Genomic Comparisons Revealed the Key Genotypes of Streptomyces sp. CB03234-GS26 to Optimize Its Growth and Relevant Production of Tiancimycins
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains and Culture Conditions
2.2. Genome Resequencing and Subsequent Bioinformatic Analyses
2.3. Construction of Different Recombinant Strains
2.4. Fermentation Production and HPLC Analysis of TNMs
2.5. Sequence Similarity Network Analysis and Streptomyces-Based Pan-Genomic Analysis
3. Results and Discussion
3.1. Genome Resequencing and Genomic Comparison of CB03234-G, CB03234-S and CB03234-GS26
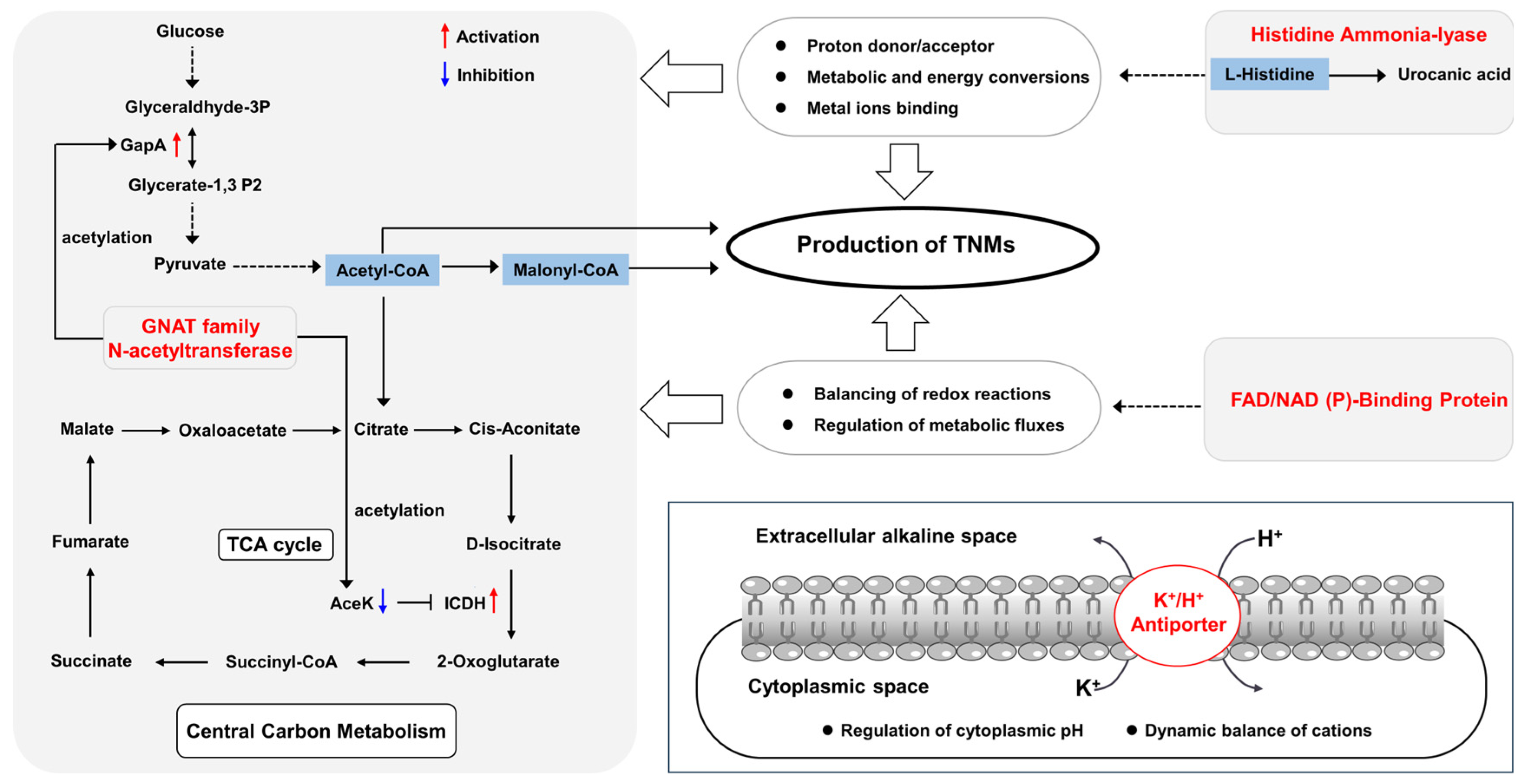
3.2. Bioinformatic Analyses and Functional Prediction of the Mutated Target Genes in CB03234-GS26
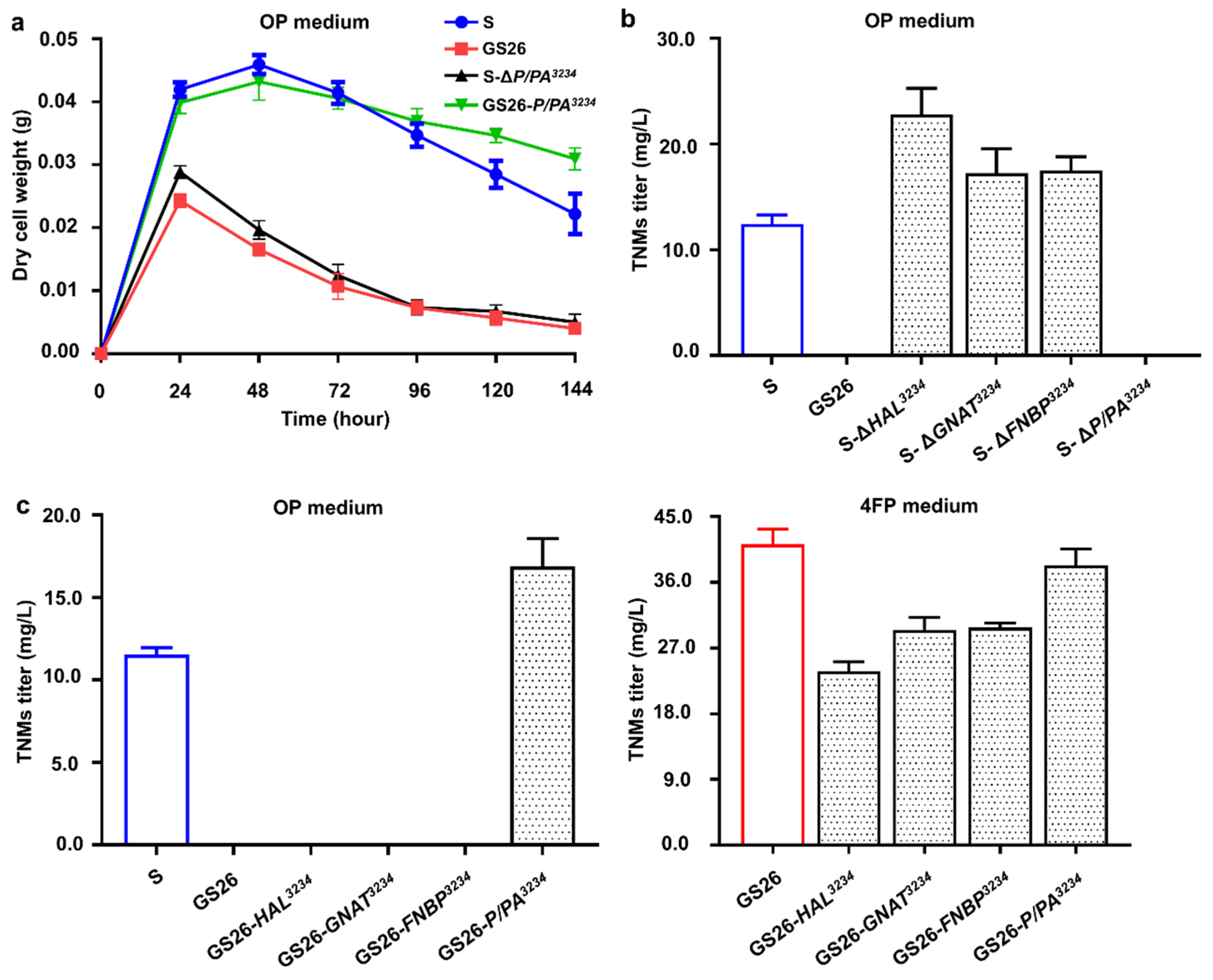
3.3. Genetic Validations of Four Target Genes in CB03234-S and CB03234-GS26
3.4. Sequence Similarity Network Analysis of P/PA Homologues and Streptomyces-Based Pan-Genomic Analyses of Target Genes
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yan, X.; Ge, H.; Huang, T.; Hindra; Yang, D.; Teng, Q.; Crnovcic, I.; Li, X.; Rudolf, J.D.; Lohman, J.R.; et al. Strain prioritization and genome mining for enediyne natural products. mBio 2016, 7, e02104–e02116. [Google Scholar] [CrossRef] [PubMed]
- Shen, B.; Hindra; Yan, X.; Huang, T.; Ge, H.; Yang, D.; Teng, Q.; Rudolf, J.D.; Lohman, J.R. Enediynes: Exploration of microbial genomics to discover new anticancer drug leads. Bioorg. Med. Chem. Lett. 2015, 25, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Yan, X. Anthraquinone-fused enediynes: Discovery, biosynthesis and development. Nat. Prod. Rep. 2022, 39, 703–728. [Google Scholar] [CrossRef] [PubMed]
- Nicolaou, K.C.; Das, D.; Lu, Y.; Rout, S.; Pitsinos, E.N.; Lyssikatos, J.; Schammel, A.; Sandoval, J.; Hammond, M.; Aujay, M.; et al. Total synthesis and biological evaluation of tiancimycins A and B, yangpumicin A, and related anthraquinone-fused enediyne antitumor antibiotics. J. Am. Chem. Soc. 2020, 142, 2549–2561. [Google Scholar] [CrossRef]
- Kamei, H.; Nishiyama, Y.; Takahashi, A.; Obi, Y.; Oki, T. Dynemicins, new antibiotics with the 1,5-diyn-3-ene and anthraquinone subunit. II. Antitumor activity of dynemicin A and its triacetyl derivative. J. Antibiot. 1991, 44, 1306–1311. [Google Scholar] [CrossRef]
- Yan, X.; Chen, J.-J.; Adhikari, A.; Teijaro, C.N.; Ge, H.; Crnovcic, I.; Chang, C.-Y.; Annaval, T.; Yang, D.; Rader, C.; et al. Comparative studies of the biosynthetic gene clusters for anthraquinone-fused enediynes shedding light into the tailoring steps of tiancimycin biosynthesis. Org. Lett. 2018, 20, 5918–5921. [Google Scholar] [CrossRef]
- Zhu, M.; Zhang, F.; Gan, T.; Lin, J.; Duan, Y.; Zhu, X. Deciphering the pathway-specific regulatory network for production of ten-membered enediyne tiancimycins in Streptomyces sp. CB03234-S. Microb. Cell Fact. 2022, 21, 188–199. [Google Scholar] [CrossRef]
- Zhuang, Z.; Jiang, C.; Zhang, F.; Huang, R.; Yi, L.; Huang, Y.; Yan, X.; Duan, Y.; Zhu, X. Streptomycin-induced ribosome engineering complemented with fermentation optimization for enhanced production of 10-membered enediynes tiancimycin-A and tiancimycin-D. Biotechnol. Bioeng. 2019, 116, 1304–1314. [Google Scholar] [CrossRef]
- Liu, H.; Jiang, C.; Lin, J.; Zhuang, Z.; Kong, W.; Liu, L.; Huang, Y.; Duan, Y.; Zhu, X. Genome shuffling based on different types of ribosome engineering mutants for enhanced production of 10-membered enediyne tiancimycin-A. Appl. Microbiol. Biotechnol. 2020, 104, 4359–4369. [Google Scholar] [CrossRef]
- Zhang, F.; Gao, D.; Lin, J.; Zhu, M.; Zhuang, Z.; Duan, Y.; Zhu, X. Construction of inducible genetic switch for the global regulator WblA to sustain both overproduction of tiancimycins and on-demand sporulation in Streptomyces sp. CB03234. ACS Synth. Biol. 2020, 9, 1460–1467. [Google Scholar] [CrossRef]
- Lin, J.; Xiao, Y.; Liu, H.; Gao, D.; Duan, Y.; Zhu, X. Combined transcriptomic and pangenomic analyses guide metabolic amelioration to enhance tiancimycins production. Appl. Microbiol. Biotechnol. 2024, 108, 18. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, Z.; Kong, W.; Wen, Z.; Tong, N.; Lin, J.; Zhang, F.; Fan, Z.; Yi, L.; Huang, Y.; Duan, Y.; et al. Combinatorial metabolic engineering of Streptomyces sp. CB03234-S for the enhanced production of anthraquinone-fused enediyne tiancimycins. Microb. Cell Fact. 2024, 23, 128–140. [Google Scholar] [CrossRef] [PubMed]
- Lacey, H.J.; Rutledge, P.J. Recently discovered secondary metabolites from Streptomyces species. Molecules 2022, 27, 887–902. [Google Scholar] [CrossRef] [PubMed]
- Newman, D.J.; Cragg, G.M. Natural products as sources of new drugs over the nearly four decades from 01/1981 to 09/2019. J. Nat. Prod. 2020, 83, 770–803. [Google Scholar] [CrossRef] [PubMed]
- Kalkreuter, E.; Pan, G.; Cepeda, A.J.; Shen, B. Targeting bacterial genomes for natural product discovery. Trends Pharmacol. Sci. 2020, 41, 13–26. [Google Scholar] [CrossRef]
- Guo, Q.; Peng, Q.-Q.; Li, Y.-W.; Yan, F.; Wang, Y.-T.; Ye, C.; Shi, T.-Q. Advances in the metabolic engineering of Saccharomyces cerevisiae and Yarrowia lipolytica for the production of β-carotene. Crit. Rev. Biotechnol. 2024, 44, 337–351. [Google Scholar] [CrossRef]
- Volk, M.J.; Tran, V.G.; Tan, S.-I.; Mishra, S.; Fatma, Z.; Boob, A.; Li, H.; Xue, P.; Martin, T.A.; Zhao, H. Metabolic engineering: Methodologies and applications. Chem. Rev. 2023, 123, 5521–5570. [Google Scholar] [CrossRef]
- Zhao, X.; Hussain, M.H.; Mohsin, A.; Liu, Z.; Xu, Z.; Li, Z.; Guo, W.; Guo, M. Mechanistic insight for improving butenyl-spinosyn production through combined ARTP/UV mutagenesis and ribosome engineering in Saccharopolyspora pogona. Front. Bioeng. Biotechnol. 2024, 11, 1329859–1329872. [Google Scholar] [CrossRef]
- Xiong, Z.; Tian, X.; Wang, G.; Song, X.; Xia, Y.; Zhang, H.; Ai, L. Development of a high-throughput screening method for exopolysaccharide-producing Streptococcus thermophilus based on Congo red. Food Res. Int. 2022, 162, 112094–112100. [Google Scholar] [CrossRef]
- Bailey, J.E.; Sburlati, A.; Hatzimanikatis, V.; Lee, K.; Renner, W.A.; Tsai, P.S. Inverse metabolic engineering: A strategy for directed genetic engineering of useful phenotypes. Biotechnol. Bioeng. 1996, 52, 109–121. [Google Scholar] [CrossRef]
- Liu, Y.; Gu, B.; Shi, J.; Fu, C.; Zhang, X.; Chen, X.; Yang, R.; Lyu, X. Inverse metabolic engineering based on metabonomics for efficient production of hydroxytyrosol by Saccharomyces cerevisiae. Bioresour. Technol. 2024, 409, 131187–131197. [Google Scholar] [CrossRef] [PubMed]
- Cai, M.; Wu, Y.; Qi, H.; He, J.; Wu, Z.; Xu, H.; Qiao, M. Improving the level of the tyrosine biosynthesis pathway in Saccharomyces cerevisiae through HTZ1 knockout and atmospheric and room temperature plasma (ARTP) mutagenesis. ACS Synth. Biol. 2021, 10, 49–62. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wang, K.; Pan, L.; Chen, X. Improved production of ε-Poly-L-lysine in Streptomyces albulus using genome shuffling and its high-yield mechanism analysis. Front. Microbiol. 2022, 13, 923526. [Google Scholar] [CrossRef] [PubMed]
- Peano, C.; Damiano, F.; Forcato, M.; Pietrelli, A.; Palumbo, C.; Corti, G.; Siculella, L.; Fuligni, F.; Tagliazucchi, G.M.; De Benedetto, G.E.; et al. Comparative genomics revealed key molecular targets to rapidly convert a reference rifamycin-producing bacterial strain into an overproducer by genetic engineering. Metab. Eng. 2014, 26, 1–16. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, H.; Li, S.; Zhang, Y.; Cheng, X.; Xiang, W.; Wang, X. Engineering of primary metabolic pathways for titer improvement of milbemycins in Streptomyces bingchenggensis. Appl. Microbiol. Biotechnol. 2021, 105, 1875–1887. [Google Scholar] [CrossRef]
- Copp, J.N.; Akiva, E.; Babbit, P.C.; Tokuriki, N. Revealing unexplored sequence-function space using sequence similarity networks. Biochemistry 2018, 57, 4651–4662. [Google Scholar] [CrossRef]
- Nikhila, K.S.; Nair, V.V. Protein sequence similarity analysis using computational techniques. Mater. Today Proc. 2018, 5, 724–731. [Google Scholar] [CrossRef]
- Atkinson, H.J.; Morris, J.H.; Ferrin, T.E.; Babbitt, P.C. Using sequence similarity networks for visualization of relationships across diverse protein superfamilies. PLoS ONE 2009, 4, e4345. [Google Scholar] [CrossRef]
- Tettelin, H.; Masignani, V.; Cieslewicz, M.J.; Donati, C.; Medini, D.; Ward, N.L.; Angiuoli, S.V.; Crabtree, J.; Jones, A.L.; Durkin, A.S.; et al. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: Implications for the microbial “pan-genome”. Proc. Natl. Acad. Sci. USA 2005, 102, 13950–13955. [Google Scholar] [CrossRef]
- Liu, N.; Liu, D.; Li, K.; Hu, S.; He, Z. Pan-genome analysis of Staphylococcus aureus reveals key factors influencing genomic plasticity. Microbiol. Spectrum 2022, 10, e0311722–e0311732. [Google Scholar] [CrossRef]
- Huang, R.; Lin, J.; Gao, D.; Zhang, F.; Yi, L.; Huang, Y.; Yan, X.; Duan, Y.; Zhu, X. Discovery of gas vesicles in Streptomyces sp. CB03234-S and potential effects of gas vesicle gene overexpression on morphological and metabolic changes in streptomycetes. Appl. Microbiol. Biotechnol. 2019, 103, 5751–5761. [Google Scholar] [CrossRef] [PubMed]
- Quevillon, E.; Silventoinen, V.; Pillai, S.; Harte, N.; Mulder, N.; Apweiler, R.; Lopez, R. InterProScan: Protein domains identifier. Nucleic Acids Res. 2005, 33, W116–W120. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, H.C. Molecular Cloning: A Laboratory Manual; CSH: Cold Spring Harbor, NY, USA, 1989. [Google Scholar] [CrossRef]
- Kohl, M.; Wiese, S.; Warscheid, B. Cytoscape: Software for visualization and analysis of biological networks. Methods Mol. Biol. 2011, 696, 291–303. [Google Scholar] [CrossRef] [PubMed]
- Miele, V.; Penel, S.; Duret, L. Ultra-fast sequence clustering from similarity networks with SILIX. BMC Bioinf. 2011, 12, 116. [Google Scholar] [CrossRef] [PubMed]
- Dyda, F.; Klein, D.C.; Hickman, A.B. GCN5-related N-acetyltransferases: A structural overview. Annu. Rev. Biophys. Biomol. Struct. 2000, 29, 81–103. [Google Scholar] [CrossRef]
- Vetting, M.W.; de Carvalho, L.P.S.; Yu, M.; Hegde, S.S.; Magnet, S.; Roderick, S.L.; Blanchard, J.S. Structure and functions of the GNAT superfamily of acetyltransferases. Arch. Biochem. Biophys. 2005, 433, 212–226. [Google Scholar] [CrossRef]
- Wang, Q.; Zhang, Y.; Yang, C.; Xiong, H.; Lin, Y.; Yao, J.; Li, H.; Xie, L.; Zhao, W.; Yao, Y.; et al. Acetylation of metabolic enzymes coordinates carbon source utilization and metabolic flux. Science 2010, 327, 1004–1007. [Google Scholar] [CrossRef]
- Holeček, M. Histidine in health and disease: Metabolism, physiological importance, and use as a supplement. Nutrients 2020, 12, 848–867. [Google Scholar] [CrossRef]
- Obata, T.; Aomine, M.; Yamanaka, Y. Protective effect of histidine on iron (II)-induced hydroxyl radical generation in rat hearts. J. Physiol. 1999, 93, 213–218. [Google Scholar] [CrossRef]
- Beliaeva, M.A.; Atac, R.; Seebeck, F.P. Bacterial degradation of Nτ-methylhistidine. ACS Chem. Biol. 2022, 17, 1989–1995. [Google Scholar] [CrossRef]
- Wang, Y.; San, K.-Y.; Bennett, G.N. Cofactor engineering for advancing chemical biotechnology. Curr. Opin. Biotechnol. 2013, 24, 994–999. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, J.R.; Weusthuis, R.A.; Huang, W.E. Growth-coupled enzyme engineering through manipulation of redox cofactor regeneration. Biotechnol. Adv. 2023, 63, 108102–108115. [Google Scholar] [CrossRef] [PubMed]
- Radchenko, M.V.; Tanaka, K.; Waditee, R.; Oshimi, S.; Matsuzaki, Y.; Fukuhara, M.; Kobayashi, H.; Takabe, T.; Nakamura, T. Potassium/proton antiport system of Escherichia coli. J. Biol. Chem. 2006, 281, 19822–19829. [Google Scholar] [CrossRef]
- Sze, H.; Chanroj, S. Plant endomembrane dynamics: Studies of K+/H+ antiporters provide insights on the effects of pH and ion homeostasis. Plant Physiol. 2018, 177, 875–895. [Google Scholar] [CrossRef]
- Nyanga-Koumou, A.P.; Ouoba, L.I.I.; Kobawila, S.C.; Louembe, D. Response mechanisms of lactic acid bacteria to alkaline environments: A review. Crit. Rev. Microbiol. 2012, 38, 185–190. [Google Scholar] [CrossRef]
- Spear, J.S.; White, K.A. Single-cell intracellular pH dynamics regulate the cell cycle by timing the G1 exit and G2 transition. J. Cell Sci. 2023, 136, jcs260458. [Google Scholar] [CrossRef]
- Simon, R.; Priefer, U.; Pühler, A. A broad host range mobilization system for in vivo genetic engineering: Transposon mutagenesis in gram negative bacteria. Nature 1983, 1, 784–791. [Google Scholar] [CrossRef]
- Bierman, M.; Logan, R.; O’Brien, K.; Seno, E.T.; Rao, R.N.; Schoner, B.E. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene 1992, 116, 43–49. [Google Scholar] [CrossRef]



| Strains | Locus_Tag | Protein Length (AA) | Mutation | Putative Functions |
|---|---|---|---|---|
| CB03234-G | AMK26_RS07435 | 288 | 295T > TG (frameshift) | zinc metalloprotease HtpX |
| AMK26_RS07635 | 398 | 329C > A(A110D) | elongation factor Tu | |
| CB03234-S | AMK26_RS10695 | 396 | 362C > A(S121T) | superoxide dismutase, Ni |
| AMK26_RS14490 | 446 | 503C > G(A168G) | uracil permease (purine permease) | |
| AMK26_RS14790 | 515 | 782G > T(S261I) 808G > A(V270I) | D-alanine glycine permease (alanine:cation symporter family protein) | |
| AMK26_RS26890 | 259 | 280C > A(L94M) | transcriptional regulator (Scr1 family TA system antitoxin-like transcriptional regulator) | |
| AMK26_RS34215 | 1081 | 103T > A(T35S) | hypothetical protein | |
| AMK26_RS07620 | 124 | 129C > G(K43N) | 30S ribosomal protein S12 | |
| CB03234-S/ GS26 | AMK26_RS22320 | 186 | 266G > C(G89A) | hypothetical protein |
| AMK26_RS34270 | 74 | 175G > C(R59G) | hypothetical protein | |
| AMK26_RS10740 | 518 | 353G > C(P118R) | alkaline phosphatase D family protein | |
| AMK26_RS16920 | 393 | 513C > A(Y171 *) | agmatine deiminase | |
| AMK26_RS17525 | 338 | 97G > T(K33Q) | hemolysin family protein | |
| AMK26_RS12260 | 194 | 582G > GC (frameshift) | ATP/GTP-binding protein | |
| AMK26_RS31245 | 63 | (141–143)GGC > G (frameshift) | hypothetical protein | |
| CB03234-GS26 | AMK26_RS08900 | 513 | 133C > A(A45S) | histidine ammonia-lyase |
| AMK26_RS16830 | 262 | (157–184) ACGGGCC GTCCGGGCCGTCC GGGCCGTC > A (codon_deletion) | GNAT family N-acetyltransferase | |
| AMK26_RS27145 | 641 | 1075C > T (G359S) | FAD/NAD (P)-binding protein | |
| AMK26_RS27375 | 487 | 235C > A (V79F) 251C > T (G84D) | potassium/proton antiporter | |
| AMK26_RS09465 | 772 | (762–295) GCTCCCC GTCGGCGACGAG CGGCCCGTCGACC CG > G (codon_deletion) | hypothetical protein | |
| AMK26_RS17285 | 132 | 339C > A(Y113 *) | hypothetical protein | |
| AMK26_RS31405 | 270 | 161A > T(V54E) | SAM-dependent methyltransferase |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, H.; Lin, J.; Huang, Y.; Duan, Y.; Zhu, X. Genomic Comparisons Revealed the Key Genotypes of Streptomyces sp. CB03234-GS26 to Optimize Its Growth and Relevant Production of Tiancimycins. Bioengineering 2024, 11, 1128. https://doi.org/10.3390/bioengineering11111128
Liu H, Lin J, Huang Y, Duan Y, Zhu X. Genomic Comparisons Revealed the Key Genotypes of Streptomyces sp. CB03234-GS26 to Optimize Its Growth and Relevant Production of Tiancimycins. Bioengineering. 2024; 11(11):1128. https://doi.org/10.3390/bioengineering11111128
Chicago/Turabian StyleLiu, Huiming, Jing Lin, Yong Huang, Yanwen Duan, and Xiangcheng Zhu. 2024. "Genomic Comparisons Revealed the Key Genotypes of Streptomyces sp. CB03234-GS26 to Optimize Its Growth and Relevant Production of Tiancimycins" Bioengineering 11, no. 11: 1128. https://doi.org/10.3390/bioengineering11111128
APA StyleLiu, H., Lin, J., Huang, Y., Duan, Y., & Zhu, X. (2024). Genomic Comparisons Revealed the Key Genotypes of Streptomyces sp. CB03234-GS26 to Optimize Its Growth and Relevant Production of Tiancimycins. Bioengineering, 11(11), 1128. https://doi.org/10.3390/bioengineering11111128






