Abstract
Stroke is a medical condition that affects around 15 million people annually. Patients and their families can face severe financial and emotional challenges as it can cause motor, speech, cognitive, and emotional impairments. Stroke lesion segmentation identifies the stroke lesion visually while providing useful anatomical information. Though different computer-aided software are available for manual segmentation, state-of-the-art deep learning makes the job much easier. This review paper explores the different deep-learning-based lesion segmentation models and the impact of different pre-processing techniques on their performance. It aims to provide a comprehensive overview of the state-of-the-art models and aims to guide future research and contribute to the development of more robust and effective stroke lesion segmentation models.
1. Introduction
Stroke is currently the second leading cause of death and the third leading cause of disability worldwide [1]. As per the World Stroke Organization, around 15 million people suffer from stroke annually; out of these 15 million, about 43% lose their lives, and of the survivors, roughly two-thirds have some disability [2]. Currently, only qualitative lesion assessment is a part of the clinical workflow, complimented by various assessments such as the National Institutes of Health Stroke Scale [3] and the Cincinnati Prehospital Stroke Severity Scale [4] to gauge stroke severity. Different medical imaging modalities, such as non-contrast computed tomography (CT) and magnetic resonance imaging (MRI), can support the subjective assessment. Both modalities have pros and cons, briefly discussed in Table 1.
In the last decade, machine and deep learning development has grown exponentially. Artificial intelligence is now being used in every field of life to reduce human effort, and medical image analysis is no different. Deep learning methods are used for automated stroke classification and rehabilitation prediction [5]. However, at the heart of these processes is lesion segmentation, a term used to define the process of tracing lesion outlines by categorising each voxel as a lesion or non-lesion in a medical image [6]. Segmented lesions can allow the model to learn the anatomical features and their impact on the prediction. The gold standard for lesion segmentation is manual segmentation [7]. Computer-aided diagnostics tools and techniques, such as 3D Slicer [8], ITK-SNAP [9], Amira [10], and MIPAV [11], are some of the open-source and commercially available tools that are helpful for lesion segmentation, analysis, and visualisation. However, some human intervention and verification is still required. It is also time-consuming and laborious to segment the lesions layer by layer [12], and the chances of error and bias are also high as lesions could be of irregular size and shape in each layer [7,13], which necessitates the development of automated techniques.
Different deep learning techniques, such as convolutional neural networks (CNNs) and transformers, are employed to perform lesion segmentation. The primary objective of this review is to critically analyse existing research on deep-learning-based lesion segmentation in stroke analysis, providing a comprehensive guide for future research directions. We do not present new findings but investigate the usage of the different deep learning models within the current literature and identify promising research avenues. We also aim to investigate the usage of different pre-processing techniques within the literature and their impact on the model’s performance.
Different databases and platforms were used to gather the literature for this study, including Google Scholar, PubMed, arxiv.org, ScienceDirect, and SpringerLink. Terms such as “Stroke Lesion Segmentation”, “Deep Learning”, “Ischaemic Stroke”, and “Neural Network” were used to search for the literature. Since much work has been carried out in deep learning and lesion segmentation, this study limited its search to between 2018 and 2023 to focus solely on the latest state-of-the-art studies. The recently introduced Must AI Criteria-10 (MAIC-10) [14] checklist was used to evaluate the quality of technical studies. The Supplementary Materials Table S1 provides further details on the scoring of each chosen study.

Table 1.
A brief side-by-side comparison of MRI and CT.
Table 1.
A brief side-by-side comparison of MRI and CT.
| Aspect | MRI | CT |
|---|---|---|
| Time Constraints | An MRI scan may require up to an hour to conclude its findings [15]. However, some medical centres have reduced the time to up to 10 min using different protocols [16]. | A CT typically takes between 5 and 15 min per scan. |
| Cost Effective | MRI costs almost double compared to a CT [17]. | A CT costs half the amount of an MRI [17]. |
| Ischaemic Lesion Detection | Since MRI scans produce detailed images, detecting small lesions is easier [18]. | CT scans are good at detecting large ischaemic lesions. It might be challenging to catch a small lesion earlier using a CT scan [18]. |
| Haemorrhage Detection | MRI scans are suitable for detecting small or chronic haemorrhages [18]. | CT scans perform well while detecting acute or larger haemorrhages. [19]. |
| Lesion Visibility | As an MRI produces a more detailed image, it is easier to detect and visualise a lesion. A lesion is more evident in the hyperintense region using a DWI map [20]. | Due to low contrast, a lesion is harder to visualise in a CT scan [20]. |
| Easier segmentation | It is easier to segment a lesion using an MRI scan manually. The different modalities, such as DWI, FLAIR, and T2-weighted, can be used to perform segmentation more accurately [21]. | Due to the low tissue contrast, it is harder to manually segment a lesion using a CT scan [21]. |
| Health Concerns | The magnetic rays emitted by the MRI scanner can disrupt the working of different implanted devices. | Since CT scanners use ionising radiation, they can cause cellular damage. |
2. Previous Literature Surveys
Since the early 2000s, considerable efforts have been made towards automating lesion segmentation and amalgamating the findings of technical studies in the form of literature and systematic surveys. With the boom of machine learning techniques in lesion segmentation, researchers have attempted to present current trends concisely. For example, Rani et al. [22] discussed the various object recognition and localisation techniques that can be applied to lesion segmentation. They included widely used techniques like region-based convolutional neural networks [23], You Only Look Once [24], Single Shot MultiBox Detector [25], and EfficientNet [26] in their discussion. One of the highlights of this study was that the authors also compared the algorithms in terms of speed and performance.
Karthik et al. [27] discussed in detail various studies that used machine and deep learning methods. The authors of this study did not limit themselves to stroke segmentation; they also expanded the study to discuss machine learning techniques for stroke classification. The study concluded that using smaller datasets made many analysed models prone to overfitting and class imbalance. To remedy the presented issues, the possible solutions given by the authors included the usage of data augmentation or employing generative adversarial networks (GANs).
Both studies, one conducted by Thiyagarajan and Murugan [28] and the other by Zhang et al. [6], presented a very in-depth analysis of the latest deep learning techniques, where Zhang et al. [6] broke down the techniques based on the dataset they used, and Thiyagarajan and Murugan [28] presented a step by step analysis of each study under discussion. Zhang et al. [6] presented a conclusion similar to the one given by Karthik et al. [27]; they compared the different factors affecting the performance of stroke lesion segmentation to brain tumour segmentation. They concluded that using GANs and employing different augmentation techniques can help with the problem of smaller datasets. Another solution presented was using features gathered from the MRI alongside the doctor’s expertise in stroke treatment as supplementary input factors for better information extraction from the image. The authors also discussed the advantages and disadvantages of using deeper or shallower models while segmenting smaller lesions. Thiyagarajan and Murugan [28] concluded that DWI modalities present overall better segmentation results for acute and sub-acute stroke, whereas T2-weighted and FLAIR perform better for chronic stroke.
Karthik and Menaka [29] discussed pre-processing techniques, lesion segmentation, and classification techniques. Their review focused on manual and deep learning segmentation techniques for lesion segmentation. Karthik and Menaka [29] concluded that segmentation techniques could be improved by incorporating structural and symmetrical properties of lesions into the models.
Wang et al. [30] discussed some state-of-the-art lesion segmentation techniques. They concluded that multi-centre data might be required to improve the performance of the AI-based models. They also presented the idea of using CT images as input as they are commonly used in clinical practice rather than MRI. Abbasi et al. [31] presented a thorough analysis of current deep learning models. They compared the different models based on image modalities, i.e., CT and MRI. Abbasi et al. [31] also presented a similar conclusion to previous studies that data augmentation techniques must be explored for better results. They also concluded that integrating multi-model imaging modalities can help better understand ischaemic stroke and improve the segmentation results.
Table 2 presents the contributions of the review papers discussed above.

Table 2.
Comprehensive analysis of the review papers.
3. Stroke Lesion Segmentation
Usually, one can divide the lesion segmentation model into three main modules. Figure 1 shows an overview of the process.
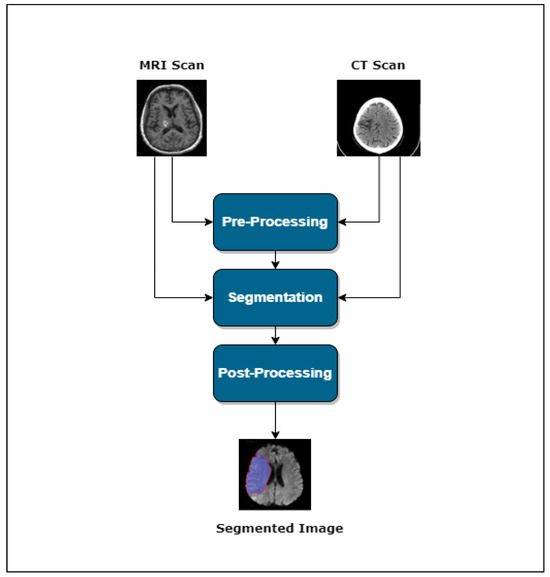
Figure 1.
Lesion segmentation pipeline.
- Pre-processing
- Segmentation
- Post-processing
This section provides a basic overview of the different techniques used in the literature for pre-processing and segmentation.
3.1. The Role of Pre-Processing in Stroke Lesion Segmentation
Pre-processing is the first step in solving any computer vision problem, and stroke lesion segmentation is no different. The purpose of adding a pre-processing layer is to remove any noise added to the image during the image acquisition phase. As deep learning models often require a considerable amount of data, the data are often collected from multiple centres. The acquisition equipment and methodology difference can often introduce inter-site and intra-site variabilities in the data [32]. The presence of these variabilities might lead to the introduction of noise and bias in the data, which can negatively impact stroke lesion segmentation. Pre-processing is crucial for accurate stroke lesion segmentation, removing noise and bias introduced by multi-centre data collection and preparing data for deep learning models despite the trend towards end-to-end learning. Hence, some studies omit the pre-processing stage and input the data directly to the network, as seen in [6,13] among others.
However, within our literature, we observed that studies still use pre-processing techniques; for example, Clèrigues et al. [33] used symmetric modality augmentation, which allowed them to learn features based on the symmetry of the brain hemisphere using image registration. Soltanpour et al. [34] applied multiple techniques to clean their data, such as intensity clipping, to ensure no high-intensity outliers were present. They also employed image registration using bilinear interpolation to replace each value from their sample image and its corresponding ground truth with the weighted average of their 2 × 2 neighbour mean. Sheng et al. [35] also used bilinear interpolation.
Image registration techniques establish a point-by-point correspondence between anatomical features present in two scans of the same organ taken from different angles [36]. This technique is beneficial as different modalities and angles are available per scan for medical images, all of which have different information. With the increased data, capturing the spatial information, such as the anatomical structures, in the scans is essential. It can be performed manually using cross-correlation-based, Fourier-transformation-based, landmark-based mapping methods [37] or by using machine learning techniques [38,39]. Within our literature, Hui et al. [40], Liu et al. [41], Wu et al. [42] applied image registration to its input data by transforming each image to MNI-152 space.
Skull stripping removes the bone structure from a medical image to ensure that only brain tissue is considered during segmentation [43]. Different methods of performing skull stripping include morphology-based [44,45], intensity-based [46,47], and atlas-based [48] methods. Traditional skull stripping methods are sensitive to noise [49,50]; hence, machine-learning-based skull stripping methods have also been introduced to cater for the sensitivity to noise [51]. Within our literature, skull stripping was applied by Anand et al. [52], Cui et al. [53], Karthik et al. [54] to remove all non-brain tissue, including the blood vessels, fat, muscles, and dura mater, making the lesion visibility better within the medical image.
Another pre-processing method favoured by studies such as Hui et al. [40], Ahmad et al. [55], Tureckova and Rodríguez-Sánchez [56] is bias correction, which removes nonuniformity from the input images. The data can introduce nonuniformity due to variations in equipment quality and spatial inhomogeneity caused by magnetic waves or X-rays [57,58]. Bias correction can be performed using multiple techniques, including filtering, surface fitting, and histogram-based techniques [59].
Though many studies favour bias correction and skull stripping, they are often followed by or used with normalisation. Normalising an image allows the model to generate a more stable output. It can be performed by changing the intensity of each pixel value to resemble the characteristics of pre-defined data [60]. The least square method, radiometric normalisation, and histogram normalisation are some of the most commonly used normalisation techniques [61]; machine learning has also been introduced for image normalisation [62,63]. Within our literature, it was observed that z-score normalisation, also called standardisation, is the most common choice, as it was used by [34,52,53,54,56,64]. Other techniques employed for normalisation include min-max normalisation used by Dolz et al. [65], and percentile clipping favoured by Wang et al. [66].
3.2. Advancements and Diverse Architectures in Automated Lesion Segmentation
The introduction of convolutional neural networks revolutionized the field of machine learning; with their inherent ability to learn features directly from the input image, they became the most commonly used machine learning technique for computer vision problems. This technique is also commonly used for lesion segmentation, especially after the U-Net [67] model was presented. GAN is another technique employed for lesion segmentation tasks; it is beneficial for low-resource models and unsupervised machine learning approaches.
3.2.1. Supervised Learning
Advancements and Variations OF U-Net Architecture
During the literature-gathering process, it was observed that many of the state-of-the-art techniques stem from the U-Net model. Some of them focused on improving the current structure, while others used the model as a base for their own. Clèrigues et al. [33] introduced residual connections in the basic U-Net model and replaced the commonly used ReLU function with PReLU; Dolz et al. [65] presented the idea of using multiple encoders for different modalities of the scan, each of which was densely connected with others for feature preservation. Karthik et al. [54] focused on how different loss, activation, and optimization functions can impact the U-Net model. They tested the model’s performance by using different combinations of these functions. Hui et al. [40] used two U-Net structures rather than one to capture the auxiliary and primary features separately. The two networks are identical, except they were trained using different loss functions.
Ou et al. [68] proposed a version of U-Net consisting of lambda layers [69] rather than convolution layers. The concept behind lambda layers is to capture the context of each value by converting it into a linear function referred to as a ”lambda”. The introduced model calculates global, local, and inter-slice lambda from a 3D feature map. All three lambdas are applied to the query, the pixel under study, to produce the final results.
Soltanpour et al. [34] presented two enhancements on a U-Net-based model called MultiResUNet [70]. Firstly, they used different filter sizes than the original model’s 3 × 3 filter. They replaced skip connections between each layer with CNN-based shortcuts where each shortcut block consisted of four 3 × 3 convolution layers, whose results were concatenated with a 1 × 1 convolution layer. The first enhancement allows the model to capture features on multiple scales, whereas the second change balances out the original model’s semantic gap. Sheng et al. [35] introduced a block called the Cross-Spatial Attention Module, which is used instead of the skip connection. Using this block rather than simple skip connections enables the feature map to capture the spatial information more accurately.
Liu et al. [71] proposed a model incorporating dense blocks instead of simple convolutions in the encoder; they also used two side-by-side encoding and decoding structures to capture all the features. Ahmad et al. [55] also presented a dense block-based structure, with a residual inception block added after the first convolution layer and used as the bottleneck layer; they also employed a deep supervision technique in the decoder for better convergence. The dense block presented in this study consisted of three convolution layers, with the first convolution layer followed by a pooling layer, whereas Liu et al. [71] followed the structure of DenseNet-121 but extended to 123 layers.
A dense block consists of multiple convolutional layers directly connected with each subsequent layer; this structure allows each layer to reuse the features from all previous layers and avoid overfitting [72]. Within the scope of the above studies, it was observed that Liu et al. [71] did not present a significant improvement in the results; however, it was the opposite case for Ahmad et al. [55] who presented a significant improvement from the baseline models. It should be noted that Ahmad et al. [55] used dense blocks combined with residual inception blocks within their decoder, which could factor in the improved results.
Tureckova and Rodríguez-Sánchez [56] experimented by replacing the convolution layers with dilated ones; they experimented by placing the dilated layers in different network modules and concluded that dilated filters in the first convolution layer produced the best results. Omarov et al. [73] also presented similar work; they introduced different optimization layers in the original U-Net model and used dilated convolutions in some layers to increase the filter’s field of view. Using dilated filters instead of simple filters allowed both studies to increase their field of view without increasing the filter size or computational power.
Liu et al. [41] presented a Multi-scale Deep Fusion unit in the bottleneck layer of U-Net; the authors employed the techniques of Atrous Spatial Pyramid Pooling (ASPP) [74] and capsules with dynamic routing [75] to capture and encode the global context. Zhang et al. [12] also employed the concept of ASPP in their bottleneck layer. The ASPP produced a fused feature map using the features produced from U-Net’s encoding layer and the feature set produced by the residual encoder. The residual encoder used multiple convolution layers to capture the low-level features. Similarly, the work proposed by Zhou et al. [64] used 2D and 3D convolutions in the encoder to maximize the information. A dimension transformation block was used in the bottleneck layer to control the number of trainable parameters. Qi et al. [76] used a feature similarity module in the bottleneck layer. They also replaced the convolution layers of the U-Net model with their proposed block called the X-block. The X-block used depth-wise convolutions to capture the contextual information present in the input scan. Using depth-wise convolutions allowed the models to perform similarly to the baseline models but with almost half the trainable parameters.
Evolution towards Capturing Global Features
The previously discussed studies focused on capturing the local features present in the image. However, some recent studies also showed the importance of capturing global features. Since every lesion is variable, capturing the contextual and spatial information present in the scan is crucial. A CNN-based model captures local features well, whereas transformers perform competently at capturing global features.
Wu et al. [42] employed both techniques in a ‘W’-like structure to capture the local and global features present in the input image. They introduced two new modules, the Boundary Deformation Module (BDM) and the Boundary Constraint Module (BCM). The proposed model first uses a U-Net to capture the local features; the calculated features are then processed through the BDM module before passing them to the transformer’s encoder. The transformer’s decoder comprises a similar model to the U-Net’s decoder; it consists of different up-scaling convolution layers, and the output of these layers is processed through the BCM layer to ensure the pixels neglected in the previous layers can also contribute to the final segmentation. The BCM uses different dilated convolution layers to capture the context of the overall image.
Wu et al. [77] proposed a similar technique where a transformer layer was introduced in the encoder network to capture the long-range relationships between the features. The authors proposed a model called MLiRA-Net, comprising a patch partition block (PPB), the MLiRA mechanism, which is the unit used for feature extraction and is called the multi-scale long-range interactive and regional attention mechanism, and lastly, the feature interpolation path (FIP), which acts as the decoder. The PPB consists of two cascading convolutional layers; it captures the local features present in each input image patch. The FIP module consists of multiple combinations of convolutional and transpose convolutional layers, each connected to its relevant encoder layer using a skip connection.
Feature Extraction Variations
Most models discussed above had an end-to-end structure, with the feature extractor embedded in the proposed model. However, some works used a separate feature extraction module, such as Wang et al. [66] who proposed a model that used a CNN-based feature extractor that captured low-level and high-level features from CT perfusion maps; these features were then used to generate a pseudo-DWI image, which was then encoded and later on decoded to extract the segmentation. The U-Net model was the base model for segmentation, with the batch normalization layers replaced with switchable normalization and a Squeeze-and-Excitation block to capture the channel-wide features.
Beyond U-Net: Novel Segmentation Techniques
Even though the U-Net structure has been heavily utilized, not all studies used it. Karthik et al. [78] proposed a fully connected CCN-based network. The network consisted of multiple layers of the Multi-Residual Attention Block, which processed mainline and input context features using multiple convolutional and down-sampling layers. The authors also proposed a masked dropout module, which was applied only on the produced mainline features to regulate the receptive field. The decoder model combines low- and high-level features with skip connections from each intermediate encoder layer. The final results are produced by integrating features of the different receptive fields. Unlike the U-Net model, the layers of the decoder work independently of each other, and majority class voting is used to produce the segmentation result. Li [13] presented a CNN-based model inspired by the human visual cortex. The proposed model consisted of three blocks called V1, V2, and V4, respectively, with a bottleneck layer. Each block of the model has a unique structure and is used to capture a specific type of information.
Liu et al. [79] proposed an encoder/decoder-based model comprising a ResNet and a global convolution network (GCN)-based encoder/decoder structure. The authors used their internal dataset, comprising MRI scans of 212 ischaemic stroke patients; DWI, ADC, and T2-weighted modalities were generated for each patient. Each modality was concatenated to a three-channel image, which acted as the input for the model. Each input image passes through a series of Res-Blocks where each block can contain an n-number of bottleneck layers. The output of each Res-Block is passed to its corresponding up-sampling layer using a skip connection comprising of a GCN and Boundary Refinement layer before it is passed to the next block. Anand et al. [52] also employed an encoder/decoder-based model; the encoder used multiple DenseNet-based layers. The input image comprises five channels, one for each modality of CT, including CBV, CBF, MTT, and .
Ou et al. [80] presented a transformer-based encoder/decoder structure called a Patcher. The encoder consisted of multiple cascading layers of patcher blocks, where each patcher block uses the combination of CNN and vision-based transformers to capture both local and global features in the image. The decoder follows the idea of the mixture of experts (MoE) technique. Firstly, multi-layer perceptrons (MLP) are used to process the feature maps produced by each encoding layer. All weights are then up-sampled and concatenated to be processed through another MLP to produce the final prediction. Though the model outperformed all baseline models, one drawback of using a vision transformer-based model would be that it is currently only limited to 2D data.
3.2.2. Semi-Supervised Learning
Accurately predicting stroke boundaries is one of the primary goals of automating stroke lesion segmentation. However, stroke lesions are often very irregular in shape, size, and form, making them harder to generalize without a large corpus of data. Though the different variants of the ATLAS [81,82] and ISLES [83,84] datasets provide various types of annotated stroke data, the models trained on them still need help with unseen data. Some studies have employed semi-supervised techniques to improve the generalization of the model. For example, Cui et al. [53] introduced a DeepMedic [85]-based student–teacher model. Both models use the same structure, and their weights are updated alternatively. The teacher model is initialized with the DeepMedic model, whereas the student model learns by minimizing the loss of the teacher model.
Another approach was presented by Zhao et al. [86], which used weakly labelled data in combination with fully labelled data in their presented model. The model comprises three modules: classification, segmentation, and inference. The VGG16-based classification module was trained on weakly labelled data and generated class activation mappings (CAM) for the inference network and a feature map for the classification network. The inference module, finally, uses the binarized segmentation produced from the segmentation model and the CAM produced from the classification module to produce the prediction.
3.2.3. Unsupervised Learning
Deep learning models require a lot of labelled data to learn real-world patterns accurately; however, labelled medical imaging data are scarce for various reasons, including but not limited to privacy issues and domain expertise required to label the data [87]. Hence, lesion segmentation has expanded to unsupervised techniques, predominantly using GANs. Each GAN comprises two modules: the generator and the discriminator. The generator generates data sample points, and the discriminator needs to determine whether the samples were from real-world data or were generated by the generator. As it is in the model’s name, both the discriminator and generator are trained in an adversarial manner, where the generator improves its ability to generate real-world data, and the discriminator improves its differentiation abilities [88].
As discussed in the preceding sections, recent research shows a prevalent use of the U-Net model. Even in unsupervised learning, most studies employ U-Net, or one of its variants, as their generator for GANs. For instance, Islam et al. [89] used a U-Net-based segmentation/generator model within the GAN framework. However, the structure of the discriminator varies from study to study, continuing with the example of Islam et al. They used an FCN-based discriminator consisting of four 2D convolutional layers followed by ReLU activation.
Another notable approach was used by Ou et al. [90]; they used a U-Net-inspired transformer model discussed beforehand called Patcher [80] as their segmentation module and used an FCN-based discriminator. Wang et al. [91] proposed an unsupervised approach called a consistent perception generative adversarial network. The proposed GAN comprises a segmentation, an assistant, and a discriminator model. The segmentation model uses the structure of U-Net in combination with a similarity connection module [76]. The assistant model assists the discriminator in assessing whether the input image is fake or the target image.
4. Results and Future Directions
Table 3 summarises the studies considered for this review, offering insights into the state-of-the-art stroke lesion segmentation techniques.

Table 3.
Overview of the articles included in the literature.
Of the chosen studies, 71% used MRI as their input modality, and 29% used CT. T1-weighted images were the preferred choice for MRI, followed closely by DWI. However, using the modalities in combination was the preferred option for CT. Most studies under consideration used supervised learning, with 53% having a U-Net-based structure. Binary cross-entropy loss and dice loss, or using them in combination with other losses, was the most common choice among the studies.
4.1. Data Dimensionality and Its Processing Techniques
Two-dimensional data or images were used by 71% of the studies as input for their model, whereas 29% used 3D data. A common trend in the studies that used 3D data was to down-sample the input. The down-sampling was performed due to the memory limitations of the GPUs. As well as down-sampling, patching was a technique that was applied to the data as shown by Qi et al. [76] and Wang et al. [91].
Future models using 3D data can explore patching further as they are less memory extensive and do not lose data like down-sampling. An added advantage of patching is that it can help cater to the class imbalance problem. In stroke data, the healthy tissue is far greater in number than the lesion, which causes the class imbalance problem, which often leads to underfitting of the model; sophisticated patching techniques can help cater for this problem as well.
4.2. Data Pre-Processing
Normalization was the most commonly used pre-processing technique, followed by skull stripping and bias correction. Of the 28 studies chosen, 46% did not mention using pre-processing techniques; however, it should be noted that they all used public datasets, which were pre-processed beforehand to normalize the data [81,82]. Of the studies that mentioned pre-processing techniques, 46% used normalization, 10% used skull stripping, and 7% used bias correction.
Since most studies used 2D data, reviewing more sophisticated pre-processing techniques, such as harmonization and super-resolution, is suggested as a future research avenue. Image harmonization normalizes and standardizes the data collected from different sources [92]. Studies have employed convolutional neural networks, transformers [93], and attention modules [94] for image harmonization. Super-resolution-based techniques improve the captured image’s resolution or quality.
Though these techniques may add a layer of complexity to the model, they can make the data more centralized and make it easier for the model to see the patterns.
4.3. Data Augmentation Trends
Another trend noted during the review was the use of data augmentation. Data augmentation using flipping, rotating, and rescaling of the image was used by 45% of the studies using 2D data—however, none of the techniques using 3D data applied augmentation. In the future, data augmentation for 3D data could also be employed; as demonstrated by Cirillo et al. [95], 3D augmentation may increase the effectiveness of the segmentation model. GANs [96,97], multi-planar image synthesis [98], and affine transformations [99] are some of the techniques that can be applied to 3D data augmentation [100]. Though it should be noted that 3D data are already memory-extensive, adding more 3D volumes could increase the training time of the models extensively.
4.4. Enhancing Segmentation Using Transfer Learning
Transfer learning is a widespread technique employed for training different machine learning models; it uses previous experiments’ knowledge to fine-tune the current problem’s features. This technique is particularly beneficial for medical information or image analysis, as the data are limited for various reasons, including the disease’s rarity and ethical and legal issues [59].
As mentioned, most studies used the U-Net or the encoder/decoder model as a base model; hence, the weights trained in a previous study can be used as a starting point rather than training the model from scratch [101]. From our current literature, only three studies employed transfer learning. ImageNet [102,103] was the standard choice for model initialization between all three studies. Wu et al. [77] used weights pre-trained on the ImageNet model to initialize their transformer layers. Anand et al. [52] used the weights to initialize their DenseNet-based encoder layer, and Zhao et al. [86] used them to initialize their VGG-16-based model.
However, it should be noted that the ImageNet model contains coloured real-world images. In contrast, the medical images are primarily in greyscale and are much noisier than natural real-world images [104]. Hence, a more appropriate choice of transfer learning could be using medical image-based datasets, such as the newly introduced RadImageNet [105], which includes weakly labelled medical images of different organs. Another technique would be to use models trained on brain tumour data, as carried out by [106]. These techniques can potentially increase the effectiveness of transfer learning in stroke lesion segmentation as they are tailored for medical images.
5. Limitations
This study has several limitations. Firstly, it only incorporated technical studies presented in English that were considered for the review. As a result, the findings may offer a partial landscape for research being conducted in lesion segmentation. Secondly, the survey was performed using a particular set of keywords. Any technical study whose title or abstract did not contain the keywords was also not considered for the study. Lastly, the selected studies only included technical studies that focused solely on lesion segmentation; techniques presented in combination with stroke classification and rehabilitation were also not considered. Hence, it narrows the scope of the study and limits it from providing a broader context about stroke lesions.
6. Conclusions
In this review, we examined the different state-of-the-art machine learning models alongside different strategies and techniques currently being utilised to improve the performance of these structures. It can be easily concluded that the medical image analysis landscape evolved significantly after the introduction of deep learning techniques. U-Net and transformer-based structures have shown great promise in accurately identifying and segmenting stroke lesions in both MRI and CT.
Since there is a lack of substantial multi-centre-based data to train deep learning models, different augmentation and pre-processing techniques were applied to increase the effectiveness of the models. However, there is a need to analyse advanced pre-processing techniques in conjunction with lesion segmentation. Additionally, improving the utilisation of transfer learning is crucial.
While this paper reviews methods for the analysis of neuroimaging data of stroke patients, it will be interesting to see how these methods can be applied even before the immediate manifestation of stroke, e.g., to individuals who have been identified as a very high risk a few hours before the onset of stroke. Methods for personalised stroke prediction a few hours ahead or even a day ahead have already been developed to identify individuals with a very high risk before stroke manifests. The data used are longitudinal, multi-modal data of environmental variables measured over several days, such as changes in temperature, solar eruptions, pollution, wind direction, geomagnetic storms [107], etc., combined with personal data, such as age, blood pressure, smoking habits, etc. [108,109,110,111,112]. These methods can predict individual stroke with an accuracy of 95% and more than a few hours or a day ahead, after which MRI data can be measured and analysed for high-risk subjects to try to prevent the onset of the event.
In closing, the performance of the deep learning models needs to be improved to replace human effort; however, they can still provide robust, accurate, adaptable solutions to help professionals and patients alike.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/bioengineering11010086/s1. Table S1: Quality Assessment of all the articles included in the literature.
Author Contributions
This paper’s conceptualization, formal analysis, and methodology were undertaken by M.M. and A.W. Visualization, data curation, and preparation of the original draft were led by M.M. Review and editing of the manuscript involved contributions from B.C., N.K.K. and A.W. Supervision and project administration responsibilities were shared among A.W., J.F., V.S. and A.W. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by the Health Research Council of New Zealand’s project 21/144, the MBIE Catalyst: Strategic Fund NZ-Singapore Data Science Research Programme UOAX2001, the Marsden Fund Project 22-UOA-120, and the Royal Society Catalyst: Seeding General Project 23-UOA-055-CSG.
Institutional Review Board Statement
Not applicable.
Acknowledgments
We extend our heartfelt appreciation to the University of Auckland and the Auckland Bioengineering Institute for their invaluable and generous support.
Conflicts of Interest
The authors declare that they have no known competing financial interests or personal relationships that could have influenced the work reported in this paper.
References
- Feigin, V.L.; Owolabi, M.O.; Abanto, C.; Addissie, A.; Adeleye, A.O.; Adilbekov, Y.; Topcuoglu, M.A. Pragmatic Solutions to Reduce the Global Burden of Stroke: A World Stroke Organization—Lancet Neurology Commission. Lancet Neurol. 2023, 10, 142–149. [Google Scholar] [CrossRef] [PubMed]
- Feigin, V.L.; Brainin, M.; Norrving, B.; Martins, S.; Sacco, R.L.; Hacke, W.; Lindsay, P. World Stroke Organization (WSO): Global Stroke Fact Sheet 2022. Int. J. Stroke 2022, 17, 18–29. [Google Scholar] [CrossRef]
- Ortiz, G.A.; Sacco, R.L. National Institutes of Health Stroke Scale (NIHSS). In Wiley Encyclopedia of Clinical Trials; D’Agostino, R.B., Sullivan, L., Massaro, J., Eds.; Wiley-Interscience: Hoboken, NJ, USA, 2008; pp. 1–9. [Google Scholar]
- Kothari, R.U.; Pancioli, A.; Liu, T.; Brott, T.; Broderick, J. Cincinnati Prehospital Stroke Scale: Reproducibility and Validity. Ann. Emerg. Med. 1999, 33, 373–378. [Google Scholar] [CrossRef]
- Wang, J.; Zhu, H.; Wang, S.H.; Zhang, Y.D. A Review of Deep Learning on Medical Image Analysis. Mob. Netw. Appl. 2021, 26, 351–380. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, S.; Li, C.; Wang, J. Application of Deep Learning Method on Ischemic Stroke Lesion Segmentation. J. Shanghai Jiaotong Univ. (Sci.) 2022, 27, 99–111. [Google Scholar] [CrossRef]
- Chen, C.; Yuan, K.; Fang, Y.; Bao, S.; Tong, R.K.Y. Hierarchically Spatial Encoding Module for Chronic Stroke Lesion Segmentation. In Proceedings of the 2021 10th International IEEE/EMBS Conference on Neural Engineering (NER), Virtual Conference, 10–13 May 2021; pp. 1000–1003. [Google Scholar]
- Pieper, S.; Halle, M.; Kikinis, R. 3D Slicer. In Proceedings of the 2004 2nd IEEE International Symposium on Biomedical Imaging: Nano to Macro (IEEE Cat No. 04EX821), Arlington, VA, USA, 18 April 2004; Volume 1, pp. 632–635. [Google Scholar]
- Yushkevich, P.A.; Gao, Y.; Gerig, G. ITK-SNAP: An interactive tool for semi-automatic segmentation of multi-modality biomedical images. In Proceedings of the 2016 38th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Orlando, FL, USA, 16–20 August 2016; pp. 3342–3345. [Google Scholar]
- Stalling, D.; Westerhoff, M.; Hege, H.C. Amira: A Highly Interactive System for Visual Data Analysis. Visualization 2005, 38, 749–767. [Google Scholar]
- McAuliffe, M.J.; Lalonde, F.M.; McGarry, D.; Gandler, W.; Csaky, K.; Trus, B.L. Medical image processing, analysis and visualization in clinical research. In Proceedings of the 14th IEEE Symposium on Computer-Based Medical Systems, CBMS 2001, Bethesda, MD, USA, 26–27 July 2001; pp. 381–386. [Google Scholar]
- Zhang, X.; Xu, H.; Liu, Y.; Liao, J.; Cai, G.; Su, J.; Song, Y. A Multiple Encoders Network for Stroke Lesion Segmentation. In Proceedings of the Chinese Conference on Pattern Recognition and Computer Vision (PRCV), Virtual Conference, 29 October–1 November 2021; Springer International Publishing: Cham, Switzerland, 2021; pp. 524–535. [Google Scholar]
- Li, C. Stroke Lesion Segmentation with Visual Cortex Anatomy Alike Neural Nets. arXiv 2021, arXiv:2105.06544. [Google Scholar]
- Cerdá-Alberich, L.; Solana, J.; Mallol, P.; Ribas, G.; García-Junco, M.; Alberich-Bayarri, A.; Marti-Bonmati, L. MAIC—10 brief quality checklist for publications using artificial intelligence and medical images. Insights Imaging 2023, 14, 11. [Google Scholar] [CrossRef]
- Birenbaum, D.; Bancroft, L.W.; Felsberg, G.J. Imaging in Acute Stroke. West. J. Emerg. Med. 2011, 12, 67–76. [Google Scholar]
- Provost, C.; Soudant, M.; Legrand, L.; Hassen, W.B.; Xie, Y.; Soize, S.; Bourcier, R.; Benzakoun, J.; Edjlali, M.; Boulouis, G.; et al. Magnetic Resonance Imaging or Computed Tomography Before Treatment in Acute Ischemic Stroke: Effect on Workflow and Functional Outcome. Stroke 2019, 50, 659–664. [Google Scholar] [CrossRef]
- Tatlisumak, T. Is CT or MRI the Method of Choice for Imaging Patients with Acute Stroke? Why Should Men Divide if Fate Has United? Stroke 2002, 33, 2144–2145. [Google Scholar] [CrossRef]
- Vitali, P.; Savoldi, F.; Segati, F.; Melazzini, L.; Zanardo, M.; Fedeli, M.P.; Benedek, A.; Di Leo, G.; Menicanti, L.; Sardanelli, F. MRI versus CT in the Detection of Brain Lesions in Patients with Infective Endocarditis Before or After Cardiac Surgery. Neuroradiology 2022, 64, 905–913. [Google Scholar] [CrossRef]
- Vymazal, J.; Rulseh, A.M.; Keller, J.; Janouskova, L. Comparison of CT and MR imaging in ischemic stroke. Insights Imaging 2012, 3, 619–627. [Google Scholar] [CrossRef] [PubMed]
- Rubin, J.; Abulnaga, S.M. CT-To-MR conditional generative adversarial networks for ischemic stroke lesion segmentation. In Proceedings of the 2019 IEEE International Conference on Healthcare Informatics (ICHI), Xi’an, China, 10–13 June 2019; pp. 1–7. [Google Scholar]
- Muir, K.W.; Buchan, A.; von Kummer, R.; Rother, J.; Baron, J.C. Imaging of Acute Stroke. Lancet Neurol. 2006, 5, 755–768. [Google Scholar] [CrossRef] [PubMed]
- Rani, S.; Singh, B.K.; Koundal, D.; Athavale, V.A. Localization of stroke lesion in MRI images using object detection techniques: A comprehensive review. Neurosci. Inform. 2022, 2, 100070. [Google Scholar] [CrossRef]
- Girshick, R.; Donahue, J.; Darrell, T.; Malik, J. Rich feature hierarchies for accurate object detection and semantic segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Columbus, OH, USA, 23–28 June 2014; pp. 580–587. [Google Scholar]
- Redmon, J.; Divvala, S.; Girshick, R.; Farhadi, A. You only look once: Unified, real-time object detection. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 779–788. [Google Scholar]
- Liu, W.; Anguelov, D.; Erhan, D.; Szegedy, C.; Reed, S.; Fu, C.Y.; Berg, A.C. SSD: Single Shot Multibox Detector. In Proceedings of the 14th European Conference on Computer Vision (ECCV 2016), Part I, Amsterdam, The Netherlands, 11–14 October 2016; Springer International Publishing: Cham, Switzerland, 2016; pp. 21–37. [Google Scholar]
- Tan, M.; Pang, R.; Le, Q.V. EfficientDet: Scalable and Efficient Object Detection. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Virtual Conference, 13–19 June 2020; pp. 10781–10790. [Google Scholar]
- Karthik, R.; Menaka, R.; Johnson, A.; Anand, S. Neuroimaging and deep learning for brain stroke detection—A review of recent advancements and future prospects. Comput. Methods Programs Biomed. 2020, 197, 105728. [Google Scholar] [CrossRef]
- Thiyagarajan, S.K.; Murugan, K. A systematic review on techniques adapted for segmentation and classification of ischemic stroke lesions from brain MR images. Wirel. Pers. Commun. 2021, 118, 1225–1244. [Google Scholar] [CrossRef]
- Karthik, R.; Menaka, R. Computer-aided detection and characterization of stroke lesion—A short review on the current state-of-the-art methods. Imaging Sci. J. 2018, 66, 1–22. [Google Scholar] [CrossRef]
- Wang, X.; Fan, Y.; Zhang, N.; Li, J.; Duan, Y.; Yang, B. Performance of machine learning for tissue outcome prediction in acute ischemic stroke: A systematic review and meta-analysis. Front. Neurol. 2022, 13, 910259. [Google Scholar] [CrossRef]
- Abbasi, H.; Orouskhani, M.; Asgari, S.; Zadeh, S.S. Automatic Brain Ischemic Stroke Segmentation with Deep Learning: A Review. Neurosci. Inform. 2023, 3, 100145. [Google Scholar] [CrossRef]
- Styner, M.A.; Charles, H.C.; Park, J.; Gerig, G. Multisite Validation of Image Analysis Methods: Assessing Intra- and Intersite Variability. In Proceedings of the Medical Imaging 2002: Image Processing, San Diego, CA, USA, 23–28 February 2002; Volume 4684, pp. 278–286. [Google Scholar]
- Clèrigues, A.; Valverde, S.; Bernal, J.; Freixenet, J.; Oliver, A.; Lladó, X. Acute and sub-acute stroke lesion segmentation from multimodal MRI. Comput. Methods Programs Biomed. 2020, 194, 105521. [Google Scholar] [CrossRef]
- Soltanpour, M.; Greiner, R.; Boulanger, P.; Buck, B. Improvement of automatic ischemic stroke lesion segmentation in CT perfusion maps using a learned deep neural network. Comput. Biol. Med. 2021, 137, 104849. [Google Scholar] [CrossRef] [PubMed]
- Sheng, M.; Xu, W.; Yang, J.; Chen, Z. Cross-Attention and Deep Supervision UNet for Lesion Segmentation of Chronic Stroke. Front. Neurosci. 2022, 16, 836412. [Google Scholar] [CrossRef]
- Goshtasby, A.A. 2-D and 3-D Image Registration: For Medical, Remote Sensing, and Industrial Applications; John Wiley & Sons: Hoboken, NJ, USA, 2005. [Google Scholar]
- Brown, L.G. A survey of image registration techniques. Acm Comput. Surv. (CSUR) 1992, 24, 325–376. [Google Scholar] [CrossRef]
- Oliveira, F.P.; Tavares, J.M.R. Medical image registration: A review. Comput. Methods Biomech. Biomed. Eng. 2014, 17, 73–93. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Lei, Y.; Wang, T.; Curran, W.J.; Liu, T.; Yang, X. Deep learning in medical image registration: A review. Phys. Med. Biol. 2020, 65, 20TR01. [Google Scholar] [CrossRef] [PubMed]
- Hui, H.; Zhang, X.; Wu, Z.; Li, F. Dual-path attention compensation U-Net for stroke lesion segmentation. Comput. Intell. Neurosci. 2021, 2021, 7552185. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Yang, H.; Qi, K.; Dong, P.; Liu, Q.; Liu, X.; Wang, R.; Wang, S. MSDF-Net: Multi-scale deep fusion network for stroke lesion segmentation. IEEE Access 2019, 7, 178486–178495. [Google Scholar] [CrossRef]
- Wu, Z.; Zhang, X.; Li, F.; Wang, S.; Huang, L.; Li, J. W-Net: A boundary-enhanced segmentation network for stroke lesions. Expert Syst. Appl. 2023, 230, 120637. [Google Scholar] [CrossRef]
- Isa, A.M.A.A.A.; Kipli, K.; Mahmood, M.H.; Jobli, A.T.; Sahari, S.K.; Muhammad, M.S.; Chong, S.K.; Al-Kharabsheh, B.N.I. A Review of MRI Acute Ischemic Stroke Lesion Segmentation. Int. J. Integr. Eng. 2020, 12, 117–127. [Google Scholar]
- Gao, Y.; Li, J.; Xu, H.; Wang, M.; Liu, C.; Cheng, Y.; Li, M.; Yang, J.; Li, X. A multi-view pyramid network for skull stripping on neonatal T1-weighted MRI. Magn. Reson. Imaging 2019, 63, 70–79. [Google Scholar] [CrossRef]
- Tsai, C.; Manjunath, B.S.; Jagadeesan, B. Automated segmentation of brain MR images. Pattern Recognit. 1995, 28, 1825–1837. [Google Scholar] [CrossRef]
- Atkins, M.S.; Mackiewich, B.T. Fully automatic segmentation of the brain in MRI. IEEE Trans. Med. Imaging 1998, 17, 98–107. [Google Scholar] [CrossRef] [PubMed]
- Cox, R.W. AFNI: Software for analysis and visualization of functional magnetic resonance neuroimages. Comput. Biomed. Res. 1996, 29, 162–173. [Google Scholar] [CrossRef] [PubMed]
- Roy, S.; Butman, J.A.; Pham, D.L.; Alzheimer’s Disease Neuroimaging Initiative. Robust skull stripping using multiple MR image contrasts insensitive to pathology. NeuroImage 2017, 146, 132–147. [Google Scholar] [CrossRef]
- Fatima, A.; Shahid, A.R.; Raza, B.; Madni, T.M.; Janjua, U.I. State-of-the-Art Traditional to the Machine-and Deep-Learning-Based Skull Stripping Techniques, Models, and Algorithms. J. Digit. Imaging 2020, 33, 1443–1464. [Google Scholar] [CrossRef]
- Hazarika, R.A.; Kharkongor, K.; Sanyal, S.; Maji, A.K. A comparative study on different skull stripping techniques from brain magnetic resonance imaging. In International Conference on Innovative Computing and Communications (ICICC) 2019, Volume 1; Springer: Singapore, 2019; pp. 279–288. [Google Scholar]
- Rehman, H.Z.U.; Hwang, H.; Lee, S. Conventional and deep learning methods for skull stripping in brain MRI. Appl. Sci. 2020, 10, 1773. [Google Scholar] [CrossRef]
- Anand, V.K.; Khened, M.; Alex, V.; Krishnamurthi, G. Fully automatic segmentation for ischemic stroke using CT perfusion maps. In Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries: 4th International Workshop, BrainLes 2018, Held in Conjunction with MICCAI 2018, Granada, Spain, 16 September 2018; Revised Selected Papers, Part I; Springer International Publishing: Cham, Switzerland, 2019; pp. 328–334. [Google Scholar]
- Cui, W.; Liu, Y.; Li, Y.; Guo, M.; Li, Y.; Li, X.; Wang, T.; Zeng, X.; Ye, C. Semi-supervised brain lesion segmentation with an adapted mean teacher model. In Information Processing in Medical Imaging, Proceedings of the 26th International Conference, IPMI 2019, Hong Kong, China, 2–7 June 2019; Proceedings 26; Springer International Publishing: Cham, Switzerland, 2019; pp. 554–565. [Google Scholar]
- Karthik, R.; Gupta, U.; Jha, A.; Rajalakshmi, R.; Menaka, R. A deep supervised approach for ischemic lesion segmentation from multimodal MRI using Fully Convolutional Network. Appl. Soft Comput. 2019, 84, 105685. [Google Scholar] [CrossRef]
- Ahmad, P.; Jin, H.; Alroobaea, R.; Qamar, S.; Zheng, R.; Alnajjar, F.; Aboudi, F. MH UNet: A multi-scale hierarchical based architecture for medical image segmentation. IEEE Access 2021, 9, 148384–148408. [Google Scholar] [CrossRef]
- Tureckova, A.; Rodríguez-Sánchez, A.J. ISLES challenge: U-shaped convolutional neural network with dilated convolution for 3D stroke lesion segmentation. In Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries, Proceedings of the 4th International Workshop, BrainLes 2018, Held in Conjunction with MICCAI 2018, Granada, Spain, 16 September 2018; Revised Selected Papers, Part I; Springer International Publishing: Cham, Switzerland, 2019; pp. 319–327. [Google Scholar]
- Song, S.; Zheng, Y.; He, Y. A review of methods for bias correction in medical images. Biomed. Eng. Rev. 2017, 1, 1–10. [Google Scholar] [CrossRef]
- Xu, Y.; Hu, S.; Du, Y. Bias correction of multiple MRI images based on an improved nonparametric maximum likelihood method. IEEE Access 2019, 7, 166762–166775. [Google Scholar] [CrossRef]
- Matsoukas, C.; Haslum, J.F.; Sorkhei, M.; Söderberg, M.; Smith, K. What makes transfer learning work for medical images: Feature reuse & other factors. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, New Orleans, LA, USA, 18–24 June 2022. [Google Scholar]
- Philipsen, R.H.; Maduskar, P.; Hogeweg, L.; Melendez, J.; Sánchez, C.I.; van Ginneken, B. Localized energy-based normalization of medical images: Application to chest radiography. IEEE Trans. Med. Imaging 2015, 34, 1965–1975. [Google Scholar] [CrossRef] [PubMed]
- Hong, G. Image Fusion, Image Registration and Radiometric Normalization for High-Resolution Image Processing. Ph.D. Thesis, University of New Brunswick, Fredericton, NB, Canada, 2023. [Google Scholar]
- Delisle, P.L.; Anctil-Robitaille, B.; Desrosiers, C.; Lombaert, H. Realistic image normalization for multi-domain segmentation. Med. Image Anal. 2021, 74, 102191. [Google Scholar] [CrossRef]
- Modanwal, G.; Vellal, A.; Mazurowski, M.A. Normalization of breast MRIs using cycle-consistent generative adversarial networks. Comput. Methods Programs Biomed. 2021, 208, 106225. [Google Scholar] [CrossRef]
- Zhou, Y.; Huang, W.; Dong, P.; Xia, Y.; Wang, S. D-UNet: A dimension-fusion U shape network for chronic stroke lesion segmentation. IEEE/ACM Trans. Comput. Biol. Bioinform. 2019, 18, 940–950. [Google Scholar] [CrossRef] [PubMed]
- Dolz, J.; Ben Ayed, I.; Desrosiers, C. Dense multi-path U-Net for ischemic stroke lesion segmentation in multiple image modalities. In Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries, Proceedings of the 4th International Workshop, BrainLes 2018, Held in Conjunction with MICCAI 2018, Granada, Spain, 16 September 2018; Revised Selected Papers, Part I; Springer International Publishing: Cham, Switzerland, 2019; pp. 271–282. [Google Scholar]
- Wang, G.; Song, T.; Dong, Q.; Cui, M.; Huang, N.; Zhang, S. Automatic ischemic stroke lesion segmentation from computed tomography perfusion images by image synthesis and attention-based deep neural networks. Med. Image Anal. 2020, 65, 101787. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Medical Image Computing and Computer-Assisted Intervention–MICCAI 2015, Proceedings of the 18th International Conference, Munich, Germany, 5–9 October 2015; Proceedings, Part III; Springer International Publishing: Cham, Switzerland, 2015; pp. 234–241. [Google Scholar]
- Ou, Y.; Yuan, Y.; Huang, X.; Wong, K.; Volpi, J.; Wang, J.Z.; Wong, S.T. Lambdaunet: 2.5D stroke lesion segmentation of diffusion-weighted MR images. In Medical Image Computing and Computer Assisted Intervention—MICCAI 2021, Proceedings of the 24th International Conference, Strasbourg, France, 27 September–1 October 2021; Proceedings, Part I; Springer International Publishing: Cham, Switzerland, 2021; pp. 731–741. [Google Scholar]
- Bello, I. Lambdanetworks: Modeling long-range interactions without attention. arXiv 2021, arXiv:2102.08602. [Google Scholar]
- Ibtehaz, N.; Rahman, M.S. MultiResUNet: Rethinking the U-Net architecture for multimodal biomedical image segmentation. Neural Netw. 2020, 121, 74–87. [Google Scholar] [CrossRef]
- Liu, L.; Yang, S.; Meng, L.; Li, M.; Wang, J. Multi-scale deep convolutional neural network for stroke lesions segmentation on CT images. In Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries, Proceedings of the 4th International Workshop, BrainLes 2018, Held in Conjunction with MICCAI 2018, Granada, Spain, 16 September 2018; Revised Selected Papers, Part I; Springer International Publishing: Cham, Switzerland, 2019; pp. 283–291. [Google Scholar]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely connected convolutional networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 4700–4708. [Google Scholar]
- Omarov, B.; Tursynova, A.; Postolache, O.; Gamry, K.; Batyrbekov, A.; Aldeshov, S.; Azhibekova, Z.; Nurtas, M.; Aliyeva, A.; Shiyapov, K. Modified UNet Model for Brain Stroke Lesion Segmentation on Computed Tomography Images. Comput. Mater. Contin. 2022, 71, 4701–4717. [Google Scholar] [CrossRef]
- Chen, L.C.; Papandreou, G.; Kokkinos, I.; Murphy, K.; Yuille, A.L. Deeplab: Semantic image segmentation with deep convolutional nets, atrous convolution, and fully connected crfs. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 40, 834–848. [Google Scholar] [CrossRef]
- Sabour, S.; Frosst, N.; Hinton, G.E. Dynamic routing between capsules. Adv. Neural Inf. Process. Syst. 2017, 30, 3856–3866. [Google Scholar]
- Qi, K.; Yang, H.; Li, C.; Liu, Z.; Wang, M.; Liu, Q.; Wang, S. X-net: Brain stroke lesion segmentation based on depthwise separable convolution and long-range dependencies. In Medical Image Computing and Computer Assisted Intervention—Proceedings of the MICCAI 2019: 22nd International Conference, Shenzhen, China, 13–17 October 2019; Proceedings, Part III; Springer International Publishing: Cham, Switzerland, 2019; pp. 247–255. [Google Scholar]
- Wu, Z.; Zhang, X.; Li, F.; Wang, S.; Huang, L. Multi-scale long-range interactive and regional attention network for stroke lesion segmentation. Comput. Electr. Eng. 2022, 103, 108345. [Google Scholar] [CrossRef]
- Karthik, R.; Menaka, R.; Hariharan, M.; Won, D. Ischemic lesion segmentation using ensemble of multi-scale region aligned CNN. Comput. Methods Programs Biomed. 2021, 200, 105831. [Google Scholar] [CrossRef]
- Liu, Z.; Cao, C.; Ding, S.; Liu, Z.; Han, T.; Liu, S. Towards clinical diagnosis: Automated stroke lesion segmentation on multi-spectral MR image using convolutional neural network. IEEE Access 2018, 6, 57006–57016. [Google Scholar] [CrossRef]
- Ou, Y.; Yuan, Y.; Huang, X.; Wong, S.T.; Volpi, J.; Wang, J.Z.; Wong, K. Patcher: Patch transformers with mixture of experts for precise medical image segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Singapore, 18–22 September 2022; Springer Nature Switzerland: Cham, Switzerland, 2022; pp. 475–484. [Google Scholar]
- Liew, S.L.; Anglin, J.M.; Banks, N.W.; Sondag, M.; Ito, K.L.; Kim, H.; Chan, J.; Ito, J.; Jung, C.; Khoshab, N.; et al. A large, open source dataset of stroke anatomical brain images and manual lesion segmentations. Sci. Data 2018, 5, 180011. [Google Scholar] [CrossRef] [PubMed]
- Liew, S.L.; Lo, B.P.; Donnelly, M.R.; Zavaliangos-Petropulu, A.; Jeong, J.N.; Barisano, G.; Hutton, A.; Simon, J.P.; Juliano, J.M.; Suri, A.; et al. A large, curated, open-source stroke neuroimaging dataset to improve lesion segmentation algorithms. Sci. Data 2022, 9, 320. [Google Scholar] [CrossRef] [PubMed]
- Maier, O.; Menze, B.H.; Von der Gablentz, J.; Häni, L.; Heinrich, M.P.; Liebrand, M.; Winzeck, S.; Basit, A.; Bentley, P.; Chen, L.; et al. ISLES 2015—A public evaluation benchmark for ischemic stroke lesion segmentation from multispectral MRI. Med. Image Anal. 2017, 35, 250–269. [Google Scholar] [CrossRef]
- Hernandez Petzsche, M.R.; de la Rosa, E.; Hanning, U.; Wiest, R.; Valenzuela, W.; Reyes, M.; Meyer, M.; Liew, S.-L.; Kofler, F.; Ezhov, I.; et al. ISLES 2022: A multi-center magnetic resonance imaging stroke lesion segmentation dataset. Sci. Data 2022, 9, 762. [Google Scholar] [CrossRef]
- Kamnitsas, K.; Ledig, C.; Newcombe, V.F.; Simpson, J.P.; Kane, A.D.; Menon, D.K.; Rueckert, D.; Glocker, B. Efficient multi-scale 3D CNN with fully connected CRF for accurate brain lesion segmentation. Med. Image Anal. 2017, 36, 61–78. [Google Scholar] [CrossRef]
- Zhao, B.; Liu, Z.; Liu, G.; Cao, C.; Jin, S.; Wu, H.; Ding, S. Deep learning-based acute ischemic stroke lesion segmentation method on multimodal MR images using a few fully labeled subjects. Comput. Math. Methods Med. 2021, 2021, 3628179. [Google Scholar] [CrossRef]
- Goceri, E. Medical image data augmentation: Techniques, comparisons and interpretations. Artif. Intell. Rev. 2023, 56, 12561–12605. [Google Scholar] [CrossRef] [PubMed]
- Goodfellow, I.; Pouget-Abadie, J.; Mirza, M.; Xu, B.; Warde-Farley, D.; Ozair, S.; Courville, A.; Bengio, Y. Generative adversarial nets. Adv. Neural Inf. Process. Syst. 2014, 27, 2672–2680. [Google Scholar]
- Islam, M.; Vaidyanathan, N.R.; Jose, V.J.M.; Ren, H. Ischemic stroke lesion segmentation using adversarial learning. In Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries, Proceedings of the 4th International Workshop, BrainLes 2018, Held in Conjunction with MICCAI 2018, Granada, Spain, 16 September 2018; Revised Selected Papers, Part I; Springer International Publishing: Cham, Switzerland, 2019; pp. 292–300. [Google Scholar]
- Ou, Y.; Huang, S.X.; Wong, K.K.; Cummock, J.; Volpi, J.; Wang, J.Z.; Wong, S.T. BBox-Guided Segmentor: Leveraging expert knowledge for accurate stroke lesion segmentation using weakly supervised bounding box prior. Comput. Med. Imaging Graph. 2023, 107, 102236. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Chen, Z.; You, S.; Wang, B.; Shen, Y.; Lei, B. Brain stroke lesion segmentation using consistent perception generative adversarial network. Neural Comput. Appl. 2022, 34, 8657–8669. [Google Scholar] [CrossRef]
- Castiglioni, I.; Rundo, L.; Codari, M.; Di Leo, G.; Salvatore, C.; Interlenghi, M.; Gallivanone, F.; Cozzi, A.; D’Amico, N.C.; Sardanelli, F. AI applications to medical images: From machine learning to deep learning. Phys. Medica 2021, 83, 9–24. [Google Scholar] [CrossRef]
- Han, D.; Yu, R.; Li, S.; Wang, J.; Yang, Y.; Zhao, Z.; Wei, Y.; Cong, S. MR Image Harmonization with Transformer. In Proceedings of the 2023 IEEE International Conference on Mechatronics and Automation (ICMA), Harbin, China, 6–9 August 2023; pp. 2448–2453. [Google Scholar]
- Yao, X.; Lou, A.; Li, H.; Hu, D.; Lu, D.; Liu, H.; Wang, J.; Stoebner, Z.; Johnson, H.; Long, J.D.; et al. Novel application of the attention mechanism on medical image harmonization. Med. Imaging 2023 Image Process. 2023, 12464, 184–194. [Google Scholar]
- Cirillo, M.D.; Abramian, D.; Eklund, A. What is the best data augmentation for 3D brain tumor segmentation? In Proceedings of the 2021 IEEE International Conference on Image Processing (ICIP), Anchorage, AK, USA, 19–22 September 2021; pp. 36–40. [Google Scholar]
- Shin, H.C.; Tenenholtz, N.A.; Rogers, J.K.; Schwarz, C.G.; Senjem, M.L.; Gunter, J.L.; Andriole, K.P.; Michalski, M. Medical image synthesis for data augmentation and anonymization using generative adversarial networks. In Simulation and Synthesis in Medical Imaging: Third International Workshop, SASHIMI 2018, Held in Conjunction with MICCAI 2018, Granada, Spain, 16 September 2018; Proceedings 3; Springer International Publishing: Cham, Switzerland, 2018; pp. 1–11. [Google Scholar]
- Sun, Y.; Yuan, P.; Sun, Y. MM-GAN: 3D MRI data augmentation for medical image segmentation via generative adversarial networks. In Proceedings of the 2020 IEEE International Conference on Knowledge Graph (ICKG), Nanjing, China, 9–11 August 2020; pp. 227–234. [Google Scholar]
- Onishi, Y.; Teramoto, A.; Tsujimoto, M.; Tsukamoto, T.; Saito, K.; Toyama, H.; Imaizumi, K.; Fujita, H. Multiplanar analysis for pulmonary nodule classification in CT images using deep convolutional neural network and generative adversarial networks. Int. J. Comput. Assist. Radiol. Surg. 2020, 15, 173–178. [Google Scholar] [CrossRef]
- Chen, Y.; Ruan, D.; Xiao, J.; Wang, L.; Sun, B.; Saouaf, R.; Yang, W.; Li, D.; Fan, Z. Fully automated multiorgan segmentation in abdominal magnetic resonance imaging with deep neural networks. Med. Phys. 2020, 47, 4971–4982. [Google Scholar] [CrossRef]
- Garcea, F.; Serra, A.; Lamberti, F.; Morra, L. Data augmentation for medical imaging: A systematic literature review. Comput. Biol. Med. 2022, 152, 106391. [Google Scholar] [CrossRef]
- Kim, H.E.; Cosa-Linan, A.; Santhanam, N.; Jannesari, M.; Maros, M.E.; Ganslandt, T. Transfer learning for medical image classification: A literature review. BMC Med. Imaging 2022, 22, 69. [Google Scholar] [CrossRef]
- Kornblith, S.; Shlens, J.; Le, Q.V. Do better ImageNet models transfer better? In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Long Beach, CA, USA, 15–20 June 2019; pp. 2661–2671. [Google Scholar]
- Yang, K.; Qinami, K.; Fei-Fei, L.; Deng, J.; Russakovsky, O. Towards fairer datasets: Filtering and balancing the distribution of the people subtree in the ImageNet hierarchy. In Proceedings of the 2020 Conference on Fairness, Accountability, and Transparency, Barcelona, Spain, 27–30 January 2020; pp. 547–558. [Google Scholar]
- Cadrin-Chênevert, A. Moving from ImageNet to RadImageNet for improved transfer learning and generalizability. Radiol. Artif. Intell. 2022, 4, e220126. [Google Scholar] [CrossRef] [PubMed]
- Mei, X.; Liu, Z.; Robson, P.M.; Marinelli, B.; Huang, M.; Doshi, A.; Jacobi, A.; Cao, C.; Link, K.E.; Yang, T.; et al. RadImageNet: An open radiologic deep learning research dataset for effective transfer learning. Radiol. Artif. Intell. 2022, 4, e210315. [Google Scholar] [CrossRef] [PubMed]
- Siddique, M.M.R.; Yang, D.; He, Y.; Xu, D.; Myronenko, A. Automated ischemic stroke lesion segmentation from 3D MRI. arXiv 2022, arXiv:2209.09546. [Google Scholar]
- Feigin, V.L.; Parmar, P.G.; Barker-Collo, S.; Bennett, D.A.; Anderson, C.S.; Thrift, A.G.; Stegmayr, B.; Rothwell, P.M.; Giroud, M.; Bejot, Y.; et al. Geomagnetic Storms Can Trigger Stroke: Evidence From 6 Large Population-Based Studies in Europe and Australasia. Stroke 2014, 45, 1639–1645. [Google Scholar] [CrossRef]
- Kasabov, N.; Feigin, V.; Hou, Z.G.; Chen, Y.; Liang, L.; Krishnamurthi, R.; Othman, M.; Parmar, P. Evolving Spiking Neural Networks for Personalised Modelling, Classification and Prediction of Spatio-Temporal Patterns with a Case Study on Stroke. Neurocomputing 2014, 134, 269–279. [Google Scholar] [CrossRef]
- Othman, M.; Kasabov, N.; Tu, E.; Feigin, V.; Krishnamurthi, R.; Hou, Z.; Chen, Y.; Hu, J. Improved predictive personalized modelling with the use of Spiking Neural Network system and a case study on stroke occurrences data. In Proceedings of the 2014 International Joint Conference on Neural Networks (IJCNN), Beijing, China, 6–11 July 2014; pp. 3197–3204. [Google Scholar]
- Kasabov, N.; Feigin, V.; Hou, Z.; Chen, Y. Improved Method and System for Predicting Outcomes Based on Spatio/Spectro-Temporal Data. PCT Patent WO2015/030606 A2. U.S. Patent US2016/0210552 A1, 21 July 2016. [Google Scholar]
- Kasabov, N.K. Time-Space, Spiking Neural Networks and Brain-Inspired Artificial Intelligence; Springer: Berlin/Heidelberg, Germany, 2019. [Google Scholar]
- Doborjeh, M.; Doborjeh, Z.; Merkin, A.; Krishnamurthi, R.; Enayatollahi, R.; Feigin, V.; Kasabov, N. Personalised Spiking Neural Network Models of Clinical and Environmental Factors to Predict Stroke. Cogn. Comput. 2022, 14, 2187–2202. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).