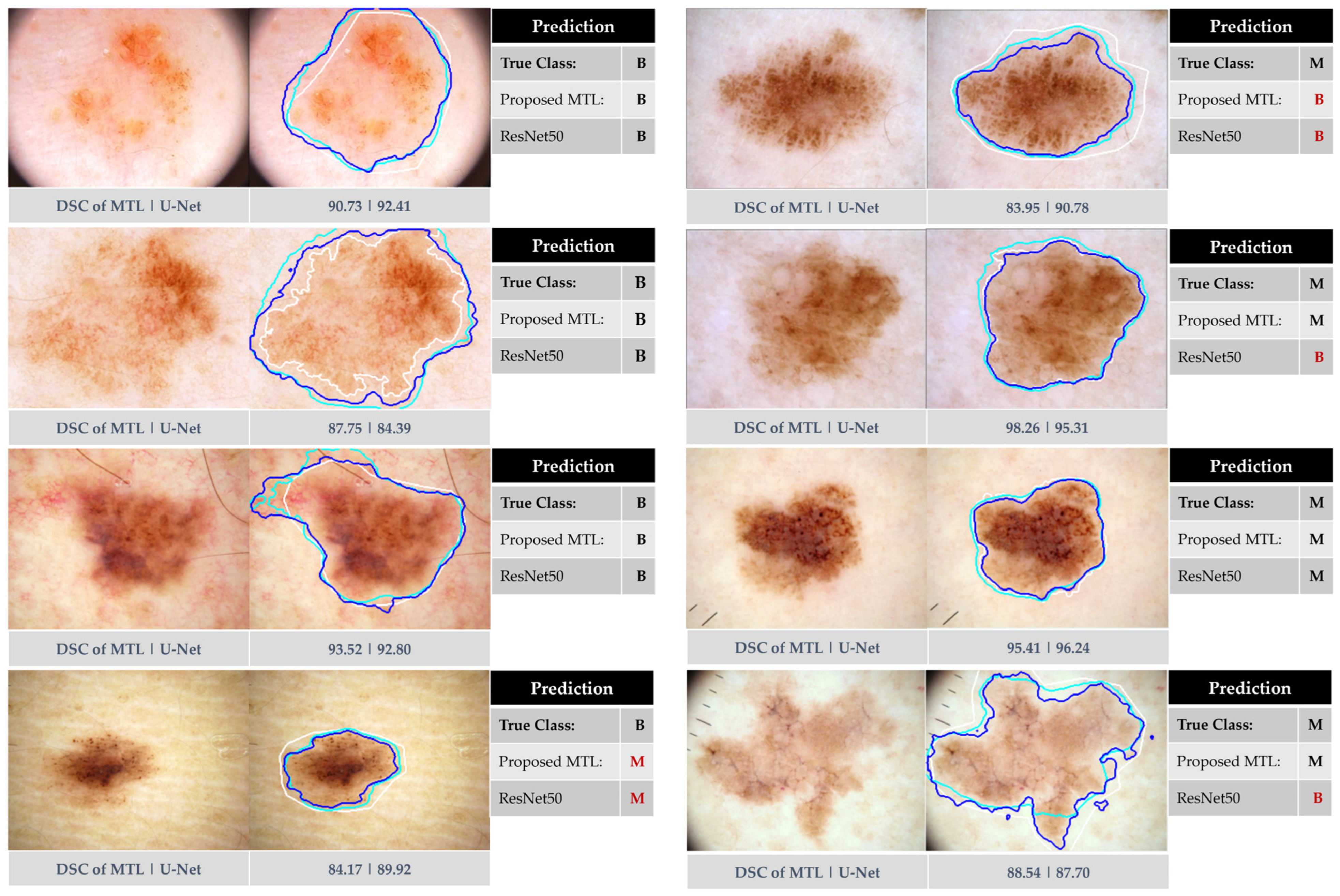
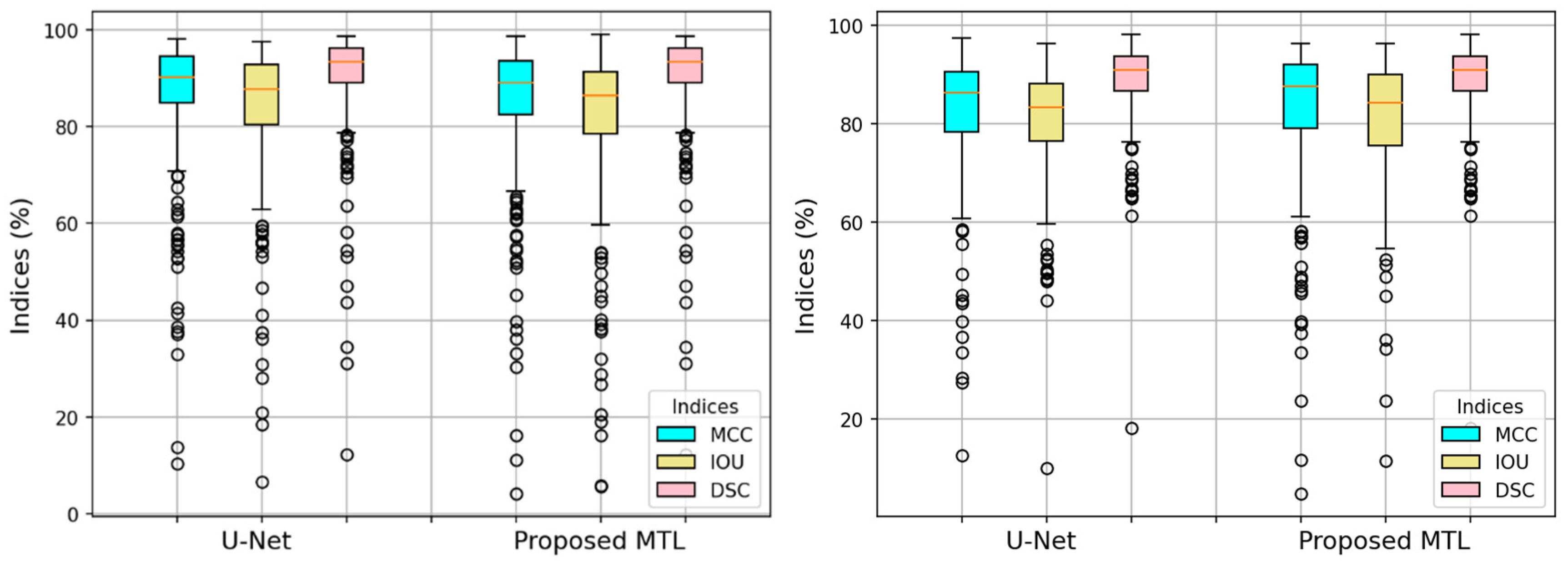
4. Experimental Results
4.1. Baseline Experiments
4.2. Ablation Study
5. Discussion
5.1. Evaluation of Additional PH2 Dataset
5.2. Comparison Against Previous Works
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Siegel, R.L.; Giaquinto, A.N.; Jemal, A. Cancer statistics, 2024. CA Cancer J. Clin. 2024, 74, 12–49. [Google Scholar] [CrossRef] [PubMed]
- Rigel, D.S.; Russak, J.; Friedman, R. The Evolution of Melanoma Diagnosis: 25 Years Beyond the ABCDs. CA Cancer J. Clin. 2010, 60, 301–316. [Google Scholar] [CrossRef] [PubMed]
- Nachbar, F.; Stolz, W.; Merkle, T.; Cognetta, A.B.; Vogt, T.; Landthaler, M.; Bilek, P.; Braunfalco, O.; Plewig, G. The Abcd Rule of Dermatoscopy—High Prospective Value in the Diagnosis of Doubtful Melanocytic Skin-Lesions. J. Am. Acad. Dermatol. 1994, 30, 551–559. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.D.; Chao, M.; Lo, Y.C. Automatic Skin Lesion Segmentation Using Deep Fully Convolutional Networks With Jaccard Distance. IEEE Trans. Med. Imaging 2017, 36, 1876–1886. [Google Scholar] [CrossRef]
- Yu, L.Q.; Chen, H.; Dou, Q.; Qin, J.; Heng, P.A. Automated Melanoma Recognition in Dermoscopy Images via Very Deep Residual Networks. IEEE Trans. Med. Imaging 2017, 36, 994–1004. [Google Scholar] [CrossRef]
- Esteva, A.; Kuprel, B.; Novoa, R.A.; Ko, J.; Swetter, S.M.; Blau, H.M.; Thrun, S. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017, 542, 115–118. [Google Scholar] [CrossRef]
- Zhang, J.P.; Xie, Y.T.; Wu, Q.; Xia, Y. Medical image classification using synergic deep learning. Med. Image Anal. 2019, 54, 10–19. [Google Scholar] [CrossRef]
- Al-masni, M.A.; Kim, D.H.; Kim, T.S. Multiple skin lesions diagnostics via integrated deep convolutional networks for segmentation and classification. Comput. Methods Programs Biomed. 2020, 190, 105351. [Google Scholar] [CrossRef]
- Yang, X.; Zeng, Z.; Yeo, S.Y.; Tan, C.; Tey, H.L.; Su, Y. A novel multi-task deep learning model for skin lesion segmentation and classification. arXiv 2017, arXiv:1703.01025. [Google Scholar]
- Chen, S.; Wang, Z.; Shi, J.; Liu, B.; Yu, N. A multi-task framework with feature passing module for skin lesion classification and segmentation. In Proceedings of the IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018), Washington, DC, USA, 24 May 2018; pp. 1126–1129. [Google Scholar]
- Zhang, Z.; Gao, J.; Li, S.; Wang, H. RMCNet: A Liver Cancer Segmentation Network Based on 3D Multi-Scale Convolution, Attention, and Residual Path. Bioengineering 2024, 11, 1073. [Google Scholar] [CrossRef]
- Xu, Y.; Quan, R.; Xu, W.; Huang, Y.; Chen, X.; Liu, F. Advances in Medical Image Segmentation: A Comprehensive Review of Traditional, Deep Learning and Hybrid Approaches. Bioengineering 2024, 11, 1034. [Google Scholar] [CrossRef] [PubMed]
- Ramakrishnan, V.; Artinger, A.; Daza Barragan, L.A.; Daza, J.; Winter, L.; Niedermair, T.; Itzel, T.; Arbelaez, P.; Teufel, A.; Cotarelo, C.L. Nuclei Detection and Segmentation of Histopathological Images Using a Feature Pyramidal Network Variant of a Mask R-CNN. Bioengineering 2024, 11, 994. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; He, X.Z.; Zhou, F.; Yu, Z.; Ni, D.; Chen, S.P.; Wang, T.F.; Lei, B.Y. Dense Deconvolutional Network for Skin Lesion Segmentation. IEEE J. Biomed. Health Inform. 2019, 23, 527–537. [Google Scholar] [CrossRef]
- Al-masni, M.A.; Al-antari, M.A.; Choi, M.T.; Han, S.M.; Kim, T.S. Skin lesion segmentation in dermoscopy images via deep full resolution convolutional networks. Comput. Methods Programs Biomed. 2018, 162, 221–231. [Google Scholar] [CrossRef] [PubMed]
- Xie, F.Y.; Yang, J.W.; Liu, J.; Jiang, Z.G.; Zheng, Y.S.; Wang, Y.K. Skin lesion segmentation using high-resolution convolutional neural network. Comput. Methods Programs Biomed. 2020, 186, 105241. [Google Scholar] [CrossRef]
- Sun, Y.H.; Dai, D.W.; Zhang, Q.N.; Wang, Y.Q.; Xu, S.H.; Lian, C.F. MSCA-Net: Multi-scale contextual attention network for skin lesion segmentation. Pattern Recogn. 2023, 139, 109524. [Google Scholar] [CrossRef]
- Wu, H.S.; Pan, J.Q.; Li, Z.Y.; Wen, Z.K.; Qin, J. Automated Skin Lesion Segmentation Via an Adaptive Dual Attention Module. IEEE Trans. Med. Imaging 2021, 40, 357–370. [Google Scholar] [CrossRef]
- Cao, W.W.; Yuan, G.; Liu, Q.; Peng, C.T.; Xie, J.; Yang, X.D.; Ni, X.Y.; Zheng, J. ICL-Net: Global and Local Inter-Pixel Correlations Learning Network for Skin Lesion Segmentation. IEEE J. Biomed. Health Inform. 2023, 27, 145–156. [Google Scholar] [CrossRef]
- Al-masni, M.A.; Kim, D.H. CMM-Net: Contextual multi-scale multi-level network for efficient biomedical image segmentation. Sci. Rep. 2021, 11, 10191. [Google Scholar] [CrossRef]
- Wu, H.S.; Chen, S.H.; Chen, G.L.; Wang, W.; Lei, B.Y.; Wen, Z.K. FAT-Net: Feature adaptive transformers for automated skin lesion segmentation. Med. Image Anal. 2022, 76, 102327. [Google Scholar] [CrossRef]
- Tschandl, P.; Sinz, C.; Kittler, H. Domain-specific classification-pretrained fully convolutional network encoders for skin lesion segmentation. Comput. Biol. Med. 2019, 104, 111–116. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.H.; Tian, J.Y.; Chen, M.Z.; Chen, L.N.; Chen, J.X. MSS-UNet: A Multi-Spatial-Shift MLP-based UNet for skin lesion segmentation. Comput. Biol. Med. 2024, 168, 107719. [Google Scholar] [CrossRef]
- Li, H.; Ding, J.; Shi, X.; Zhang, Q.; Yu, P.; Li, H. D-SAT: Dual semantic aggregation transformer with dual attention for medical image segmentation. Phys. Med. Biol. 2024, 69, 015013. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Li, J.; Zou, Y.; Wang, T. ETU-Net: Edge enhancement-guided U-Net with transformer for skin lesion segmentation. Phys. Med. Biol. 2024, 69, 015001. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.; Lin, J.Y.; Feng, C.M.; Lu, G.M. GAN inversion-based semi-supervised learning for medical image segmentation. Biomed. Signal Process. Control. 2024, 88, 105536. [Google Scholar] [CrossRef]
- Li, X.F.; Peng, B.; Hu, J.; Ma, C.Y.; Yang, D.P.; Xie, Z.Y. USL-Net: Uncertainty self-learning network for unsupervised skin lesion segmentation. Biomed. Signal Process. Control 2024, 89, 105769. [Google Scholar] [CrossRef]
- Cheong, K.H.; Tang, K.J.W.; Zhao, X.; Koh, J.E.W.; Faust, O.; Gururajan, R.; Ciaccio, E.J.; Rajinikanth, V.; Acharya, U.R. An automated skin melanoma detection system with melanoma-index based on entropy features. Biocybern. Biomed. Eng. 2021, 41, 997–1012. [Google Scholar] [CrossRef]
- Zakeri, A.; Hokmabadi, A. Improvement in the diagnosis of melanoma and dysplastic lesions by introducing ABCD-PDT features and a hybrid classifier. Biocybern. Biomed. Eng. 2018, 38, 456–466. [Google Scholar] [CrossRef]
- Hameed, N.; Shabut, A.; Hossain, M.A. A Computer-aided diagnosis system for classifying prominent skin lesions using machine learning. In Proceedings of the 10th Computer Science and Electronic Engineering (CEEC), Colchester, UK, 19–21 September 2018; pp. 186–191. [Google Scholar]
- Hameed, N.; Shabut, A.M.; Ghosh, M.K.; Hossain, M.A. Multi-class multi-level classification algorithm for skin lesions classification using machine learning techniques. Expert. Syst. Appl. 2020, 141, 112961. [Google Scholar] [CrossRef]
- Xie, F.Y.; Fan, H.D.; Li, Y.; Jiang, Z.G.; Meng, R.S.; Bovik, A. Melanoma Classification on Dermoscopy Images Using a Neural Network Ensemble Model. IEEE Trans. Med. Imaging 2017, 36, 849–858. [Google Scholar] [CrossRef]
- Abbes, W.; Sellami, D.; Marc-Zwecker, S.; Zanni-Merk, C. Fuzzy decision ontology for melanoma diagnosis using KNN classifier. Multimed. Tools Appl. 2021, 80, 25517–25538. [Google Scholar] [CrossRef]
- Sun, J.; Yao, K.; Huang, G.Y.; Zhang, C.R.; Leach, M.; Huang, K.Z.; Yang, X. Machine Learning Methods in Skin Disease Recognition: A Systematic Review. Processes 2023, 11, 1003. [Google Scholar] [CrossRef]
- Ali, M.S.; Miah, M.S.; Haque, J.; Rahman, M.M.; Islam, M.K. An enhanced technique of skin cancer classification using deep convolutional neural network with transfer learning models. Mach. Learn. Appl. 2021, 5, 100036. [Google Scholar] [CrossRef]
- Patil, R.; Bellary, S. Machine learning approach in melanoma cancer stage detection. J. King Saud Univ. Comput. Inf. Sci. 2022, 34, 3285–3293. [Google Scholar] [CrossRef]
- Riaz, S.; Naeem, A.; Malik, H.; Naqvi, R.A.; Loh, W.K. Federated and Transfer Learning Methods for the Classification of Melanoma and Nonmelanoma Skin Cancers: A Prospective Study. Sensors 2023, 23, 8457. [Google Scholar] [CrossRef]
- Mahbod, A.; Tschandl, P.; Langs, G.; Ecker, R.; Ellinger, I. The effects of skin lesion segmentation on the performance of dermatoscopic image classification. Comput. Methods Programs Biomed. 2020, 197, 105725. [Google Scholar] [CrossRef]
- Dhivyaa, C.R.; Sangeetha, K.; Balamurugan, M.; Amaran, S.; Vetriselvi, T.; Johnpaul, P. Skin lesion classification using decision trees and random forest algorithms. J. Ambient. Intell. Humaniz. Comput. 2020, 1–13. [Google Scholar] [CrossRef]
- Balaji, V.R.; Suganthi, S.T.; Rajadevi, R.; Kumar, V.K.; Balaji, B.S.; Pandiyan, S. Skin disease detection and segmentation using dynamic graph cut algorithm and classification through Naive Bayes classifier. Measurement 2020, 163, 107922. [Google Scholar] [CrossRef]
- Batista, L.G.; Bugatti, P.H.; Saito, P.T.M. Classification of Skin Lesion through Active Learning Strategies. Comput. Methods Programs Biomed. 2022, 226, 107122. [Google Scholar] [CrossRef]
- Gonzalez-Diaz, I. DermaKNet: Incorporating the Knowledge of Dermatologists to Convolutional Neural Networks for Skin Lesion Diagnosis. IEEE J. Biomed. Health Inform. 2019, 23, 547–559. [Google Scholar] [CrossRef]
- Kadirappa, R.; Deivalakshmi, S.; Pandeeswari, R.; Ko, S.B. An automated multi-class skin lesion diagnosis by embedding local and global features of Dermoscopy images. Multimed. Tools Appl. 2023, 82, 34885–34912. [Google Scholar] [CrossRef]
- Xie, Y.T.; Zhang, J.P.; Xia, Y.; Shen, C.H. A Mutual Bootstrapping Model for Automated Skin Lesion Segmentation and Classification. IEEE Trans. Med. Imaging 2020, 39, 2482–2493. [Google Scholar] [CrossRef] [PubMed]
- Klepaczko, A.; Majos, M.; Stefanczyk, L.; Eikefjord, E.; Lundervold, A. Whole kidney and renal cortex segmentation in contrast-enhanced MRI using a joint classification and segmentation convolutional neural network. Biocybern. Biomed. Eng. 2022, 42, 295–311. [Google Scholar] [CrossRef]
- Oliveira, B.; Torres, H.R.; Morais, P.; Veloso, F.; Baptista, A.L.; Fonseca, J.C.; Vilaça, J.L. A multi-task convolutional neural network for classification and segmentation of chronic venous disorders. Sci. Rep. 2023, 13, 761. [Google Scholar] [CrossRef]
- Al-masni, M.A.; Al-Shamiri, A.K. Joint segmentation and recognition of melanoma skin lesions via multi-task learning. In Proceedings of the International Conference on Green Energy, Computing and Intelligent Technology (GEn-CITy 2023), Iskandar Puteri, Malaysia, 10–12 July 2023; pp. 1–5. [Google Scholar]
- Jin, Q.G.; Cui, H.; Sun, C.M.; Meng, Z.P.; Su, R. Cascade knowledge diffusion network for skin lesion diagnosis and segmentation. Appl. Soft Comput. 2021, 99, 106881. [Google Scholar] [CrossRef]
- Song, L.; Lin, J.Z.; Wang, Z.J.; Wang, H.Q. An End-to-End Multi-Task Deep Learning Framework for Skin Lesion Analysis. IEEE J. Biomed. Health Inform. 2020, 24, 2912–2921. [Google Scholar] [CrossRef]
- He, X.; Wang, Y.; Zhao, S.; Chen, X. Joint segmentation and classification of skin lesions via a multi-task learning convolutional neural network. Expert. Syst. Appl. 2023, 230, 120174. [Google Scholar] [CrossRef]
- Song, L.; Wang, H.Q.; Wang, Z.J. Decoupling multi-task causality for improved skin lesion segmentation and classification. Pattern Recogn. 2023, 133, 108995. [Google Scholar] [CrossRef]
- Cataldo, A.; De Benedetto, E.; Schiavoni, R.; Monti, G.; Tedesco, A.; Masciullo, A.; Piuzzi, E.; Tarricone, L. Portable microwave reflectometry system for skin sensing. IEEE Trans. Instrum. Meas. 2022, 71, 1–8. [Google Scholar] [CrossRef]
- Cataldo, A.; De Benedetto, E.; Angrisani, L.; Cannazza, G.; Piuzzi, E. A microwave measuring system for detecting and localizing anomalies in metallic pipelines. IEEE Trans. Instrum. Meas. 2020, 70, 1–11. [Google Scholar] [CrossRef]
- Rana, S.P.; Dey, M.; Tiberi, G.; Sani, L.; Vispa, A.; Raspa, G.; Duranti, M.; Ghavami, M.; Dudley, S. Machine learning approaches for automated lesion detection in microwave breast imaging clinical data. Sci. Rep. 2019, 9, 10510. [Google Scholar] [CrossRef] [PubMed]
- Khalid, N.; Zubair, M.; Mehmood, M.Q.; Massoud, Y. Emerging paradigms in microwave imaging technology for biomedical applications: Unleashing the power of artificial intelligence. npj Imaging 2024, 2, 13. [Google Scholar] [CrossRef]
- Cataldo, A.; Cino, L.; Distante, C.; Maietta, G.; Masciullo, A.; Mazzeo, P.L.; Schiavoni, R. Integrating microwave reflectometry and deep learning imaging for in-vivo skin cancer diagnostics. Measurement 2024, 235, 114911. [Google Scholar] [CrossRef]
- Gutman, D.; Codella, N.C.F.; Celebi, E.; Helba, B.; Marchetti, M.; Mishra, N.; Halpern, A. Skin Lesion Analysis toward Melanoma Detection: A Challenge at the International Symposium on Biomedical Imaging (ISBI) 2016, hosted by the International Skin Imaging Collaboration (ISIC). arXiv 2016, arXiv:1605.01397. [Google Scholar]
- Mendonça, T.; Ferreira, P.M.; Marques, J.; Marcal, A.R.S.; Rozeira, J. PH²—A dermoscopic image database for research and benchmarking. In Proceedings of the 35th International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Osaka, Japan, 3–7 July 2013; pp. 5437–5440. [Google Scholar]
- Caruana, R. Multitask Learning. Mach. Learn. 1997, 28, 41–75. [Google Scholar] [CrossRef]
- Fifty, C.; Amid, E.; Zhao, Z.; Yu, T.; Anil, R.; Finn, C. Efficiently identifying task groupings for multi-task learning. Adv. Neural Inf. Process. Syst. 2021, 34, 27503–27516. [Google Scholar]
- Crawshaw, M. Multi-task learning with deep neural networks: A survey. arXiv 2020, arXiv:2009.09796. [Google Scholar]
- Zhao, Y.; Wang, X.; Che, T.; Bao, G.; Li, S. Multi-task deep learning for medical image computing and analysis: A review. Comput. Biol. Med. 2023, 153, 106496. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In Proceedings of the 18th International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI), Munich, Germany, 5–9 October 2015; pp. 234–241. [Google Scholar]
- Isensee, F.; Petersen, J.; Klein, A.; Zimmerer, D.; Jaeger, P.F.; Kohl, S.; Wasserthal, J.; Koehler, G.; Norajitra, T.; Wirkert, S.; et al. nnU-Net: Self-adapting Framework for U-Net-Based Medical Image Segmentation. arXiv 2018, arXiv:1809.10486. [Google Scholar]
- Springenberg, J.T.; Dosovitskiy, A.; Brox, T.; Riedmiller, M. Striving for simplicity: The all convolutional net. arXiv 2015, arXiv:1412.6806. [Google Scholar]
- Woo, S.; Park, J.; Lee, J.-Y.; Kweon, I.S. CBAM: Convolutional Block Attention Module. In Proceedings of the 15th Proceedings of the European Conference on Computer Vision (ECCV), Munich, Germany, 8–14 September 2018; pp. 3–19. [Google Scholar]
- Hu, J.; Shen, L.; Sun, G. Squeeze-and-excitation networks. In Proceedings of the IEEE conference on Computer Vision and Pattern Recognition (CVPR), Salt Lake City, UT, USA, 18–23 June 2018; pp. 7132–7141. [Google Scholar]
- Al-masni, M.A.; Lee, S.; Yi, J.; Kim, S.; Gho, S.M.; Choi, Y.H.; Kim, D.H. Stacked U-Nets with Self-Assisted Priors Towards Robust Correction of Rigid Motion Artifact in Brain MRI. Neuroimage 2022, 259, 119411. [Google Scholar] [CrossRef] [PubMed]
- Perera, V.; Chung, T.; Kollar, T.; Strubell, E. Multi-Task Learning for Parsing the Alexa Meaning Representation Language. In Proceedings of the 32 AAAI Conference on Artificial Intelligence, New Orleans, LA, USA, 2–7 February 2018; pp. 5390–5397. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–23 June 2016; pp. 770–778. [Google Scholar]
- Wang, Y.; Su, J.; Xu, Q.Y.; Zhong, Y.X. A Collaborative Learning Model for Skin Lesion Segmentation and Classification. Diagnostics 2023, 13, 912. [Google Scholar] [CrossRef] [PubMed]
- Tang, P.; Liang, Q.K.; Yan, X.T.; Xiang, S.; Zhang, D. GP-CNN-DTEL: Global-Part CNN Model With Data-Transformed Ensemble Learning for Skin Lesion Classification. IEEE J. Biomed. Health Inform. 2020, 24, 2870–2882. [Google Scholar] [CrossRef] [PubMed]

| Dataset | Augmentation | Training Set | Testing Set | Total | ||||
|---|---|---|---|---|---|---|---|---|
| B * | M * | Total | B | M | Total | |||
| ISIC 2016 | × | 727 | 173 | 900 | 304 | 75 | 379 | 1279 |
| ISIC 2016 | ○ | 2908 | 2768 | 5676 | 304 | 75 | 379 | 6055 |
| PH2 | × | 160 | 40 | 200 | 200 | |||
| ID | Experiment Details | Param. [M] | Classification Measurements | Segmentation Measurements | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| B | M | TPR | TNR | F1 score | MCC | IOU | DSC | ||||
| Cls | Baseline Classification via ResNet50 [70] | 23.84 | B | 261 | 43 | 78.10 | 85.86 | 78.26 | - | - | - |
| 85.86% | 14.14% | ||||||||||
| M | 40 | 35 | |||||||||
| 53.33% | 46.67% | ||||||||||
| Seg | Baseline Segmentation via U-Net [63] | 10.71 | B | - | - | - | - | - | 87.06 | 84.62 | 91.04 |
| - | - | ||||||||||
| M | - | - | |||||||||
| - | - | ||||||||||
| MTL0 | Multi-Task Learning (Base) | 10.76 | B | 274 | 30 | 78.10 | 90.13 | 76.52 | 86.10 | 83.15 | 90.03 |
| 90.13% | 9.87% | ||||||||||
| M | 53 | 22 | |||||||||
| 70.67% | 29.33 | ||||||||||
| MTL1 | Integrating Segmentation Decoder Path Features into Classification Sub-Model | 10.80 | B | 274 | 30 | 79.16 | 90.13 | 77.96 | 84.89 | 81.56 | 88.86 |
| 90.13% | 9.87% | ||||||||||
| M | 49 | 26 | |||||||||
| 65.33% | 34.67% | ||||||||||
| MTL2 | Joint Reverse Learning from Classification to Segmentation | 10.86 | B | 277 | 27 | 81.53 | 91.12 | 80.66 | 84.74 | 81.95 | 89.15 |
| 91.12% | 8.88% | ||||||||||
| M | 43 | 32 | |||||||||
| 57.33% | 42.67% | ||||||||||
| MTL3 | CBAM Attention Module | 10.90 | B | 277 | 27 | 81.79 | 91.12 | 81.79 | 85.07 | 82.08 | 89.37 |
| 91.12% | 8.88% | ||||||||||
| M | 42 | 33 | |||||||||
| 56.0% | 44.0% | ||||||||||
| MTL4 | More Melanoma Data | 10.90 | B | 248 | 56 | 80.74 | 81.58 | 82.07 | 85.46 | 82.46 | 89.48 |
| 81.58% | 18.42% | ||||||||||
| M | 17 | 58 | |||||||||
| 22.67% | 77.33% | ||||||||||
| Method | Classification Measurements | Segmentation Measurements | |||||||
|---|---|---|---|---|---|---|---|---|---|
| B | M | TPR | TNR | F1 Score | MCC | IOU | DSC | ||
| Baseline Classification via ResNet50 [70] | B | 152 | 8 | 84.0 | 95.0 | 82.38 | - | - | - |
| 95.0% | 5.0% | ||||||||
| M | 24 | 16 | |||||||
| 60.0% | 40.0% | ||||||||
| Baseline Segmentation via U-Net [63] | B | - | - | - | - | - | 82.10 | 80.44 | 88.56 |
| - | - | ||||||||
| M | - | - | |||||||
| - | - | ||||||||
| Proposed Multi-Task Learning | B | 135 | 25 | 84.50 | 84.38 | 85.50 | 82.27 | 81.0 | 88.81 |
| 84.38% | 15.63% | ||||||||
| M | 6 | 34 | |||||||
| 15.0% | 85.0% | ||||||||
| Method | Number of Stages | TPR (%) | TNR (%) |
|---|---|---|---|
| CUMED [5] (1st) | non-end-to-end two stages (learned independently) | 50.7 | 94.1 |
| GTDL (2nd) | single stage (VGG-19) | 57.3 | 87.2 |
| BF-TB (3rd) | single stage (N.A.) | 32.0 | 96.1 |
| ThrunLab (4th) | single stage (Inception v3) | 66.7 | 81.6 |
| Jordan Yap (5th) | single stage (N.A.) | 24.0 | 99.3 |
| ResNet50 [70] | single stage | 46.7 | 85.9 |
| GP-CNN-DTEL [72] | non-end-to-end two stages (learned independently) | 32.0 | 99.7 |
| MTL [47] | end-to-end two stages (joint learning) | 89.5 | 44.0 |
| MTL-CNN [50] | end-to-end two stages (joint learning) | 67.3 | 96.3 |
| Proposed MTL | end-to-end two stages (joint learning) | 77.3 | 81.6 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Al-masni, M.A.; Al-Shamiri, A.K.; Hussain, D.; Gu, Y.H. A Unified Multi-Task Learning Model with Joint Reverse Optimization for Simultaneous Skin Lesion Segmentation and Diagnosis. Bioengineering 2024, 11, 1173. https://doi.org/10.3390/bioengineering11111173
Al-masni MA, Al-Shamiri AK, Hussain D, Gu YH. A Unified Multi-Task Learning Model with Joint Reverse Optimization for Simultaneous Skin Lesion Segmentation and Diagnosis. Bioengineering. 2024; 11(11):1173. https://doi.org/10.3390/bioengineering11111173
Chicago/Turabian StyleAl-masni, Mohammed A., Abobakr Khalil Al-Shamiri, Dildar Hussain, and Yeong Hyeon Gu. 2024. "A Unified Multi-Task Learning Model with Joint Reverse Optimization for Simultaneous Skin Lesion Segmentation and Diagnosis" Bioengineering 11, no. 11: 1173. https://doi.org/10.3390/bioengineering11111173
APA StyleAl-masni, M. A., Al-Shamiri, A. K., Hussain, D., & Gu, Y. H. (2024). A Unified Multi-Task Learning Model with Joint Reverse Optimization for Simultaneous Skin Lesion Segmentation and Diagnosis. Bioengineering, 11(11), 1173. https://doi.org/10.3390/bioengineering11111173










